Abstract
VPS4B, an AAA ATPase (ATPase associated with various cellular activities), participates in vesicular trafficking and autophagosome maturation in mammalian cells. In solid tumors, hypoxia is a common feature and an indicator of poor treatment outcome. Our studies demonstrate that exogenous or endogenous (assessed with anchorage-independent three-dimensional multicellular spheroid culture) hypoxia induces VPS4B downregulation by the ubiquitin-proteasome system. Inhibition of VPS4B function by short hairpin VPS4B (sh-VPS4B) or expression of dominant negative VPS4B(E235Q) promotes anchorage-independent breast cancer cell growth and resistance to gefitinib, U0126, and genotoxicity. Biochemically, hyperactivation of epidermal growth factor receptor (EGFR), a receptor tyrosine kinase essential for cell proliferation and survival, accompanied by increased EGFR accumulation and altered intracellular compartmentalization, is observed in cells with compromised VPS4B. Furthermore, enhanced FOS/JUN induction and AP-1 promoter activation are noted in EGF-treated cells with VPS4B knockdown. However, VPS4B depletion does not affect EGFRvIII stability or its associated signaling. An inverse correlation between VPS4B expression and EGFR abundance is observed in breast tumors, and high-grade or recurrent breast carcinomas exhibit lower VPS4B expression. Together, our findings highlight a potentially critical role of VPS4B downregulation or chronic-hypoxia-induced VPS4B degradation in promoting tumor progression, unveiling a nongenomic mechanism for EGFR overproduction in human breast cancer.
INTRODUCTION
Members of the epidermal growth factor receptor (EGFR) family transmit signals from the extracellular to the intracellular environment, leading to various cellular responses involved in cell proliferation, differentiation, and death (49). The EGFR family is comprised of four transmembrane receptor tyrosine kinases: EGFR (ErbB1), HER2 (ErbB2), ErbB3, and ErbB4. EGFR is internalized and degraded upon ligand EGF binding. The endocytosed, activated EGFR, destined for lysosomal degradation, accumulates on the intraluminal vesicle of multivesicular bodies (MVBs, or late endosomes), whereas a receptor destined for recycling remains on the perimeter membrane of MVBs, from where it is returned to the plasma membrane. Following removal of recycled proteins, MVBs containing EGFR fuse directly with lysosomes, resulting in EGFR degradation and a dramatic decrease in steady-state levels of EGFR protein (55). The process of EGF-induced EGFR downregulation is the major negative feedback regulatory mechanism that controls the intensity and duration of EGFR signaling (53). Clinically, increased levels of EGFR expression are observed in a variety of cancers, conferring an adverse prognosis. While gene amplification of EGFR contributes to the resultant EGFR overexpression, many cancers display EGFR overexpression without gene amplification (reviewed in reference 47). However, the pathological events that lead to nongenomic divergence in EGFR expression in solid tumors are poorly understood.
Endocytosis is one of the major inactivation pathways for EGFR. For example, EGFRvIII is the most common EGFR mutant, with an in-frame deletion of exons 2 to 7, and the lack of the corresponding amino acid residues 6 to 273 renders EGFRvIII constitutively tyrosine phosphorylated in a ligand-independent manner (reviewed in reference 19). Compared with EGFR, EGFRvIII has a low rate of endocytosis and thus escapes downregulation (18). During endocytic process, four ESCRT (endosomal sorting complexes required for transport) protein complexes are utilized to culminate in lysosomal degradation of activated EGFR (reviewed in references 13, 21, and 46). VPS4, an AAA ATPase (ATPase associated with various cellular activities) protein complex (12), is responsible for the disassembly of the ESCRT-III complex, required for recycling of membrane-associated proteins in mammalian cells (66). There are two VPS4 isoforms, VPS4A and VPS4B, and VPS4B shares an overall 60% sequence identity to yeast Vps4 and is able to complement the MVB sorting defect in Vps4-null yeast cells (48). The endocytic pathway is reportedly involved in lysosomal degradation of proteins and organelles via autophagy in some cases (23). Recent studies demonstrated that VPS4 and ESCRT-III are required for autophagosomes to fuse with MVBs and for the fusion of the resultant amphisomes with lysosomes during autophagic flux (44). Given the essential role of VPS4B in MVB maturation, we therefore examined whether the loss of VPS4B function would affect the duration and signaling of the membrane receptors.
As oxygen is able to diffuse only 100 to 180 μm from a capillary vessel to cells, cells located farther than this from a capillary vessel will be exposed to hypoxia (38). Consequently, hypoxia is present in 90% of solid tumors because the vascular system is not able to supply the growing tumor mass with adequate amounts of oxygen. Although the role of functional MVB in membrane receptor trafficking has been extensively investigated (40, 42), little is known about how a faulty MVB pathway affects tumor cells via dysregulating membrane receptor signaling under hypoxic stress. To address this, we explored the effects of hypoxic stress on EGFR signaling using two-dimensional (2D) monolayer cultures and anchorage-independent three-dimensional (3D) multicellular spheroid cultures. We found that chronic exposure to exogenous or endogenous hypoxia destabilizes VPS4B through the ubiquitin-proteasome system. Loss of VPS4B function, by expression of a dominant negative form of VPS4B [VPS4B(EQ), harboring a Glu (E)-to-Gln (Q) mutation at amino acid residue 235] (9, 39, 44) or VPS4B knockdown, increases EGFR stability and accentuates its endosomal signaling. Concomitantly, inhibition of VPS4B function also promotes breast cancer cell anchorage-independent growth and renders cells resistant to treatment with gefitinib (an EGFR inhibitor), U0126 (a selective MEK inhibitor), or doxorubicin (a genotoxicity inducer). An inverse correlation between VPS4B expression and EGFR abundance in human breast tumors was established, suggesting clinical relevance. Collectively, our data provide evidence suggesting a novel pathway in which hypoxia promotes tumorigenesis through VPS4B degradation, unveiling a previously unrecognized role for VPS4B in human cancers.
MATERIALS AND METHODS
3D spheroid culture.
The 3D spheroid culture was performed as described by Yuhas et al. (69). Briefly, a thin layer of 1.5% agar was added to the bottom of wells in 96-well culture plates (5,000 to 10,000 cells/well) to prevent cell adhesion to the culture surface, resulting in cell aggregation and formation of spheroids with a radius of 200 to 350 μm. Cells from eight wells were pooled for each time point analyses.
Generation of inducible cell lines and short hairpin RNA (shRNA) knockdown cell lines.
Pa-4/TR/VPS4B(wt), Pa-4/TR/VPS4B(EQ), MCF7/TR/VPS4B(wt), and MCF7/TR/VPS4B(EQ) cells were generated as follows. Coding sequences for wild-type VPS4B [VPS4B(wt)] and the EQ mutant [VPS4B(EQ)] were amplified by PCR from VPS4B(wt)- and VPS4B(EQ)-DsRed, respectively, and cloned into the multiple cloning site (HindIII/XbaI) of pTRE-Myc (Clontech). Myc-tagged VPS4B sequences were subsequently amplified by PCR and inserted into the entry vector pENTR/D-TOPO. An LR recombination reaction was performed between the entry clones and expression vector pLenti4/TO/V5-DEST (Invitrogen) to generate VPS4B(wt)- and VPS4B(EQ)-pLenti4/TO/V5-DEST constructs. Tetracycline-inducible expression of VPS4B(wt) or VPS4B(EQ) was achieved by transducing cells with Tet repressor (TR)-containing lentiviruses of interest. Lentiviral stock was made by cotransfecting pLenti6/TR plasmid (Invitrogen) and virus packaging mix into the 293FT virus producing cell line, as previously described (36). Pa-4 and MCF7 cells were transduced with these packaged lentiviruses and selected by blasticidin (10 μg/ml) for Tet repressor-expressing cells. Pa-4 and MCF7 cells that expressed Tet repressor were then stably transfected with VPS4B(wt)- or VPS4B(EQ)-pLenti4/TO/V5-DEST and selected with zeocin (100 μg/ml) for VPS4B-expressing cells. Expression of VPS4B in these cells was induced by doxycycline (1 μg/ml), which was added to culture medium 16 h prior to the experiment.
NR6-EGFRvIII/sh-VPS4B, MDA-MB-231/sh-VPS4B, and SKBR3/sh-VPS4B cells were generated by transducing NR6-EGFRvIII, MDA-MB-231, and SKBR3 cells, respectively, with lentiviruses harboring shRNA against VPS4B. HEK293/DsRed-VPS4B:tGFP and HEK293/EGFP-LC3:tRFP cells were engineered by infecting HEK293 cells with lentiviruses harboring DsRed-tagged VPS4B in a bicistronic vector with internal ribosome entry site (IRES)-driven TurboGFP (tGFP) or EGFP-tagged LC3 with IRES-driven tRFP, respectively. The IRES-driven tGFP or tRFP serves as a control for the expression of the tagged VPS4B or LC3 prior to and after stress treatment. The package of self-inactivating lentivirus harboring various constructs and the following transduction to host cells were performed as previously described (36). sh-VPS4B-transduced cells were selected and maintained in growth medium containing puromycin (2 μg/ml); pools of stable cells were used in studies. Transduced cells positive for both GFP and RFP cells were enriched via sorting through a MoFlo sorter (Beckman Coulter) by the Analytical Cytometry Core Facility at City of Hope.
Cell lines and reagents.
Rat parotid epithelial Pa-4 cells were cultured as previously described (9). Cells of the human breast tumor cell lines MCF7, MDA-MB-231, HEK293, HEK293/DsRed-VPS4B:tGFP, HEK293/EGFP-LC3:tRFP, and MEF/GFP-LC3 were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) plus penicillin (100 U/ml)-streptomycin (100 μg/ml) and grown at 37°C. The human breast cancer cell line SKBR3 was grown in McCoy's 5A medium containing 10% FBS and antibiotics. The tetracycline (Tet)-inducible stably VPS4B expressing cell lines Pa-4/TR/VPS4B(wt) and Pa-4/TR/VPS4B(EQ) were cultured in DMEM/F-12 medium with 2.4% fetal bovine serum, insulin (5 μg/ml), l-glutamine (2.3 mM), transferrin (5 μg/ml), epidermal growth factor (25 ng/ml), hydrocortisone (1.1 μM), glutamate (5 mM), T3 (3,3′,5-triiodo-l-thyronine) (1.7 nM), kanamycin (94 μg/ml), amphotericin B (Fungizone) (47 μg/ml), zeocin (100 μg/ml), and blasticidin (10 μg/ml) and grown at 35°C. MCF7/TR/VPS4B(wt) and MCF7/TR/VPS4B(EQ) cells were cultured in 10% FBS-supplemented DMEM with zeocin (100 μg/ml) and blasticidin (10 μg/ml). NR6 cells that stably expressed EGFRvIII (NR6/EGFRvIII) were maintained in DMEM supplemented with 10% FBS plus penicillin (100 μ/ml)-streptomycin (100 μ/ml) and G418 (750 μg/ml). Desferrioxamine (DFO), bafilomycin A1 (BafA1), and Earle's balanced salt solution without phenol red (EBSS) were purchased from Sigma (St. Louis, MO), MG-132 was from Calbiochem, doxorubicin (Dox) was from Ben Venue Labs Inc. (Bedford, OH), gefitinib was from LC Laboratories (Woburn, MA), U0126 was from Cell Signaling (Danvers, MA), and doxycycline and EGF were from BD Biosciences. Hypoxic treatment was performed with an OxyCycler system (model C42; BioSpherix, Redfield, NY) as per the manufacturer's instructions. Expression constructs for GFP-HER2, EGFR, and HA-HER2 (wt, constitutively active, or kinase dead) (61) were a generous gift from Shao-Chun Wang.
Western analyses.
For protein expression level and phosphorylation studies, whole-cell lysates were extracted with sodium dodecyl sulfate (SDS) lysis buffer as previously described (9, 11), supplemented with both complete protease inhibitor cocktail (Roche) and PhosSTOP phosphatase inhibitor (Roche). Equal amounts of whole-cell lysates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and subsequently immunoblotted with antibodies recognizing VPS4B (Abnova), p62/SQSTM1 (American Research Products, Inc.), BECLIN-1 and UVRAG (Abcam), hemagglutinin (HA) and Myc tags (Santa Cruz Biotechnologies), LC3 (MBL International Co.), pERK1/2, pJNK1/2, EGFR, pY1068-EGFR (recognizes human pY1092), pY1173-EGFR (recognizes human pY1197), and pY845-EGFR (recognizes human pY869) (all from Cell Signaling), and actin (Millipore). Blots were visualized with an enhanced chemiluminescence detection kit (ECL-Plus; GE Healthcare) and a VersaDoc 5000 imaging system (Bio-Rad). Signal intensities of the captured images were analyzed with Quantity One software (Bio-Rad). The results of Western analyses shown are representative of two to four independent experiments. The relative protein levels altered by various treatments were normalized to the control, which was set as 1, assuming equal variances.
Immunoprecipitation.
Whole-cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris, 125 mM NaCl, 1% Nonidet P-40 [NP-40], 0.1% SDS, 0.5% sodium deoxycholate, 0.004% sodium azide [pH 8.0]) containing complete protease inhibitor cocktail. Protein concentrations were determined with a bicinchoninic acid (BCA) assay (Bio-Rad). Whole-cell lysates (1 mg) were incubated (4°C, 2 h) with 1 to 5 μg/ml of a specific antibody for the Myc tag or the HA tag, followed by incubation (4°C, overnight) with 20 μl protein A/G Plus agarose beads (Santa Cruz Biotechnologies) to capture immune complexes, which were then washed three times with PBS containing 1% NP-40, 0.5% sodium deoxycholate, and complete protease inhibitor cocktail, eluted from the beads by adding SDS-PAGE loading buffer, boiled for 6 min, and subjected to Western analyses.
Measurement of GFP-LC3 intensity by fluorescence activated cell sorting (FACS) analysis and fluorescence microscopy.
MEF cells stably expressing GFP-LC3 or Pa-4/TR/VPS4B(EQ) cells transduced with lentiviral GFP-LC3 were pretreated with doxycycline (doxy; 1 μg/ml) for transgene induction and seeded subconfluently in six-well plates (for the 2D experiment) or on agarose-coated 96-well plates (for the 3D experiment) prior to the indicated treatment (see. Fig. 2A). Cells were subsequently harvested with trypsin-EDTA, washed with PBS, fixed at room temperature for 30 min with 2% paraformaldehyde in PBS, and then washed with PBS twice prior to FACS analysis (1 × 105 cells per sample) using a CyAn ADP nine-color flow cytometer (Dako; performed at the Analytic Cytometry Core facility at the City of Hope Medical Center), and viable cell counts were plotted as GFP fluorescence intensity by FlowJo software (Tree Star, Inc.), as previously described (9). The relative level of GFP-LC3 intensity in each treatment was calculated from at least three independent experiments after normalization to the level of resting, vehicle-treated controls, and values reported are means and standard deviations (SD).
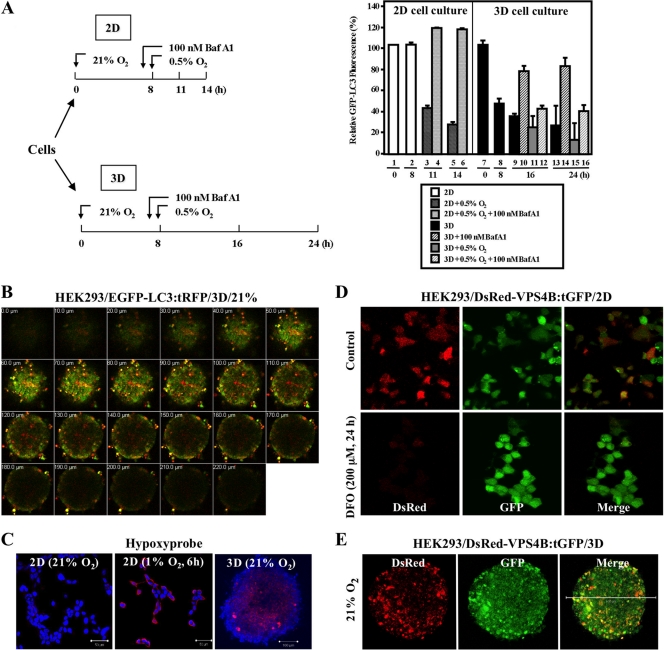
Fig 2.
Hypoxic stress is detected in multicellular spheroids. (A) (Left) Diagram of experimental design. (Right) Equal numbers of MEF/GFP-LC3 cells were seeded as 2D monolayer cultures and as 3D spheroids under 21% O2 for 8 h and then exposed to 0.5% O2 for 3 or 6 h for monolayers (columns 3 to 6) and 8 or 16 h for 3D cultures (columns 9 to 16). BafA1 was added to cultures (columns 4, 6, 10, 12, 14, and 16) 1 h prior to the start of hypoxic exposure. The relative GFP-LC3 intensities were analyzed by FACS, and the fluorescence intensity of control samples was set at 100%. (B) A series of images of a HEK293/EGFP-LC3:tRFP spheroid coexpressing EGFP-tagged LC3 and tRFP were collected at 10-μm intervals along the z axis, from top to bottom of the spheroid. (C) HEK293 cells were grown as monolayers during normoxia (21% O2) and hypoxia (1% O2) or as spheroids. Alexa-546 (red) staining marks detection of Hypoxyprobe. Nuclei were counterstained with DAPI (blue). (D) Reduced DsRed-VPS4B expression level in DFO-treated (200 μM, 24 h) HEK293/DsRed-VPS4B:tGFP monolayer cells (bottom) was evident, compared to that of the untreated control (top). Magnification, ×40. (E) Attenuated VPS4B signal is observed in the center of the spheroid. Maximum-intensity projection images of a HEK293/DsRed-VPS4B:tGFP spheroid under 21% O2 show DsRed (left), GFP (middle), and a merge of the two (right). Bar, 500 μm; magnification, ×10.
Luciferase assays.
Cells were cotransfected with AP-1-Luc(wt), AP-1-Luc(mt), or Gal4TATA-Luc reporter combined with Gal4-Elk-1 expression construct together with a Renilla luciferase control reporter construct, pRL-TK (for normalization), using Lipofectamine 2000 (Invitrogen). Luciferase assays were carried out with a Dual-Glo luciferase assay kit (Promega). Luciferase activity was normalized to cotransfected Renilla luciferase activity.
Electron microscopy.
Transmission electron microscopy with an FEI Tecnai 12 Twin equipped with a Gatan 894 Ultrascan 1000 charge-coupled device (CCD) camera, a cryo-stage, a cryo-holder, and a tomography holder, was performed at the City of Hope Electron Microscopy Core facility. Cells were fixed in 2% paraformaldehyde–1.5% glutaraldehyde in 0.1 M cacodylate buffer with 2% sucrose (pH 7.2), spun down, postfixed in 1% OsO4, washed, dehydrated with graded alcohol, and embedded in Eponate. Thin sections (60 nm) were cut with a diamond knife and stained (15 min) with 5% aqueous uranyl acetate, followed by a 2-min staining with Reynolds lead stain.
Hypoxyprobe staining.
HEK293 cells were seeded on cover glasses (2D) or 96-well plates coated with agar (3D) for 1 day, and 2D monolayers were exposed to normoxia or hypoxia (1% O2) for 6 h. The hypoxic local environment was visualized using Hypoxyprobe (HPI, Inc.) staining according to the manufacturer's instructions with modifications. Briefly, Hypoxyprobe-1 (pimonidazole HCl) was added to the medium of 2D and 3D cultures at 6 h prior to staining; culture medium with Hypoxyprobe was then removed, and cells were fixed in 100% methanol by incubation at −20°C overnight. Methanol was subsequently removed, and cells were washed three times with PBS. The 3D spheroids were transferred to hybridization slide chambers. Slides containing cells from both 2D and 3D cultures were blocked with 10% FBS–PBS solution for 6 h and washed three times with PBS, and the primary antibody (1:25 dilution in 5% BSA–PBS) was added for overnight incubation. After removal of primary antibody, slides were washed three times with PBS and incubated with secondary antibody conjugated with Alexa-546 (1:1,000 in 5% BSA–PBS) overnight. DAPI (4′,6-diamidino-2-phenylindole) was added as a nuclear counterstain for 10 min. The slides were washed three times with PBS, mounted, and examined by confocal microscopy.
Confocal fluorescence microscopy.
SKBR3 and SKBR3/sh-VPS4B cells grown on coverslips were serum starved overnight and then incubated (0 or 2 h, 37°C) with EGF (100 ng/ml), fixed (3.7% paraformaldehyde [Sigma] in PBS), quenched and permeabilized (5 min) with 0.2% saponin, blocked with 10% FBS in PBS, and incubated with antibodies against EGFR (BD Pharmingen) and Rab7 (Santa Cruz Biotechnology, Inc.). Secondary antibodies, Alexa Fluor 488–goat anti-mouse and Alexa Fluor 568–goat anti-rabbit antibodies, were from Molecular Probes. Samples were mounted using a ProLong antifade kit (Molecular Probes). Images were collected from different fields using a confocal laser scanning microscope (LSM 510 Meta NLO imaging system; Zeiss) equipped with argon, red HeNe, and green HeNe lasers and a Ti-Sapphire tunable Chameleon laser (Coherent, Inc.; 720 to 950 nm) for multiphoton excitation of DAPI. Profiles for the green channel along the x and y axes were generated using the 2.5-dimensional function available with the Zeiss LSM 510 software. Fluorescent images of the HEK293/DsRed-VPS4B:tGFP and HEK293/EGFP-LC3:tRFP cells were acquired using a Zeiss LSM 510 Meta inverted microscope. For the 3D spheroid culture, a series of images at intervals of 3 to 10 μm along the z axis were collected. All images were compiled using Adobe Photoshop software (version 7.0; Adobe Systems, Mountain View, CA). Maximum-intensity projection (MIP) images were created using Zeiss LSM Image Browser software by taking all the z sections and collapsing them into a single image.
MTS cell viability assays.
For 2D experiments, cells were seeded onto 12-well plates, in the presence and absence of doxycycline (1 μg/ml), to reach 70% confluence on the day of the experiment. Cells were treated with increasing concentrations of doxorubicin for up to 120 h; media were then replaced by fresh media with 40 μl of 3-[4,5-dimethylthiazol-2-yl]-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) (CellTiter 96 aqueous one-solution cell proliferation assay; Promega, Madison, WI), and cells were incubated for 4 h at 37°C. The MTS solution was then transferred to 96-well plates, and absorbance was read immediately at 490 nm using a scanning microplate reader. For 3D spheroids, 1 × 105 cells in 200 μl medium with or without doxycycline (1 μg/ml) were seeded onto 1.5% agarose-coated 96-well plates. At 24 h postseeding, cells were treated with various concentrations of doxorubicin for up to 120 h, and MTS assays were performed as described above. Results are presented as percent cell viability and are means and SD from three independent experiments performed in triplicate.
Soft-agar colony formation.
For soft-agar assays, pairs of 5,000 MDA-MB-231 cells were mixed in 0.35% agarose–complete medium and were plated on 0.7% agarose–complete medium bottom layers in each well of 6-well plates. The culture media containing EGF (10 ng/ml) alone, with U0126 (10 and 20 μM), or with gefitinib (10 and 20 μM) were changed every 2 to 3 days during the 2 to 4 weeks of cell growth. Colonies were stained with 0.2 mg/ml MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide; Sigma] and were counted with colony-counting software using a VersaDoc imaging system (Bio-Rad). Each experiment was performed at least twice in triplicate wells.
Patient tissue samples, RNA extraction, and quantitative reverse transcription-PCR (RT-PCR).
Subjects comprised 28 patients with stage II-III (n = 18) and stage IV (n = 10) breast cancer at City of Hope National Medical Center. Upon inclusion in the resources (with informed consent under institutional review board [IRB]-approved protocol 06206), all cases underwent pathological characterization of tissues and review of medical records. Three normal breast tissue samples were obtained from tissue reduction as surgical waste. Total RNA was extracted from breast tissue samples using a RNeasy lipid tissue minikit (Qiagen) and from NR6-EGFRvIII (parental and sh-VPS4B) or SKBR3 (parental and sh-VPS4B) cells using an RNeasy minikit (Qiagen), according to the manufacturer's instructions. Subsequent cDNA synthesis was carried out with an iScript cDNA synthesis kit (Bio-Rad); the targeted sequence was amplified with IQ SYBR green Supermix and specific primer pairs using a My IQ real-time PCR detection system (Bio-Rad). Relative mRNA expression levels were calculated using the ΔCT method against 18S rRNA (for tissue samples) or the ΔΔCT method against GAPDH (for cells). The primer pairs used are shown in Table S1 in the supplemental material.
Immunohistochemical (IHC) analysis.
Breast cancer tissue specimens were obtained from an archival paraffin block inventory under IRB protocol 06206 of the City of Hope National Medical Center. IHC staining was performed on 5-μm-thick sections prepared from formalin-fixed, paraffin-embedded tissue. Tissue sections were deparaffinized in xylene followed by 100% ethanol. Samples were then quenched in 3% hydrogen peroxide and pretreated with steam with EDTA solution to promote antigen retrieval. After antigen retrieval, slides were incubated in Protein Block for 5 min. Then slides were incubated with primary anti-EGFR antibody (Zymed catalog no. 28-0005) at room temperature, washed in Dako buffer, and incubated with Dako EnVision+ mouse polymer antibody for 30 min. After three washes in Dako buffer, slides were incubated with the chromogen diaminobenzidine tetrahydrochloride (DAB), counterstained with hematoxylin, and mounted. IHC staining was assessed independently by X.L. and Y.Y., and the staining intensity was graded as 0+ (negative), 1+ (weak and incomplete membrane staining), 2+ (weak but complete circumferential membrane staining), or 3+ (strong and complete circumferential membrane staining).
Statistical analysis.
Data were processed by Microsoft Excel and analyzed by using GraphPad Prism 5.0. The nonparametric test was employed to determine the significant differences in ΔCT of VPS4B between the stage IV and stage II-III subgroups. Linear regression analysis was used to determine correlation between expression levels of genes. A P value of <0.05 was considered significant. For the differentially expressed gene analysis, we obtained gene expression data from a study by Ivshina et al. (20), which reported microarray profiling of a cohort of 289 breast tumor samples using Affymetrix U133A and U133AB gene chips (GEO accession number GSE4922). Two-group t tests comparing grade III (n = 55) and grade I and II (n = 234) tumors were performed on the logarithm-transformed VPS4B expression levels.
RESULTS
Hypoxic stress induces VPS4B reduction.
While hypoxia triggers autophagy (9, 10, 31, 41), little is known about the effect of hypoxia on the stability of key members in autophagy pathway, such as VPS4B, BECLIN-1, p62/SQSTM1 (referred to herein as p62), and UVRAG. We therefore evaluated steady-state protein levels of these molecules under hypoxic stress. First, Pa-4 cells were treated with desferrioxamine (DFO), known to induce a hypoxic environment (9, 11). Consistent with recent reports by us and others (10, 41), sustained hypoxic stress decreased steady-state levels of BECLIN-1 and p62, while the UVRAG level increased during chronic hypoxic exposure (Fig. 1A). Notably, DFO treatment also induced a decrease in steady-state levels of VPS4B in a time-dependent manner. Adding MG132, a proteasome inhibitor, led to a partial reversal of hypoxic-stress-induced degradation of VPS4B and p62 but not BECLIN-1. Treatment with the genotoxicity inducer doxorubicin induced only a negligible VPS4B downregulation (see Fig. S1A in the supplemental material). These results indicated that VPS4B is selectively downregulated under hypoxic conditions through proteasome degradation. Based on the intricate role of VPS4B in the regulation of membrane receptor endocytosis and trafficking network as well as autophagy, we further explored the mechanism underlying hypoxia-mediated VPS4B downregulation and the subsequent biological consequences of loss of VPS4B function.
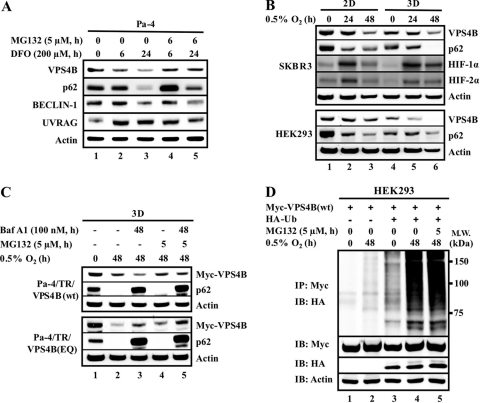
Fig 1.
Hypoxia-induced VPS4B degradation. (A) Pa-4 cells were treated with desferrioxamine (DFO; 200 μM) for the indicated times in the presence or absence of MG132 (5 μM) during the last 6 h prior to harvesting. (B) SKBR3 and HEK293 cells were grown as either monolayer cultures (2D) or spheroids (3D) prior to hypoxia exposure. (C) Pa-4/TR/VPS4B(wt) and Pa-4/TR/VPS4B(EQ) cells were grown to form spheroids, which were exposed to 0.5% O2 in the presence of bafilomycin A1 (Baf A1; 100 nM, 48 h) and MG132 (5 μM, last 5 h), as indicated. (D) HEK293 cells were transfected with Myc-VPS4B(wt) and HA-Ub, as indicated. At 48 h posttransfection, cells were exposed to 0.5% O2 and treated with MG132 (5 μM, last 5 h of hypoxic exposure) prior to harvesting. Equal amounts of total cellular lysates were subjected to SDS-PAGE (A to C) or immunoprecipitation (D), followed by Western analysis with the indicated antibodies.
We utilized two-dimensional (2D) monolayer culture and anchorage-independent three-dimensional (3D) multicellular spheroid culture to compare the effect of hypoxic (0.5%) O2 on endogenous and exogenous VPS4B levels. In both 2D and 3D cultures, while hypoxia stabilized the steady-state levels of HIF-1α and HIF-2α, a decrease of endogenous VPS4B upon chronic hypoxic exposure was observed (Fig. 1B). Notably, the steady-state levels of VPS4B, prior to hypoxic exposure, in cells in 3D cultures were significantly lower than those in cells in 2D cultures. To confirm that the observed decreased VPS4B level was mediated through protein degradation, we established doxycycline-inducible VPS4B(wt) or dominant negative VPS4B(EQ) cells (see Fig. S1B in the supplemental material). VPS4B steady-state levels were measured in 3D cultures treated with a combination of bafilomycin A1 (BafA1, an inhibitor of lysosomal vacuolar H+-ATPase [22]) and MG132, with or without exogenous hypoxic stress (Fig. 1C). During hypoxia, BafA1 alone exacerbated the degradation of ectopically expressed VPS4B(wt), whereas it retarded degradation of VPS4B(EQ) (Fig. 1C). In contrast, treatment with MG132 partially reversed the degradation of VPS4B(wt) and VPS4B(EQ) in 3D spheroids. While it is likely that VPS4B(wt) and VPS4B(EQ) utilized different protein degradation pathways under hypoxic conditions, the underlying mechanism is unclear. Treatment with both BafA1 and MG132 had little effect on the steady-state levels of VPS4B(wt or EQ) during normoxia (see Fig. S1C in the supplemental material), supporting the idea that the stability of VPS4B(wt) and VPS4B(EQ) is subjected to regulation by the O2 environment.
To confirm the involvement of ubiquitylation in hypoxia-mediated VPS4B degradation, we next overexpressed HA-tagged ubiquitin (HA-Ub) in HEK293 cells during hypoxic or normoxic exposure in 2D and 3D cultures. Upon exogenous Ub expression, an accumulation of high-molecular-weight ubiquitylated VPS4B was observed, which was more apparent following hypoxic exposure (Fig. 1D). Comparatively, the accumulation of ubiquitylated-VPS4B species was more pronounced in 3D spheroids than 2D monolayers (see Fig. S1D in the supplemental material). As expected, overexpression of Ub accelerated the degradation of ectopically expressed VPS4B(wt) and VPS4B(EQ) after hypoxic exposure in 2D monolayer cultures (see Fig. S1E). Together, these results support the involvement of the ubiquitin-proteasome system in both 3D culture-associated and hypoxia-mediated VPS4B degradation.
Hypoxic stress is generated within spheroids.
The decrease in the steady-state level of p62 is widely used as an indicator for autophagic flux via interactions with both LC3 and ubiquitylated proteins (3, 4). Accordingly, the accelerated decrease in p62 levels in 3D spheroids (Fig. 1B) suggested that there was more active autophagy-mediated destabilization of p62 in 3D spheroids than in 2D cultures, presumably through hypoxic stress within 3D spheroids. To address whether the accelerated VPS4B or p62 decrease is caused by an endogenous hypoxic stress in spheroids, we used a quantitative assay developed by Shvets et al. (52) to monitor GFP-LC3 turnover by fluorescence-activated cell sorting (FACS) analysis using the experimental protocol shown in Fig. 2A (left). Briefly, the assay monitored the reduction in GFP-LC3 fluorescence, which reflected the delivery of GFP-LC3 into lysosomes for degradation. A decrease of GFP-LC3 fluorescence in MEFs was observed 8 h after the formation of spheroids (Fig. 2A, right). Upon further incubation, the GFP-LC3 fluorescence in the spheroids remained at reduced levels, and additional exogenous hypoxic exposure elicited a modest, if any, further decrease in GFP-LC3 fluorescence. In contrast, no change in GFP-LC3 intensity was observed in cells cultured as a monolayer until hypoxic exposure. Together with the observation that BafA1 pretreatment prevented the decrease in GFP-LC3 fluorescence in 3D as well as 2D cultures, our data suggest that the 3D spheroid culture is a useful model for studying endogenous hypoxic stress.
Next, we utilized three different approaches to further confirm that the endogenous hypoxic stress within 3D spheroids decreased VPS4B levels in spheroids. First, we generated HEK293 cells with stably integrated EGFP-LC3:tRFP, a bicistronic unit with a CAG-driven EGFP-LC3 and an IRES-translated tRFP, to monitor autophagy in spheroids. As our results indicated that lysosomal turnover of LC3 is indicative of hypoxic stress, we monitored EGFP and RFP levels in a series of z sections of HEK293/EGFP-LC3:tRFP spheroids by confocal microscopy (Fig. 2B). Consistent with a previous report that there were greater oxygen gradients in spheroids than 2D growth (24), a gradual decrease in EGFP-LC3 fluorescence with a relatively constant RFP expression was observed from the outer surface toward the center of the spheroid cultured in nutrient-rich medium during normoxia (Fig. 2B). Second, Hypoxyprobe staining confirmed that cells in the inner layers of spheroids were under hypoxic stress even when the spheroids were cultured in a normoxic environment of 21% O2 (Fig. 2C; also, see Fig. S2 in the supplemental material), despite the insignificant levels of HIF-1α and HIF-2α detected in 3D spheroids prior to hypoxic exposure by Western analyses (Fig. 1B). Third, analysis of engineered HEK293/DsRed-VPS4B:tGFP cells revealed that both DsRed and GFP signals from bicistronic DsRed-VPS4B:tGFP were equally detected in monolayer cells cultured under normoxic conditions, whereas hypoxic exposure of 2D-cultured cells decreased DsRed-VPS4B signals without markedly affecting GFP levels (Fig. 2D). As expected, a reduced DsRed-VPS4B intensity was observed in the inner layers of the spheroids, where cells are likely to undergo hypoxia, with a relatively uniform GFP intensity throughout the spheroids (Fig. 2E; also, see Fig. S3 in the supplemental material). Altogether, our results suggested that hypoxic stress associated with spheroid formation results in the destabilization of VPS4B, which is independent of induction of HIF-1α or HIF-2α.
Loss of VPS4B function increases EGFR stability and prolongs EGF-induced phosphorylation.
Although VPS4B plays a role in regulating basal autophagy (see Fig. S4A in the supplemental material), overexpression of dominant negative VPS4B(EQ) elicited a limited effect on hypoxia-induced autophagy in two different cell types (see Fig. S4B and C). We therefore searched for biological consequences associated with hypoxia-mediated VPS4B degradation. Given the prominent role of endocytic trafficking in degradation of membrane receptors (reviewed in reference 42) and the observation that chronic hypoxia upregulates EGFR protein level (see Fig. S5A in the supplemental material) (15), we speculated that hypoxia-mediated VPS4B destabilization might dysregulate EGFR protein stability and/or signaling. Three breast cancer cell lines, MCF7, SKBR3, and MDA-MB-231, with distinct expression levels of EGFR and HER2 (see Fig. S5B) (33) were used to analyze VPS4B-dependent consequences for EGFR signaling.
First, EGF-induced EGFR signaling and receptor downregulation were examined in 2D and 3D culture systems of EGFR/HER2-expressing SKBR3 breast cancer cells. As localized hypoxia accumulated within spheroids, a more robust EGFR activation, based on the evaluation of Tyr phosphorylation with commercially available antibodies, was observed, with an attenuated EGF-induced EGFR degradation in 3D spheroids compared with that obtained in 2D monolayers (Fig. 3A). Knockdown of VPS4B by sh-VPS4B in SKBR3 2D monolayer cultures resulted in a prolonged Tyr phosphorylation of EGFR upon EGF treatment, which was further accentuated in 3D spheroids of SKBR3/sh-VPS4B cells (Fig. 3A). These data suggested that a decrease in VPS4B level brought about by VPS4B knockdown and/or spheroid formation results in an increased stability of Tyr-phosphorylated EGFR. Comparable results were observed in EGF-treated, EGFR-positive, HER2-negative MDA-MB-231 cells (see Fig. S5C in the supplemental material). Similarly, we investigated whether dominant negative VPS4B(EQ) would interfere with EGFR downregulation. As shown in Fig. S5D in the supplemental material, expression of VPS4B(EQ) resulted in a delay of EGFR degradation following EGF treatment in MCF7 cells, compared to MCF7 cells expressing VPS4B(wt).
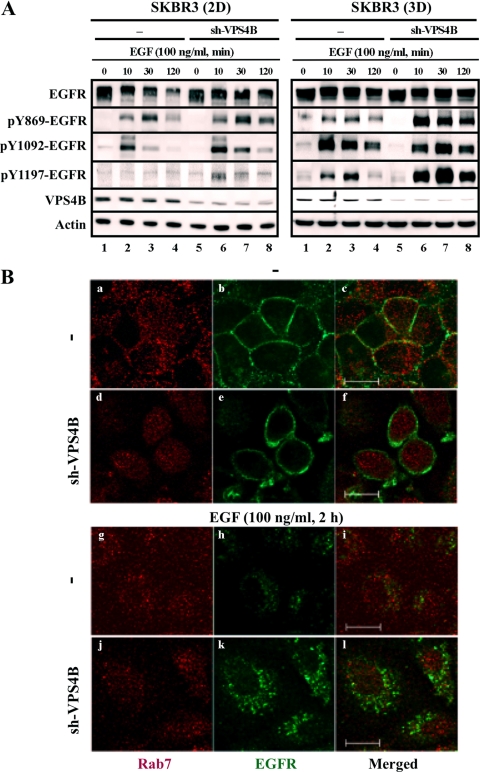
Fig 3.
VPS4B depletion prolongs EGFR expression and signaling in EGF-treated SKBR3 cells. (A) Parental and VPS4B-knockdown (sh-VPS4B) SKBR3 cells in 2D (left) and 3D (right) cultures were serum starved for 16 h. Cells were subsequently treated with EGF (100 ng/ml) for the indicted times prior to harvesting. Equal amounts of whole-cell lysates were subjected to Western blot analyses with the indicated antibodies. (B) SKBR3 and SKBR3/sh-VPS4B cells were treated with EGF as for panel A and immunostained with primary antibody and the corresponding fluorescent secondary antibodies to detect EGFR (green) and Rab7 (red). Bar, 10 μm.
Overexpression of HER2 reportedly inhibits EGF-induced EGFR downregulation (25, 63); however, our results on the HER2-expressing SKBR3 (Fig. 3A) and HER2-negative MDA-MB-231 cells (see Fig. S5C in the supplemental material) indicated that the lessening of EGF-induced EGFR downregulation mediated by the reduction of VPS4B was irrespective of HER2 context. We further investigated the effects of various HER2 mutants on EGFR turnover and signaling in the presence of dominant negative VPS4B(EQ) in MCF7 cells. Clearly, expression of HER2, either wild type, kinase dead, or constitutively active, did not affect the ability of VPS4B(EQ) to enhance EGF-induced EGFR Tyr phosphorylation in MCF7 cells (see Fig. S5E). In addition, compared to cells expressing VPS4B(wt), increased HER2 Tyr phosphorylation was observed in MCF7 cells expressing VPS4B(EQ) (see Fig. S5E). Altogether, these observations led us to speculate that compromised VPS4B function not only impairs EGF-induced EGFR downregulation irrespective of HER2 context, it may also promote HER2 Tyr phosphorylation.
Next, confocal microscopy was used to visualize trafficking of ligand-bound EGFR in SKBR3 cells with attenuated VPS4B expression. As expected, EGFR was localized at the plasma membranes of both control and knockdown cells prior to EGF stimulation, and EGF treatment triggered EGFR internalization (Fig. 3B). Although the Rab7-containing endosomal compartment is reportedly involved in EGFR trafficking (34), no significant colocalization between EGFR and Rab7 signals was observed either prior to or 2 h after EGF stimulation in SKBR3 cells. Consistent with a previous report that expression of SKD1(EQ), a mouse homologue of VPS4B(EQ), impaired the degradation of internalized EGFR-ligand EGF as a result of an accumulation of perinuclear “E235Q compartments” (68), there were clearly more EGFR signals visualized in SKBR3/sh-VPS4B cells at 2 h after EGF treatment. In contrast, EGFR signals largely disappeared in control cells at the same time point (Fig. 3B). Quantification of EGFR pixel intensity profiles verified that following EGF stimulation, EGFR pixel intensity was 4-fold lower in SKBR3 cells than in SKBR3/sh-VPS4B cells (see Fig. S6A in the supplemental material). These results suggested that VPS4B knockdown attenuated EGF-induced EGFR downregulation. Moreover, the inhibitory effect of gefitinib on EGFR phosphorylation was less robust in 3D spheroids of SKBR3 and SKBR3/sh-VPS4B than the corresponding 2D cultures (see Fig. S6B). Likewise, a series of time-dependent changes in the intensity level of green pixels of GFP-HER2 were observed in EGF-treated control cells but not in EGF-treated, VPS4B(EQ)-induced MCF7 cells (see Fig. S7 in the supplemental material), indicating a lack of GFP-HER2 trafficking in VPS4B(EQ)-expressing cells along the x axis (indicated by arrows in Fig. S7A; tracings are summarized in Fig. S7B). Colocalization of GFP-HER2 with membrane was more notable in EGF-treated control MCF7 cells than that of the VPS4B(EQ)-expressing cells (yellow membrane signals in Fig. S7A). Collectively, we concluded that VPS4B depletion or inactivation alters the subcellular localization and/or compartmentalization of activated EGFR and HER2, perhaps through modified endocytic/recycling pathways, thus affecting their degradation and phosphorylation profile.
Decreased VPS4B expression alters EGFR but not EGFRvIII downstream signaling.
To further delineate the effect of loss of VPS4B function on EGFR signaling, we examined whether VPS4B knockdown affected the downstream signaling of the EGFR variant, EGFRvIII. Consistent with previous findings (19), NR6-EGFRvIII-expressing cells exhibited ligand-independent, constitutive Tyr phosphorylation of EGFRvIII in the absence of EGF (Fig. 4A). Notably, VPS4B knockdown in the NR6-EGFRvIII/sh-VPS4B cells did not affect either the stability or the phosphorylation state of EGFRvIII. Since AP-1 and Elk-1 are well-characterized downstream transcription factor targets of EGFR, AP-1- and Elk-1-dependent promoter-reporter assays were used to assess whether VPS4B knockdown affected their activities in EGFR- and EGFRvIII-expressing cells. EGF-stimulated AP-1-dependent promoter activation was enhanced by VPS4B underexpression in SKBR3 cells (Fig. 4B, left). However, Elk-1-mediated promoter activation was suppressed by VPS4B knockdown in EGF-treated SKBR3 cells (Fig. 4B, right). In addition, while an upregulation of pERK1/2 and JNK1/2 signaling was observed in EGF-treated SKBR3 cells upon VPS4B downregulation, the same effect in NR6-EGFRvIII cells was modest (see Fig. S8 in the supplemental material). Conceivably, the observed promotion effect on AP-1 transactivation may be a result of the sequestration of EGFR in late endosomes, as reported by Taub et al. (56).
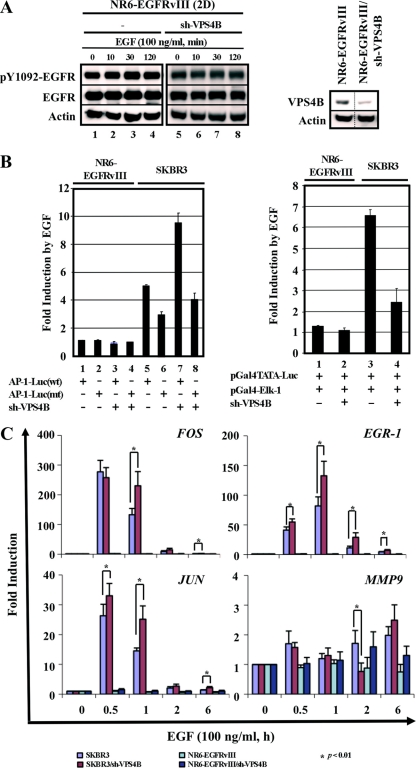
Fig 4.
sh-VPS4B enhances AP-1 function in SKBR3 cells. (A) NR6-EGFRvIII and NR6-EGFRvIII/sh-VPS4B cells in 2D monolayers were serum starved and treated with EGF (100 ng/ml) for the indicated times, as described for Fig. 3A. Equal amounts of total cell lysates were subjected to SDS-PAGE, followed by Western analysis with the indicated antibodies (left). Knockdown by sh-VPS4B in NR6-EGFRvIII was confirmed by Western analysis (right). (B) For AP-1 transactivation assays (left), pairs of SKBR3:SKBR3/sh-VPS4B and NR6-EGFRvIII:NR6-EGFRvIII/sh-VPS4B cells were transfected with either AP-1-Luc(wt) or AP-1-Luc(mt) and treated with vehicle or EGF (100 ng/ml) at 24 h posttransfection for 6 h. For Elk-1 transactivation assays (right), pairs of SKBR3:SKBR3/sh-VPS4B and NR6-EGFRvIII:NR6-EGFRvIII/sh-VPS4B cells were transfected with pGal4TATA-Luc and pGal4-Elk-1 fusion protein expression constructs and treated as described for AP-1 assays. Cell lysates were analyzed and normalized to cotransfected Renilla luciferase. Values for induction by EGF are means and SD (n = 3) of duplicate experiments; normalized luciferase activities of vehicle-treated SKBR3 and NR6-EGFRvIII cells were set at 1. (C) For quantitative RT-PCR analyses, pairs of SKBR3:SKBR3/sh-VPS4B and NR6-EGFRvIII:NR6-EGFRvIII/sh-VPS4B cells were serum starved and treated with vehicle or EGF (100 ng/ml) for the indicated times prior to RNA isolation. RNA isolation, cDNA synthesis, and quantitative PCR were performed as described in Materials and Methods. The relative change was calculated such that each transcript level in cells before EGF treatment (0 h), normalized to that of GAPDH, was set to 1. Results are means and SD from three independent experiments.
To further analyze the differential effect of VPS4B on EGF-triggered AP-1 transactivation, we performed quantitative RT-PCR analyses on the expression profiles of endogenous FOS, JUN, EGR-1, and MMP9 in SKBR3 and NR6-EGFRvIII cells with distinct VPS4B contexts upon EGF stimulation. As shown in Fig. 4C, the expression of FOS and JUN, components of the AP-1 transcription factor, was rapidly and transiently induced with a maximal induction between 30 min and 1 h after EGF treatment in SKBR3 cells. The induction at 1 h posttreatment was significantly higher for VPS4B knockdown cells than the parental SKBR3 cells. In contrast, NR6-EGFRvIII cells, irrespective of VPS4B context, were refractory to EGF treatment and exhibited no appreciable induction (Fig. 4C). This was consistent with the AP-1-Luc assays performed in SKBR3 and NR6-EGFRvIII cells with distinct VPS4B contexts (Fig. 4B). Interestingly, knockdown of VPS4B in SKBR3 cells led to the opposite effect over the time course on two of the ELK-1 targets, MMP9 and EGR-1. This could be explained by the observations that JUN was robustly induced in SKBR3 (parental and sh-VPS4B) cells (Fig. 4C) and that JUN regulates EGR-1 expression via binding directly to the AP-1 element and indirect recruitment of phosphorylated Elk-1 to the serum response elements (17). This further exemplified the complexity of EGFR/ELK-1 signaling. Together, these results demonstrated that VPS4B regulates the stability and downstream signaling of EGFR but not those of its mutant variant, EGFRvIII.
Functional consequence of VPS4B-mediated EGFR regulation in breast cancer cells.
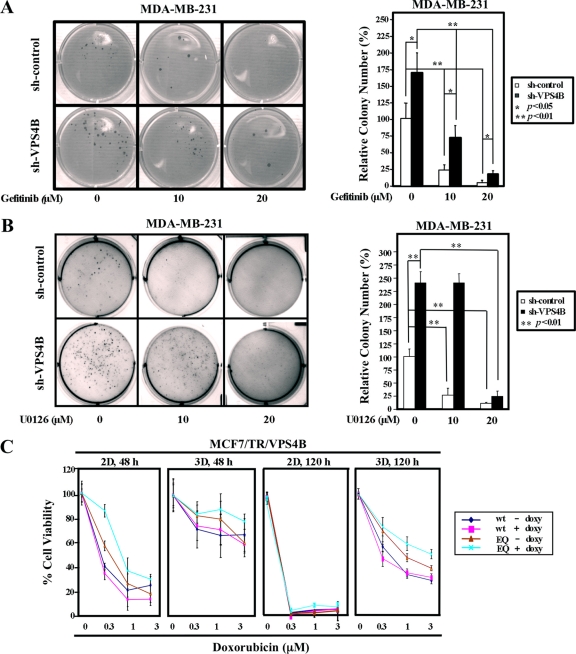
To further understand the biological consequences from compromised VPS4B function in breast cancer cells, we examined the effect of VPS4B knockdown on anchorage-independent growth in the presence and absence of the EGFR inhibitor gefitinib or the MEK inhibitor U0126 in MDA-MB-231 cells (Fig. 5A and B). First, higher numbers of colonies were observed in VPS4B-underexpressing MDA-MB-231 cells than in the corresponding parental cells (Fig. 5A and B). Larger colonies were formed from MDA-MB-231/sh-VPS4B cells than from MDA-MB-231 cells (see Fig. S9A [left] in the supplemental material). As EGFR/ERK/AP-1 signaling pathway was enhanced by VPS4B knockdown (Fig. 4C; also, see Fig. S8 in the supplemental material), both the EGFR inhibitor gefitinib and the MEK inhibitor U0126 dampened the anchorage-independent growth more effectively in parental MDA-MB-231 cells than in VPS4B-underexpressing cells (Fig. 5A and B). These observations further support the functional roles of VPS4B-knockdown in promoting EGFR/ERK1/AP-1 signaling and thus tumorigenesis.
Fig 5.
Loss of VPS4B function increases resistance to gefitinib, U0126, and doxorubicin in breast cancer cells. (A and B) Knockdown of VPS4B attenuated the ability of gefitinib or U0126 to inhibit colony formation. Pairs of MDA-MB-231 cells with or without VPS4B knockdown were analyzed for anchorage-independent growth in the presence of EGF (10 ng/ml) and indicated concentrations of gefitinib or U0126 by soft-agar assays. The bar graph shows the mean foci numbers in tested wells. Values are means and SD (n = 3) from triplicate experiments. **, P < 0.01. Images of a representative assay plate are shown (left). Relative colony number (%) is calculated with the mean colony number of MDA-MB-231/sh-control cells in the absence of gefitinib and U0126 set to 1. (C) MCF7/TR/VPS4B(wt) and MCF7/TR/VPS4B(EQ) cells in 2D monolayers and 3D spheroids were treated with doxorubicin for 48 h or 120 h and subsequently harvested for the MTS assay. Expression of VPS4B was induced with doxycycline (doxy; 1 μg/ml). The percentage of viable cells is calculated relative to vehicle-treated control cells; values are means and SD (n = 3) of triplicate experiments.
MCF7 cells are known to be sensitive to genotoxic doxorubicin treatment and to express EGFR at relatively low level (see Fig. S5B in the supplemental material). We therefore investigated whether MCF7 cells, with or without compromised VPS4B function, would respond to doxorubicin differently when grown in 2D cultures and 3D spheroids. Treatment with increasing concentrations of doxorubicin for 48 h elicited a concentration-dependent cell growth inhibition in 2D cultures of MCF7/VPS4B(wt) and MCF7/VPS4B(EQ) cells (Fig. 5C). In contrast, 3D spheroid proliferation was only slightly reduced at 48 h posttreatment, suggesting that cells become more resistant to doxorubicin in spheroids. A concentration-dependent cell growth inhibition was observed at 120 h posttreatment in spheroid 3D cultures, yet no significant number of viable cells was detected after the same treatment for 120 h in 2D monolayers. In addition, VPS4B(EQ)-expressing MCF7 cells were more resistant to doxorubicin treatment than VPS4B(wt)-expressing cells in both 2D and 3D cultures (Fig. 5C). These results suggested that functional loss of VPS4B either from 3D-induced VPS4B degradation or expression of dominant negative VPS4B(EQ) contributes to the enhanced doxorubicin resistance.
Moreover, we examined the effect of doxorubicin treatment on p62 level and ERK1/2 signaling when VPS4B function was compromised. As expected, a higher level of p62 and an augmented activation of pERK1/2 were observed in VPS4B(EQ)-expressing cells relative to those in VPS4B(wt)-expressing MCF7 cells in response to doxorubicin treatment, both in 2D cultures and in 3D spheroids (see Fig. S9B in the supplemental material). This is especially the case for doxorubicin treatment at higher concentrations (1 and 3 μM). Collectively, these results suggested that compromised VPS4B function prolongs cell survival against doxorubicin even in the low-EGFR-expression MCF7 cells.
VPS4B mRNA expression is reduced in high-grade and recurrent breast carcinomas.
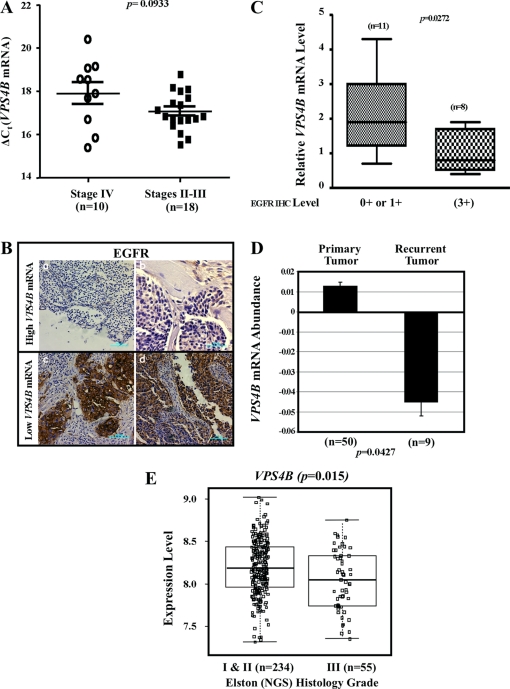
Lastly, we speculated that VPS4B level was downregulated in EGFR-overexpressing, high-grade solid tumors, based on the established role of enhanced EGFR activation in tumor progression together with the observed enhancement of EGFR stability and signaling as a result of attenuated VPS4B. Due to the lack of proper anti-VPS4B antibody for immunohistochemical (IHC) analyses, we resorted to measuring VPS4B mRNA amounts in stage IV (n = 10) and stage II-III (n = 18) breast tumor specimens by quantitative RT-PCR. Overall, VPS4B mRNA levels were approximately 1.8-fold lower in stage IV (ΔCT: 17.9 ± 0.5) than in stage II-III (ΔCT, 17.0 ± 0.2) (Fig. 6A). Analysis of three normal breast tissues revealed that VPS4B mRNA levels (ΔCT, 14.0, 14.4, and 14.5) were significantly higher than those observed in the breast cancer tissues. To determine whether these decreases in VPS4B mRNA expression correlated with changes in EGFR expression, we examined EGFR expression in 19 human breast tumor specimens by IHC analysis (Fig. 6B). A reverse correlation between VPS4B mRNA level and EGFR protein levels in breast samples was observed (P = 0.0272) (Fig. 6C). Lastly, we determined the potential relevance of VPS4B expression levels to human breast cancer recurrence by retrospectively analyzing VPS4B mRNA expression in the GEO DataSets (http://www.ncbi.nlm.nih.gov/gds). Analysis of data set GDS2415 showed that VPS4B mRNA expression was significantly lower in the samples of recurrent tumors (n = 9) than in primary tumors (n = 50) (Fig. 6D; P = 0.042). To validate this observation in a large cohort of samples, we further analyzed VPS4B mRNA expression in a larger human breast tumor cohort reported by Ivshina et al. (GEO accession number GSE4922) (20). VPS4B mRNAs were significantly lower in samples from high-grade breast tumors (n = 55) than in samples from low-grade tumors (n = 234) (Fig. 6E; P = 0.015). Collectively, these results indicated that decreased VPS4B expression is associated with tumor aggressiveness for patients with breast cancer and that this is correlated with increased EGFR abundance.
Fig 6.
Downregulation of VPS4B mRNA levels in high-grade or recurrent breast tumors. (A) Comparison of VPS4B mRNA expression levels between stage IV (n = 10) and stage II-III (n = 18) breast cancer, using a nonparametric test. Bars represent means and standard errors of the mean (SEM). (B) EGFR immunostaining in breast tumor specimens with low VPS4B mRNA levels. Two representative images of samples with 0+ EGFR levels (a and b) and two representative images of samples with 3+ EGFR levels (c and d) are shown. Bar, 100 μm (a, c, and d) and 50 μm (b). (C) Comparison of relative VPS4B expression and EGFR level. Relative VPS4B expression is significantly lower when EGFR is high (IHC, 3+; n = 8) than when it is low (IHC, 0+ or 1+; n = 11). P = 0.0272 using a two-tailed unpaired t test. The means and SEM were 2.064 ± 0.3228 for low-EGFR tumors and 1.038 ± 0.2211 for high-EGFR tumors. (D) Retrospective analysis of a gene expression data set for 50 primary breast tumors and 9 recurrent tumors using Affymetrix GeneChip expression analysis. VPS4B abundance is reported as measurements extracted from original submitter-supplied GEO sample records and reflects the measured level of abundance of VPS4B across the samples that make up the data set. Values are presented in arbitrary units (log2 ratio). VPS4B expression is significantly downregulated in recurrent tumors. P = 0.0427. The means and SEM are 0.0129 ± 0.006 for primary tumors and −0.0448 ± 0.005 for recurrent tumors. (E) Retrospective analysis of gene expression data set of 234 Elston (Nottingham grading system [NGS]) histology grade I and II breast tumors and 55 grade III tumors using Affymetrix GeneChip expression analysis. VPS4B expression levels are measurements extracted from the original report by Ivshina et al. (20) and reflect the measured level of abundance of VPS4B across the samples that make up the data set. Values are in arbitrary units (log intensity). VPS4B expression is significantly downregulated in grade III tumors. P = 0.015. The means and SEM are 8.04 ± 0.05 for grade III tumors and 8.18 ± 0.02 for grade I and II tumors. Box plots represent quartiles and extremes of expression values.
DISCUSSION
Hypoxia and overproduction of EGFR are two known factors that contribute to poor prognosis and poor response to cancer therapies. In this report, we provide evidence for a novel link between the hypoxia-induced reduction in VPS4B protein levels and altered compartmentalization of activated EGFR and its downstream signaling in breast cancer cells. To adapt to hypoxic conditions, cells typically activate HIF-1 to mediate gene transcription of a large number of genes and in particular those regulating cell metabolism (reviewed in reference 5). Accumulating reports have also suggested that there are hypoxic adaptations other than the HIF response (6, 54, 64, 67). For example, there is evidence to support a functional role for the induction of a lysosomal degradation pathway known as autophagy in response to hypoxic stress (31, 41). The molecular mechanism by which chronic hypoxia regulates genome- and autophagy-independent adaptive responses via protein degradation is largely unknown. To our knowledge, this report represents the first demonstration that chronic hypoxia results in VPS4B degradation via the ubiquitin-proteasome system. However, it is equally possible that metabolic stress, associated with chronic hypoxia, could account for the observed VPS4B destabilization. Nonetheless, signal transduction through EGFR governs a variety of biological responses, including proliferation and survival of many cancer cells. For these reasons, dysregulating EGFR abundance and compartmentalization can have dramatic effect on tumorigenesis and treatment resistance. Our findings identify hypoxia-induced VPS4B reduction as a previously undisclosed means to potentiate EGFR signaling.
Consistent with previous reports that EGFR remains activated after being endocytosed and that some, but not all, EGFR-activated signaling pathways require endocytosis for their maximal activation (7, 57, 59), our results indicated that EGF-mediated AP-1-dependent reporter activation is augmented upon VPS4B knockdown (Fig. 4B and C). It is conceivable that this preferential EGFR/ERK/AP-1 signaling event results from distinct intracellular compartmentalization of activated EGFR. It is also possible that additional EGFR-mediated signaling events or other membrane receptors, such as HER2, could be affected by loss of VPS4B function. However, the mutant variant form of EGFR, EGFRvIII, which does not bind to ligand or become internalized, does not respond to loss of VPS4B as EGFR or HER2 did. Taken together, these findings support an intriguing role for reduced VPS4B protein and its associated function in pathogenesis of solid human tumors through the promotion of EGFR/HER2 signaling during tumor hypoxia. Clearly, further studies are needed to delineate the machinery for VPS4B degradation and the underlying mechanism during normoxia and chronic hypoxia.
Using soft-agar assays and a multicellular 3D spheroid model, we showed that lower VPS4B expression promotes resistance to gefitinib, U0126, or doxorubicin (Fig. 5). Notably, mutations or amplifications of the EGFR gene are less widespread than its overexpression profiles (47). Recently, Cao et al. reported that breast cancer cells having no or low HER2 expression may become resistant to therapy due to enhanced HER2 gene expression following irradiation (8). By the same analogy, it is conceivable that in solid human tumors that do not display genetic alterations of the EGFR gene, microenvironments such as hypoxia may trigger VPS4B downregulation, resulting in elevated EGFR abundance and signaling. Accordingly, the decrease in VPS4B level may act as a universal trigger to phenotypically mimic EGFR amplification, contributing to the ability of cancer cells to survive under hypoxic conditions or to develop resistance to cancer therapy. Our findings on hypoxia-mediated VPS4B reduction and EGFR regulation are corroborated by findings from several studies. For instance, the depletion of other ESCRT or autophagy components, such as Hrs, STAM1/2, Tsg101, Vps24, Vps25, UVRAG, or Rubicon, reportedly affects EGFR degradation or signaling (1, 2, 26–28, 30, 58). Knockdown of individual ESCRT-0 through -3 components constitutively delays or inhibits EGFR degradation; however, it affects EGFR signaling differentially (32). While knockdown components of ESCRT-0 and -1 prolong EGF-induced EGFR signaling, degradation and signaling are uncoupled in ESCRT-2 and -3 knockdown cells (2, 29). Additional reports that VPS4 is essential for the degradation of KFERQ motif-containing proteins through MVB (45) and that EGFR contains a KFERQ-like motif (51) strongly support our observations. The uncovering of additional pathways, such as that by which hypoxia downregulates the transcription of rabaptin-5, a critical effector for Rab5-mediated endosome fusion, to impede EGFR endocytosis (62), further attests to the importance of EGFR signaling control during hypoxia. Interestingly, our results suggest that loss of VPS4B function also alters GFP-HER2 trafficking (see Fig. S7 in the supplemental material). The effect of rabaptin-5 on HER2 trafficking is yet to be demonstrated. Moreover, knockdown of VPS4 has been demonstrated to affect microautophagy (45), and a functional MVB pathway is required for autophagic clearance of protein aggregates, such as those associated with neurodegenerative disease (14).
Our analyses further reveal that lower VPS4B mRNA levels, which are inversely correlated with high EGFR expression, are associated with high-grade or recurrent breast tumors (Fig. 6). Although we do not have direct evidence of altered VPS4B transcriptional control in tumor cells, the decreased VPS4B mRNA level would presumably result in even greater VPS4B downregulation during chronic hypoxia. Dysregulated EGFR signaling has been implicated in the pathogenesis of many human cancers, including breast cancer, colorectal cancer, and glioblastoma multiforme, and this correlates with disease progression and poor prognosis (37). In addition to EGF-mediated signaling, EGFR is also activated in a ligand-independent manner by radiation, initiating signaling events which are the same as or similar to those induced by EGF (16, 50). Many reports have further suggested that enhanced EGFR signaling plays a role in fostering DNA double-strand break repair induced by either irradiation or chemotherapy (16, 35, 43, 60) and in increasing glucose transport and ATP production in cancer cells in a kinase-independent manner (65). Altogether, our findings distinguish VPS4B from other ESCRT components by performing a putative “tumor suppressor” function, suggesting its potential clinical relevance. Collectively, our results allow us to speculate that VPS4B has potential as a biomarker for predicting resistance to anticancer therapy.
In summary, our current work supports a model wherein accumulation of hypoxic stress triggers destabilization of VPS4B, thereby promoting tumorigenesis. Based on our observations, VPS4B appears to be a critical component that connects hypoxia signaling with endosomal trafficking. As such, VPS4B reduction may represent a common denominator for the aberrant EGFR signaling often observed in solid tumors. Clearly, identifying additional mechanisms that positively or negatively regulate EGFR could have profound implications for treating human diseases. Further studies directed at identifying the regulators of VPS4B degradation will be essential to understand how hypoxia results in the nongenomic adaptive response program.
Supplementary Material
ACKNOWLEDGMENTS
We are sincerely grateful to Chih-Pin Liu, Mei-Ling Kuo, Susan Kane, Shao-Chun Wang, and Yi-Rong Chen for their valuable reagents, helpful suggestions, and critical reading of the manuscript. We also thank Marcia M. Miller and Mariana Tihova of the Electron Microscopy Core at City of Hope for transmission electron microscopy analyses, Brian Armstrong of the Light Microscopy Digital Imaging Core at City of Hope for confocal microscopy analyses, Yate-Ching Yuan of the Bioinformatics Core at City of Hope for advice on Affymetrix array data analyses, Sofia Loera of the Pathology Core at City of Hope for immunohistochemical analyses, the RNAi Consortium at Academia Sinica for providing the lentiviral constructs against human VPS4B, the Translational Research Laboratory at City of Hope for providing breast cancer tissue specimens, members of David Ann's laboratory for helpful discussions, and Silvia R. da Costa and Keely Walker for editing.
This work was supported in part by National Institutes of Health Research Grants R01DE10742 and R01DE14183 (to D.K.A.), R01AG025323 (to A.Y.), R01CA74138 (to D.J.), and R01CA127541 (to Y.Y.) and by National Science Council Grant 97-2321-B-001-107 (to H.-M.S.).
Footnotes
Published ahead of print 17 January 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Bache KG, Slagsvold T, Stenmark H. 2004. Defective downregulation of receptor tyrosine kinases in cancer. EMBO J. 23:2707–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bache KG, et al. 2006. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol. Biol. Cell 17:2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bjorkoy G, et al. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bjorkoy G, Lamark T, Johansen T. 2006. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2:138–139 [DOI] [PubMed] [Google Scholar]
- 5. Brahimi-Horn MC, Bellot G, Pouyssegur J. 2011. Hypoxia and energetic tumour metabolism. Curr. Opin. Genet. Dev. 21:67–72 [DOI] [PubMed] [Google Scholar]
- 6. Buchberger A, Bukau B, Sommer T. 2010. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell 40:238–252 [DOI] [PubMed] [Google Scholar]
- 7. Burke P, Schooler K, Wiley HS. 2001. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol. Biol. Cell 12:1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao N, et al. 2009. NF-κB-mediated HER2 overexpression in radiation-adaptive resistance. Radiat. Res. 171:9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen JL, et al. 2008. Novel roles for protein kinase Cδ-dependent signaling pathways in acute hypoxic stress-induced autophagy. J. Biol. Chem. 283:34432–34444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen JL, et al. 2009. PKC delta signaling: a dual role in regulating hypoxic stress-induced autophagy and apoptosis. Autophagy 5:244–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clavijo C, Chen JL, Kim KJ, Reyland ME, Ann DK. 2007. Protein kinase Cδ-dependent and -independent signaling in genotoxic response to treatment of desferroxamine, a hypoxia-mimetic agent. Am. J. Physiol. Cell Physiol. 292:C2150–C2160 [DOI] [PubMed] [Google Scholar]
- 12. Esclatine A, Chaumorcel M, Codogno P. 2009. Macroautophagy signaling and regulation. Curr. Top. Microbiol. Immunol. 335:33–70 [DOI] [PubMed] [Google Scholar]
- 13. Fader CM, Colombo MI. 2009. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 16:70–78 [DOI] [PubMed] [Google Scholar]
- 14. Filimonenko M, et al. 2007. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179:485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Franovic A, et al. 2007. Translational up-regulation of the EGFR by tumor hypoxia provides a nonmutational explanation for its overexpression in human cancer. Proc. Natl. Acad. Sci. U. S. A. 104:13092–13097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Golding SE, et al. 2009. Pro-survival AKT and ERK signaling from EGFR and mutant EGFRvIII enhances DNA double-strand break repair in human glioma cells. Cancer Biol. Ther. 8:730–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoffmann E, et al. 2008. Transcriptional regulation of EGR-1 by the interleukin-1-JNK-MKK7-c-Jun pathway. J. Biol. Chem. 283:12120–12128 [DOI] [PubMed] [Google Scholar]
- 18. Huang HS, et al. 1997. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J. Biol. Chem. 272:2927–2935 [DOI] [PubMed] [Google Scholar]
- 19. Huang PH, Xu AM, White FM. 2009. Oncogenic EGFR signaling networks in glioma. Sci. Signal. 2:re6. [DOI] [PubMed] [Google Scholar]
- 20. Ivshina AV, et al. 2006. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 66:10292–10301 [DOI] [PubMed] [Google Scholar]
- 21. Katzmann DJ, Odorizzi G, Emr SD. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893–905 [DOI] [PubMed] [Google Scholar]
- 22. Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. 2008. Does bafilomycin A(1) block the fusion of autophagosomes with lysosomes? Autophagy 4:849–950 [DOI] [PubMed] [Google Scholar]
- 23. Klionsky DJ, Emr SD. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290:1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leek RD, Stratford I, Harris AL. 2005. The role of hypoxia-inducible factor-1 in three-dimensional tumor growth, apoptosis, and regulation by the insulin-signaling pathway. Cancer Res. 65:4147–4152 [DOI] [PubMed] [Google Scholar]
- 25. Lenferink AE, et al. 1998. Differential endocytic routing of homo- and hetero-dimeric ErbB tyrosine kinases confers signaling superiority to receptor heterodimers. EMBO J. 17:3385–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li L, Cohen SN. 1996. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell 85:319–329 [DOI] [PubMed] [Google Scholar]
- 27. Liang C, et al. 2008. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10:776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lloyd TE, et al. 2002. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell 108:261–269 [DOI] [PubMed] [Google Scholar]
- 29. Malerod L, Stuffers S, Brech A, Stenmark H. 2007. Vps22/EAP30 in ESCRT-II mediates endosomal sorting of growth factor and chemokine receptors destined for lysosomal degradation. Traffic 8:1617–1629 [DOI] [PubMed] [Google Scholar]
- 30. Matsunaga K, et al. 2009. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11:385–396 [DOI] [PubMed] [Google Scholar]
- 31. Mazure NM, Pouyssegur J. 2010. Hypoxia-induced autophagy: cell death or cell survival? Curr. Opin. Cell Biol. 22:177–180 [DOI] [PubMed] [Google Scholar]
- 32. Metcalf D, Isaacs AM. 2010. The role of ESCRT proteins in fusion events involving lysosomes, endosomes and autophagosomes. Biochem. Soc. Trans. 38:1469–1473 [DOI] [PubMed] [Google Scholar]
- 33. Moasser MM, Basso A, Averbuch SD, Rosen N. 2001. The tyrosine kinase inhibitor ZD1839 (“Iressa”) inhibits HER2-driven signaling and suppresses the growth of HER2-overexpressing tumor cells. Cancer Res. 61:7184–7188 [PubMed] [Google Scholar]
- 34. Mosesson Y, Mills GB, Yarden Y. 2008. Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer. 8:835–850 [DOI] [PubMed] [Google Scholar]
- 35. Mukherjee B, et al. 2009. EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res. 69:4252–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nguyen HV, et al. 2006. SUMOylation attenuates sensitivity toward hypoxia- or desferroxamine-induced injury by modulating adaptive responses in salivary epithelial cells. Am. J. Pathol. 168:1452–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nicholson RI, Gee JM, Harper ME. 2001. EGFR and cancer prognosis. Eur. J. Cancer 37 Suppl. 4:S9–S15 [DOI] [PubMed] [Google Scholar]
- 38. O'Reilly MS, Holmgren L, Chen C, Folkman J. 1996. Angiostatin induces and sustains dormancy of human primary tumors in mice. Nat. Med. 2:689–692 [DOI] [PubMed] [Google Scholar]
- 39. Papandreou I, Lim AL, Laderoute K, Denko NC. 2008. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ. 15:1572–1581 [DOI] [PubMed] [Google Scholar]
- 40. Piper RC, Katzmann DJ. 2007. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23:519–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pursiheimo JP, Rantanen K, Heikkinen PT, Johansen T, Jaakkola PM. 2009. Hypoxia-activated autophagy accelerates degradation of SQSTM1/p62. Oncogene 28:334–344 [DOI] [PubMed] [Google Scholar]
- 42. Raiborg C, Stenmark H. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458:445–452 [DOI] [PubMed] [Google Scholar]
- 43. Rodemann HP, Dittmann K, Toulany M. 2007. Radiation-induced EGFR-signaling and control of DNA-damage repair. Int. J. Radiat. Biol. 83:781–791 [DOI] [PubMed] [Google Scholar]
- 44. Rusten TE, et al. 2007. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 17:1817–1825 [DOI] [PubMed] [Google Scholar]
- 45. Sahu R, et al. 2011. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell 20:131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. 2009. Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136:97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. 2010. A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer 10:59–64 [DOI] [PubMed] [Google Scholar]
- 48. Scheuring S, et al. 2001. Mammalian cells express two VPS4 proteins both of which are involved in intracellular protein trafficking. J. Mol. Biol. 312:469–480 [DOI] [PubMed] [Google Scholar]
- 49. Schlessinger J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103:211–225 [DOI] [PubMed] [Google Scholar]
- 50. Schmidt-Ullrich RK, Contessa JN, Lammering G, Amorino G, Lin PS. 2003. ERBB receptor tyrosine kinases and cellular radiation responses. Oncogene 22:5855–5865 [DOI] [PubMed] [Google Scholar]
- 51. Shen S, et al. 2009. Cyclodepsipeptide toxin promotes the degradation of Hsp90 client proteins through chaperone-mediated autophagy. J. Cell Biol. 185:629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shvets E, Fass E, Elazar Z. 2008. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy 4:621–628 [DOI] [PubMed] [Google Scholar]
- 53. Sorkin A, Von Zastrow M. 2002. Signal transduction and endocytosis: close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 3:600–614 [DOI] [PubMed] [Google Scholar]
- 54. Spriggs KA, Bushell M, Willis AE. 2010. Translational regulation of gene expression during conditions of cell stress. Mol. Cell 40:228–237 [DOI] [PubMed] [Google Scholar]
- 55. Stoscheck CM, Carpenter G. 1984. Down regulation of epidermal growth factor receptors: direct demonstration of receptor degradation in human fibroblasts. J. Cell Biol. 98:1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Taub N, Teis D, Ebner HL, Hess MW, Huber LA. 2007. Late endosomal traffic of the epidermal growth factor receptor ensures spatial and temporal fidelity of mitogen-activated protein kinase signaling. Mol. Biol. Cell 18:4698–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Teis D, Wunderlich W, Huber LA. 2002. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell 3:803–814 [DOI] [PubMed] [Google Scholar]
- 58. Thompson BJ, et al. 2005. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev. Cell 9:711–720 [DOI] [PubMed] [Google Scholar]
- 59. Vieira AV, Lamaze C, Schmid SL. 1996. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science 274:2086–2089 [DOI] [PubMed] [Google Scholar]
- 60. Vigneron A, Gamelin E, Coqueret O. 2008. The EGFR-STAT3 oncogenic pathway up-regulates the Eme1 endonuclease to reduce DNA damage after topoisomerase I inhibition. Cancer Res. 68:815–825 [DOI] [PubMed] [Google Scholar]
- 61. Wang SC, et al. 2004. Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell 6:251–261 [DOI] [PubMed] [Google Scholar]
- 62. Wang Y, et al. 2009. Regulation of endocytosis via the oxygen-sensing pathway. Nat. Med. 15:319–324 [DOI] [PubMed] [Google Scholar]
- 63. Wang Z, Zhang L, Yeung TK, Chen X. 1999. Endocytosis deficiency of epidermal growth factor (EGF) receptor-ErbB2 heterodimers in response to EGF stimulation. Mol. Biol. Cell 10:1621–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Webb JD, Coleman ML, Pugh CW. 2009. Hypoxia, hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen sensing. Cell Mol. Life Sci. 66:3539–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Weihua Z, et al. 2008. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell 13:385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. White SR, Lauring B. 2007. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic 8:1657–1667 [DOI] [PubMed] [Google Scholar]
- 67. Wouters BG, Koritzinsky M. 2008. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat. Rev. Cancer 8:851–864 [DOI] [PubMed] [Google Scholar]
- 68. Yoshimori T, et al. 2000. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell 11:747–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yuhas JM, Li AP, Martinez AO, Ladman AJ. 1977. A simplified method for production and growth of multicellular tumor spheroids. Cancer Res. 37:3639–3643 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.