Abstract
A temporal framework linking circadian rhythms and clocks to aging rates identifies a specific window of target of rapamycin (TOR) signaling associated with growth hormone (GH) and insulin-like growth factor (IGF-1) (largely exclusive of insulin) in early sleep. IGF-1 signaling is released by growth hormone secretory peaks and downregulation of IGF-1 binding protein-1 resulting in activation of the mitogen activated protein kinase/extracellular signal response kinase (MAPK/ERK) and phosphoinositide 3-kinase-protein kinase B (PI3K-PKB/Akt) signaling pathways. Phosphorylation of Akt activates TOR which mediates the protein synthesis and growth functions of the GH axis. TOR activity is also associated with downregulated stress resistance, faster aging and reduced lifespan. IGF-1 signaling is terminated by falling GH and upregulation of IGF-1 binding proteins mediated by somatostatin and rising corticosteroids in later sleep. This suppresses PI3K-Akt signaling, thus activating the forkhead transcription factors (FOXOs) and stress-resistance pathways involved in promoting longevity. Thus, sleep appears to encompass both pathways currently identified as most relevant to aging and they toggle successively on the phosphorylation status of Akt. I propose a modified version of Pearl’s rate of living theory emphasizing the hard-wired antagonism of growth (TOR) and stress resistance (FOXO). The sleep association of TOR and FOXO in temporally separated windows and their sequential temporal deployment may change much of the way we think about aging and how to manipulate it.
Keywords: Forkhead transcription factors, target of rapamycin, clocks, energy metabolism, rate of living, stress resistance, growth, sleep
“One Ring to rule them all, One Ring to find them, One Ring to bring them all and in the darkness bind them” J.R.R. Tolkien [1]
A highly conserved regulatory structure that critically modulates aging centers on insulin and IGF-1 signaling via the phosphoinositide 3-kinase pathway (PI3K). This pathway, and another IGF-1/growth-factor pathway, MAPK/ERK, also regulates anabolic processes such as protein synthesis and growth [2]. Among mammals, body size is strongly associated with longevity. Intra-specifically, those growing fastest to larger size generally live shorter lives, whereas inter-specifically, long-lived larger animals are associated with slower but prolonged growth [3–5]. Key models of extended longevity include mice with defective growth hormone or IGF-1 signaling such as the Ames and Snell dwarfs, GH receptor knockout mice, IGF-1 heterozygous receptor knockout mice, mice with deletion of the growth factor adaptor protein, p66shc, or deletion of either insulin receptor substrate-1 or 2 [6–11]. Alternatively, mice with upregulated growth, such as GH transgenic mice (TGM) express a progeroid aging syndrome [12–14]. Our research program with TGM found that their rapid growth was associated with highly elevated free radical processes and accelerated aging. This suggested that enforced allocation of resources to growth may compromise “longevity assurance” investments [4, 14, 15]. These ideas were strongly supported by more recent recognition that the antagonism between growth (largely mediated by target of rapamycin [TOR] pathways) and stress resistance relevant to aging (mediated by forkhead transcription factors [FOXOs]) is not simply a resource tradeoff, it reflects a hard-wired regulatory dichotomy conserved across eukaryote phylogenies spanning yeast to vertebrates [16–20]. TOR and FOXO occupy opposite and antagonistic arms of the PI3K pathway.
Fibroblasts from long-lived Ames, Snell and GH receptor knockout mice were exceptionally resistant to multiple stressors (e.g., reactive oxygen species (ROS), cadmium, ultraviolet light, heat) but treatment of Ames dwarf mice with GH abolished such resistance [2, 21]. Alternatively transgenic GH mice have increased sensitivity to paraquat [2]. Cells obtained from Snell dwarf mice within one week of birth had similar stress responses as cells from normal mice. Young adult donors, however, had cells exceptionally resistant to multiple stressors, suggesting a hormonal/developmental mechanism. Reduced levels of IGF-1 may contribute to stress resistance in mammalian cells [21].
We found that TGM had dramatically increased sleep that was modulated by energy supply. A predominant role of the GH axis in sleep regulation is now well established and we developed a framework suggesting that regulation of the genome and metabolome involved relegation of anabolic processes (e.g., synthesis, growth, repair and recharging) to sleep under control of the GH axis. This would then facilitate maximization of performance of niche-interfacing functions crucial to competitive fitness during waking [4, 15, 22–24]. These ideas further evolved into a synthesis recognizing “electroplasmic cycles” as a fundamental and highly conserved temporal regulatory framework encompassing the regulation of living systems across organismal to genomic levels [4, 20, 25].
Recently, a suite of elegant studies in yeast demonstrated that their genomic-metabolic organization reflects cycles of linked energy metabolism and redox as predicted [19, 26–33]. An unexpected feature, however, was that rather than a simple catabolic-anabolic dichotomy, yeast express three distinct temporal phases. Oxidative metabolism and generation of ATP constitute one aspect, a window dedicated to synthesis and growth constitutes another and a third phase is associated with functions requiring reducing conditions such as heme synthesis and antioxidant recharging. A literature synthesis confirmed that the circadian organization of vertebrates is also associated with three distinct functional phases [20]. Remarkably, an anabolic window dominated by GH-IGF-1-TOR signaling occurs in early sleep, whereas late sleep emerges as the realm of fasting-associated FOXO. Thus, both key pathways currently considered most critical to aging may occur in sleep despite the fact that they are strongly antagonistic and temporally, mutually exclusive. The range of critical functions linked to these pathways is extensive, and suggests that chronic downregulation of either may have undesirable impacts (Table 1). This must fundamentally alter our understanding and management of aging.
Table 1.
The Target of Rapamycin (TOR) and Forkhead transcription factors (FOXO) are strongly antagonistic in their regulation of numerous processes relevant to health and disease.
| Process or mechanism | Impact of the target of rapamycin factors | Impact of forkhead transcription |
|---|---|---|
| Protein synthesis | + | − |
| Cellular growth | + | − |
| Cell proliferation | + | − |
| Cell differentiation | + | − |
| Organismal growth | + | − |
| Stem cell proliferatio | + | − |
| Stem cell maintenance | − | + |
| Immunity, immunocyte proliferation | + | − |
| Autoimmunity | +? | −? |
| Wound healing | + | − |
| Cancer | + | − |
| Artherosclerosis | + | − |
| Type II diabetes/insulin resistance | +? | −? |
| Memory consolidation | + | − |
| Primordial follicle maintenance | − | +? |
| Sperm count | +? | −? |
| Proteasome Function | − | + |
| Sperm count | +? | −? |
| Proteasome Function | − | + |
| Autophagy | − | + |
| Apoptosis | − | + |
| Muscle mass | + | − |
| Gluconeogenesis | − | + |
| IGFBP-1 | − | + |
| Antioxidant defenses | − | + |
| Xenobiotic detoxification | − | + |
| Chaperone functions | −? | +? |
| Oxidative conditions | + | − |
| General stress resistance | − | + |
| Aging rate | + | − |
THE CIRCADIAN CLOCK
Recognition that the organization of redox and energy metabolism constitutes temporal cycles highlights the value of clocks. Accurate timekeeping is essential to synchronize crucial activities and life history aspects with external circadian and seasonal variation. Internally, clocks allow coordination among diverse organs and tissues, and integration across hierarchical levels of organization spanning populations to genes [20, 34–37].
The mammalian master clock resides in paired suprachiasmatic nuclei (SCN) located in the hypothalamus – a structure constituting a central microprocessor that senses and determines resource allocation and that coordinates global functioning across circadian and seasonal cycles. The basic structure of circadian clocks is phylogenetically conserved and involves recursive cycles of transcription, translation, nuclear-cytoplasmic transport and protein degradation. The core mechanism involves stimulation of transcription of the Cryptochrome (Cry) and Period (Per) genes by heterodimers formed of CLOCK (circadian locomotor output cycles kaput) and BMAL-1 (Brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT-like)). Translated CRY and PER proteins themselves form heterodimers that translocate to the nucleus where they inhibit their own transcription. This yields a fundamental timing loop. Input from light via the retinohypothalamic tract provides extrinsic entrainment. The clock has diverse outputs including direct and indirect linkages to hypothalamic circuits regulating stress, hunger, metabolism, growth, thermoregulation and reproduction. Crucial linkage to the pineal gland controls nocturnal release of melatonin, a hormone and antioxidant that enters the circulation and that can signal circadian time and daylength (seasons).
Besides the central SCN clock, peripheral tissues maintain their own clocks that are variously linked to the central timekeeper. Surprisingly, despite the clock elements themselves being largely similar among tissues, gene arrays find remarkably little commonality of other genes expressing circadian expression across tissues [38, 39]. This likely reflects a critical role of clocks in cell differentiation itself. As our understanding of clocks increases, what might be considered to be part of the clock or important indirect targets progressively expands. Feedback from energy sensors such as NAMPT (nicotinamide phosphoribosyltransferase) AMPK (AMP activated protein kinase), and linkage of the clock to couples reflecting both redox and energy (e.g., NAD(P)/ NAD(P)H, ADP/ATP) are highlighted (NADPH = nicotinamide adenine dinucleotide phosphate). Of profound importance, a critical component of the clock linked to NAD(P)/NAD(P)H is the deacetylase, Sirtuin-1 (SIRT1 = silent mating type information regulation 2 homolog). SIRT1 has been highlighted as the target of the red wine flavinoid, resveratrol that proves to have pervasive benefits for health and aging rates (perhaps as a dietary restriction mimetic). AMPK is indirectly linked to both the target of rapamycin (TOR) and forkhead (FOXO) functions (see below) and SIRT1 links AMPK, PPARs (peroxisome proliferator-activated receptors) and FOXOs to the clock.
A role of clocks in tissue differentiation and tissue-specific control of redox/metabolic processes involves intimate interfacing and regulation of clock elements to nuclear receptors. At least 49 nuclear receptors are pervasively linked to energy metabolism, steroidogenesis, development, mitochondrial function and xenobiotic responses. Their complex regulation and interactions may represent a combinatorial code relevant to tissue-specific functions and even organismal-level integration [40–42]. Clocks regulate numerous nuclear receptors, but these receptors also feed back to the clock. Direct interactions and feedback between clock elements and nuclear receptors provides seamless integration of clocks to redox and metabolic states [42].
Aspects of metabolism including gluconeogenesis, insulin sensitivity, lipid metabolism, heme synthesis, mitochondrial activity and ATP production show pronounced circadian rhythmicity. Alterations in the clock elements Clock or Bmal1 result in significant metabolic disturbance [38, 43–45]. The charging-reductive phase of yeast involves numerous enzymes involved in carbohydrate and ethanol metabolism that anticipate the impending oxidative phase. This phase is associated with accumulation of acetyl-CoA units, the key substrate for respiration [33]. Although elements contributing to gluconeogenesis, glycolysis and fatty acid metabolism peak during early waking associated with feeding, the circadian rise in such processes begins in the late sleep-associated phase [38]. Clock function is altered by diet (e.g., dietary restriction (DR), high fat diets, glucose intake and meal timing) as well as feeding regulatory signals like insulin, glucocorticoids, cyclic AMP, leptin and ghrelin. Thus, clocks are tightly linked to energy metabolism [37, 46–48].
PER2 (but not PER1) physically interacts with multiple nuclear receptors including REV-ERBα (reverse erbα), TRα (thyroid hormone receptor α), PPARα, and HNF4α (hepatocyte nuclear factor 4α) to co-regulate transcriptional targets (e.g., Bmal1, Hnf1α and Glucose-6-phosphatase) [49]. Loss of function in Per2 or Rev-Erbα disrupts rhythmicity in nuclear receptor targets such as PCK-1 (phosphoenolpyruvate carboxykinase), glucose-6-phosphatase and glycogen as well as other energy regulating genes such as Glut2(glucose transporter type 2) [49]. Rev-erbα is particularly highlighted in linking the clock to metabolic processes, including both glucose and lipid metabolism [50]. Such mechanisms demonstrate that core clock genes can influence transcription of nuclear receptor targets. Heme itself did not significantly alter the interaction of PER2 with REV-ERBα. Bmal1 and Rev-erbα expression are stimulated by binding of the retinoic acid orphan nuclear receptors RORα and RORγ to promoter RORE response elements. PGC-1α (peroxisome proliferator-activated receptor γ coactivator 1 α) is a cofactor for RORs [37]. Clock-nuclear receptor integration dissolves the boundary between metabolic and clock functions and suggests that clocks regulate robust homeorhesis in tissue-specific nutritional and metabolic processes [42].
Nuclear receptors include those for aldosterone, thyroid hormone, estrogens, androgens, progesterone, glucocorticoids, fatty acids (PPARs), cholesterol, vitamin D3, retinoic acids (RAR, ROR), xenobiotics (CAR = constitutive androstane receptor), bile acids and linoleic acid. Despite their importance, ligands for many nuclear receptors remain unknown. Nearly all show strong circadian rhythmicity that varies among tissues [40. 51] and many are already recognized components or close associates of the clock. The nuclear Rev-erb receptor, an established clock component, was recently shown to bind heme [52]. This undoubtedly provides redox-sensing capacity to the clock. Stimulation of Ucp-1 (uncoupling protein 1) transcription by adrenergic β receptors (associated with increased c-AMP) involves binding of the orphan nuclear receptor NOR-1 (neuron derived orphan receptor 1) to the promoter region of Ucp-1[53]. Adrenergic receptor β2 signaling in skeletal muscle activated genes promoting fatty acid and pyruvate utilization including PGC-1, lipin-1, FOXO1 and PDK4 (pyruvate dehydrogenase kinase isozyme 4). NOR-1 was also induced. NOR-1 binds lipin-1 and PDK-4 promoters and is instrumental in regulating muscle metabolism [54]. NOR-1 siRNA also disrupted expression of UCP-2 and UCP-3 in skeletal muscle suggesting a capacity to regulate mitochondrial coupling.
NOR-1 showed strong peaks of expression in the early resting/photophase for muscle and brown adipose tissue and secondary peaks in the early waking phase for brown adipose tissue and during the mid-activity period in skeletal muscle [40]. Binding of the essential fatty acid linoleic acid by the HNF4α receptor is drastically reduced by fasting. HNF4α expression rises across the activity cycle and declines across the resting photophase in mouse liver. Although arhythmic in muscle, some of its actions may involve non-transcriptional mechanisms [40, 51].
The hypothalamic-adrenal axis (HPA) is activated in late sleep and is associated with increasing levels of rapid eye movement sleep and the transition to waking. The HPA then dominates waking functions and appears to be a primary endocrine mediator of clock signaling. Wake-associated functions important to niche-related fitness have priority over sleep. Rhythmic production of catecholamines and corticosteroid under control of the adrenal clock likely represent an extended hand or even an important adjunct to the SCN clock. The primary HPA effector, corticosteroid, is synthesized by adrenal genes expressing circadian rhythmicity (notably steroidogenic acute regulatory protein (StAR)) and knockout of Bmal1 in the adrenal abolishes rhythmic glucocorticoid production and disrupts mouse behavior [42, 55, 56].
Mice lacking Per2 lack corticosterone rhythms but responses to feeding, ACTH (adrenocorticotropic hormone) and stress remain intact [57]. Rhythmic expression of synthetic genes in the mouse adrenal peaked at the day-night/rest-wake transition [55]. Catecholamine metabolism and steroid synthesis both showed strong circadian rhythmicity. Interestingly, the peak in catecholamine metabolism was associated with late sleep and the beginnings of arousal (∼ZT:10) (ZT = zeitgeber time where lights on is ZT 0). The nuclear glucocorticoid receptor has pervasive impacts on tissues throughout the body. Many clock genes (e.g., Per) have glucocorticoid response elements in their promoters indicating bi-directional integration of clocks and glucocorticoids [42].
THE PHOSPHOINOSITIDE PI3K
Insulin, IGF-1 and other growth factors signal via the PI3K pathway that regulates energy, redox and diverse aspects of aging [2, 58–60]. PI3K activity is associated with NAD(P)H oxidase [NOX] activation [58] that inhibits tyrosine phosphatases (particularly PTEN (phosphatase and tensin homolog)) via oxidative modification of redox-regulated cysteine residues. Antioxidants and peroxiredoxin are inhibitory [58]. Mutation, or downregulation of PI3K elements that bias signaling pathways away from TOR and toward activation of FOXO generally slow aging and extend lifespan [11, 61].
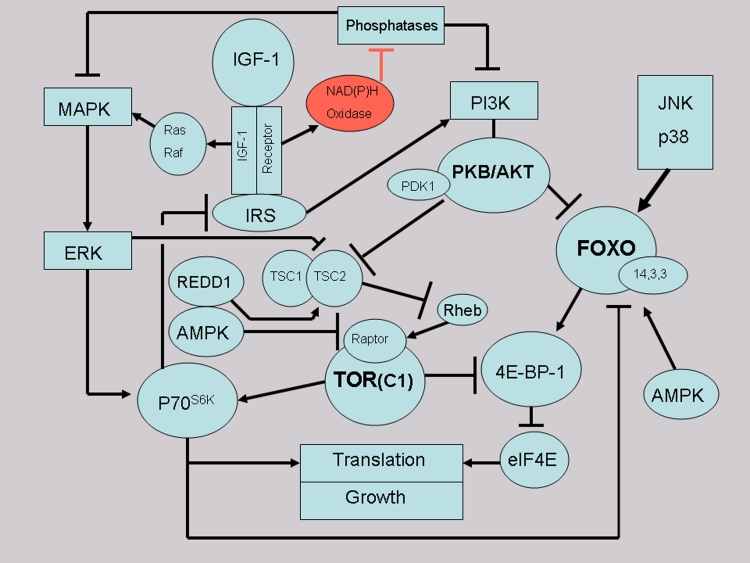
The bifurcation point directing signal flow to TOR versus FOXO toggles on protein kinase B (PKB/Akt). There are three Akt isoforms. Akt1 is widely distributed (and is most relevant to aging), Akt2 may particularly serve insulin, and Akt3 occurs in testes and brain. Akt1 controls growth, protein and glycogen synthesis, immunity and aging (i.e., largely TOR functions) whereas Akt2 mainly modulates glucose transport and fat deposition [62–64]. Phosphorylation and activation of Akt directs signaling to TOR. Alternatively, Akt actively inhibits FOXO and consequently reduced Akt phosphorylation activates FOXO via disinhibition (Fig. 1). This bifurcating regulatory structure imposing hard-wired mutual antagonism between stress resistance (FOXO) and growth (TOR) consolidates the tradeoff between growth and aging.
Figure 1.
Insulin-like growth factor (IGF-1) signaling relevant to the target of rapamycin (TOR) and forkhead transcription factors (FOXO). Growth hormone (GH) upregulates IGF-1 in early sleep both directly and by downregulating IGF-1 binding protein 1 (IGFBP-1). IGF-1 activates two pathways (PI3K-AKT and MAPK/ERK) that both regulate protein synthesis and growth processes. IGF-1 activity generates free radicals via membrane-bound NAD(P)H oxidases. Oxidation of phosphatases disinhibits kinase signaling and both IGF-1 signaling pathways. The toggle switch regulating TOR-FOXO antagonism is bifurcation of PI3K signaling according to the phosphorylation status of Protein Kinase B (PKB/AKT). Activated PKB/AKT inhibits FOXO and activates TOR via disinhibitory signaling to tuberous sclerosis factor 2 (TSC2). Inhibition of TSC2 releases activity of Rheb (Ras homolog enriched in brain) which in turn activates TOR. TOR signaling then activates eukaryotic initiation factor (elF4E) and S6 protein kinase (S6K) activity required for protein synthesis. Stress, amino acid availability and energy supply strongly modulate TOR activity. REDD1 (regulated in development and DNA damage-1) signals status of diverse stressors and AMP activated protein kinase (AMPK) reflects energy associated with AMP/ATP status. S6K is also a sensor of amino acids and energy and is implicated in insulin resistance via impacts on the insulin receptor substrate. Lack of insulin and IGF-1 signaling, termination of oxidase activity, activation of phosphatases (PTEN and PP2A) and unphosphorylated PKB/Akt inhibits TOR and releases FOXO nuclear localization and activation in late sleep. Modified after Rollo [20].
A GH-IGF-TOR WINDOW IN EARLY SLEEP
The target of rapamycin (TOR) associates in two complexes defined by the presence of either Raptor or Rictor. Downregulation of TOR or its target, ribosomal S6 protein kinase (S6K), strongly impacts aging and extends longevity of yeast, nematodes, flies and mammals [60, 61, 65–69]. Mechanisms include autophagy, mRNA translation and mitochondrial metabolism. Autophagy appears important in DR and fasting and may provide resitance to neurodegenerative diseases involving accumulation of misfolded proteins. Rapamycin impacts on longevity are mainly via TOR complex I. TOR is activated by growth factors and amino acids and is downregulated by AMPK [68].
Insulin and IGF-1 both signal via PI3K but a temporal framework highlights that insulin secretion is associated with meals so it is largely excluded from sleep. Although IGF-1 may also signal during ultradian meal cycles, its strongest signaling likely occurs independently of insulin during a specifically designated window in early sleep. IGF-I is the key effector of GH but does not exhibit plasma secretory spikes like those of GH. IGF-1 levels are considered a reliable biomarker of general nitrogen metabolism and it permissively regulates and coordinates other growth factors [70]. IGF-1signaling via both MAPK/ERK and PI3K converge on TOR which critically mediates GH-IGF-1 regulation of protein synthesis, cell growth and cell proliferation (including that of cancers) (Fig. 1).
The MAPK/ERK pathway that mediates IGF-1 signaling in cooperation with PI3K is active in sleep [71]. In fact, ERK protein levels show strong circadian rhythmicity in the mouse hippocampus with a peak at ∼ZT:04 (early mouse sleep/rest period) and low levels during waking. This rhythm covaries with activity of cyclic adenosine monophosphate (cAMP) and cAMP response element binding protein (CREB). Ca2+-activated adenylyl cyclases were important in maintaining rhythmicity [72]. Although most GH secretion occurs shortly after sleep entry, rhythmicity in the MAPK pathway was maintained in constant darkness, suggesting direct clock control as well [72]. Interestingly, light signals to the mouse SCN evoke ERK-dependent TOR signaling and this contributed to clock phase shifting via impacts on PER1 and PER2 expression [73, 74].
IGF-1 availability is complexly controlled by GH-regulated transcription, six IGF binding proteins (IGFBPs) and relatively specific IGFBP proteases [75]. IGFBP-3 chaperones circulating IGF-1 and likely determines stability whereas IGFBP-1 and IGFBP-4 negatively regulate availability of the bioactive pool. Because IGF-1 is already in serum, responses can be relatively rapid. The power of these mechanisms became apparent when a protein highly expressed in pregnancy (Pregnancy- associated plasma protein A: PAPP-A) proved to be the protease that cleaves IGFBP-4, thereby increasing IGF-1 availability [76]. Loss of PAPP-A inhibits growth of the placenta and young are born dwarfed [75].
Remarkably, hepatic Igfbp-1 shows 22-fold changes in circadian expression that even exceed the amplitude of the core clock genes [77]. If IGFBP-1 is not part of the clock it may be the clock’s best servant. Expression of Igfbp-1 was lowest during early sleep indicating that maximal IGF-1 availability coincides with greatest GH signaling. Besides stimulating IGF-1 transcription, GH suppresses IGFBP-1 function independently of insulin, which is also inhibitory [78]. IGF-1-Akt signaling activates TOR via impacts on TSC2 (tuberous sclerosis protein 2), and IGF-1 and TOR cooperatively stimulate cell proliferation and inhibit apoptosis [79]. Conversely, IGFBP-1 inhibits proliferation in breast cancer [80].
GH inhibition of IGFBP-1 activity would release IGF-1 and activate both MAPK-ERK and PI3K-TOR. IGFBP-1 is also negatively regulated by TOR itself and like TOR, by amino acid availability [81, 82]. Further reinforcing this window is reduction of the strong positive regulation of IGFBP-1 by corticosteroids (CORT). CORT generally acts antagonistically to GH and expresses a nadir of activity at the time of GH peaks. Coupled stimulation of IGF-1 transcription and release of bioactive IGF-1 by decreased IGFBP-1 (and IGFBP-4?) ensures maximal signaling of IGF-1 in early sleep in association with GH secretory peaks. Consistent with GH-IGF-1 regulation of TOR, Ames dwarf mice with low GH axis activity have downregulated TOR signaling [83] whereas GH transgenic mice have elevated Akt and TOR activity [84]. GH also induced PI3K-Akt and activated TOR in hepatoma cells [85].
Further linkage of GH, sleep and TOR is the close association of GH secretory peaks with slow wave sleep. Brain protein synthesis (i.e., TOR function) was associated with slow wave sleep in rats [86] and sleep was associated with positive regulators of protein translation (71, 87, 88]. Alternatively, sleep deprivation reduces protein synthesis and demand for protein synthesis strongly stimulates slow-wave sleep [71, 89]. Thus, GH/IGF-1 regulates protein synthesis and growth via TOR in a dedicated synthetic window localized in early sleep. Interestingly, peak expression of many genes associated with nucleosome assembly and histone proteins also peak at the night-day/activity-rest transition in the adrenal, SCN, liver and kidney. Thus, chromatin alterations appear particularly associated with the GH-IGF-1-TOR window [55].
PAPP-A is best studied with respect to IGFBP-4, but also may also function with IGFBP-2 and 5 [90]. The linkage of PAPP-A to IGF-1 and growth processes (e.g., wound healing, bone remodeling, placental and fetal development, atherosclerotic plaques) suggests that PAPP-A may be associated with the TOR window. Given the linkage of IGF-1, TOR and growth to longevity, one might expect that elevated IGFBP-1 might extend life span. Indeed, deletion of PAPP-A and downregulation of IGF-1 (via elevated IGFBP-4) extended maximal longevity of mice by ∼35% and delayed age-related tumorigenesis [75, 91].
A key feature of mitogen-TOR signaling is ROS generation via membrane-bound NAD(P)H oxidases (NOX). This would suggest that early sleep is not strongly reducing as one might assume from reductions in temperature and metabolic rate. Is it possible that mitochondrial function might actually be reduced to accommodate NOX oxidative signaling? In that case, increased mitochondrial efficiency could be engaged by increased mitochondrial coupling while reducing overall activity to reduce ROS. Such aspects require examination. Ultimately, TOR-related anabolism associated with the sleep-associated fasting could activate AMPK and this may be a key signal activating SIRT and FOXO (see below). Remarkably, AMPK can also be activated by oxidative stress suggesting that it might respond directly to NOX or changes in NAD(P)/NAD(P)H generated by NOX. The fact that AMPK inhibits NOX is consistent with this speculation since in temporal organization, elements coming on line generally suppress their inducers [20].
A LATE-SLEEP FOXO WINDOW
Late sleep is the most reducing circadian period and also that most reflecting the longest sleep-associated fast. This period also represents a time of preparation in anticipation of the increasing energy demands, redox stress and possible xenobiotic exposure associated with waking and feeding [20]. Thus, late sleep is associated with fasting responses and the marshaling of stress resistance elements. Early events may encompass mitochondrial biogenesis and heme production whereas later processes include proteasome activity, autophagy and DNA repair. This compartment is associated with sirtuins, PGC-1α and FOXO known to regulate stress and fasting responses including those involved in extension of longevity by dietary restriction. Increasing somatostatin (SRIF) stimulated by high levels of GH and IGF-1 acts to terminate production of GH peaks in early sleep, and together with SRIF-mediated induction of IGFBP-1 and rising corticosteroid, closes the GH-IGF-1-TOR window at multiple levels [78, 92].
Fasting-induced insulin inhibition and termination of the IGF-1-PI3K signaling at ∼ZT:04 leaves no activating signals for PI3K-Akt for the remaining sleep period. Associated reducing conditions likely favor phosphatases like PTEN over kinases. Increased phosphatase activity and lack of insulin-IGF-1 signaling constitutes the prescription for down-regulation of TOR and activation of FOXO in association with FOXO translocation to the nucleus (Fig. 1). PTEN may inhibit overall PI3K activity whereas the phosphatase PP2A appears to directly dephosphorylate FOXO and release its apoptotic potential [93].
Corticosteroids inhibit PI3K signaling by either insulin or IGF-1 and are closely associated with FOXO activity [94, 95]. Consequently, rising corticosteroids in late sleep would reinforce closure of the TOR window and contribute to activation of FOXO. The glucocorticoid nuclear receptor is consequently available to interact with FOXO at this time. Cooperation between FOXO and the glucocorticoid receptor was demonstrated in muscle where a ubiquitin E3 ligase (MuRF1) strongly contributes to wasting conditions. This gene contains both glucocorticoid and FOXO1 response elements in its promoter and glucocorticoids and FOXO synergize to enhance MuRF1 expression. IGF-1 was suppressive [96]. IGFBP-1 protein and corticosteroid levels are tightly linked whereas insulin and glucose are negatively associated with IGFBP-1 [97–99]. Following a nadir in early sleep, IGFBP-1 levels rise across late sleep to peak in early waking (likely reflecting rising corticosteroid and FOXO activation).
Hypothalamic SRIF projects to the clock and SRIF in the SCN peaks about 4 h into the sleep period [38,100]. GHRH phase advanced the rat SCN during the resting-photophase suggesting further linkage of the GH axis to the clock [101]. IGFBP-1 is upregulated by fasting, amino acid deprivation and by FOXO, all of which are antagonistic to TOR [81, 102]. FOXO also upregulated IGFBP-3 in thymocytes [103] suggesting that FOXO may reduce IGF-1 signaling by manipulating multiple IGFBPs. Cytokine-mediated induction of PAPP-A and proteolysis of IGFBP-4 are suppressed by resveratrol [104]. This suggests the possibility that the FOXO window partly inhibits IGF-1 signaling via down-regulation of PAPP-A (or other IGFBP proteases).
The IGFBP-1 promoter contains closely juxtaposed glucocorticoid receptor (GR) and FOXO response elements and FOXO1 and FOXO3 interact with the p300/CBP acetyltransferase and GR to enhance transcription of IGFBP-1 [96, 97, 105, 106]. One mechanism of FOXO action may involve interactions with promoter region insulin response sequences [107]. Reduced GH and IGF-1 signaling in dwarf mice is associated with a 7-fold increase in IGFBP1 and a 1.6-fold increase in IGFBP-2. DR of itself also increased expression of IGFBP-1 (1.5-fold) and IGFBP-2 (1.7-fold). In dietary restricted dwarfs IGFBP-1 was increased nearly 12-fold while IGFBP-2 was upregulated by ∼3-fold [108].
Late sleep is associated with absence of meal-associated insulin, suppression of IGF-1 signaling by SRIF and elevations in IGFBPs. This ensures a temporal nadir of Akt phosphorylation known to suppress TOR and activate FOXO. FOXO activation might be further enhanced by SIRT1 activity and rising glucocorticoids during late sleep. This period also represents a nadir in metabolic rate likely to promote reducing conditions permissive of FOXO activity and co-localization with GRs in the nucleus.
Heme synthesis requires reducing conditions and consequently might be expected to be restricted to mid sleep. Heme critically contributes to mitochondrial function and must be maintained at optimal levels [109]. The ligand for the nuclear receptor and clock component Rev-erbα, is heme. Rev-erbα negatively regulates Bmal1 in the clock and also regulates metabolic genes such as glucose-6-phosphatase. Induction of the rate limiting enzyme for heme synthesis, aminolevulinate synthase 1 (Alas-1) is negatively regulated by insulin and positively regulated by PGC-1α, FOXO1 and NRF-1 (nuclear respiratory factor 1)[110]. Maximal expression of mouse Alas mRNA occurred between ZT:08-ZT:12 (late resting photophase) [111]. Rev-erbα recruits the deacetylase HDAC3 to directly suppress Pgc-1α transcription and associated heme synthesis [109]. Increasing levels of Rev-erbα reduces mitochondrial respiration and inhibits the cell cycle. Thus, Rev-erbα provides negative feedback to its own ligand and mediates clock regulation of mitochondrial energy metabolism [109].
PGC-1α also strongly contributes to mitochondrial biogenesis. Consequently, control by REV-Erbα and linkage to heme suggests a mid-to-late sleep associated time frame for these processes. Generation of heme is then likely to be terminated in late sleep by REV-Erbα negative feedback to Pgc-1α. Indeed, Rev-erbα and Rev-erbβ transcription show mid-to-late sleep peaks in muscle, liver and brown and white adipose tissues. In skeletal muscle, however, a second peak of Rev-erbβ occurs in the early wake period that then declines dramatically after ∼ZT:17 [40].
Despite identical DNA binding domains, various FOXOs yield different regulatory impacts. This traces partly to tissue-specific expression patterns but also the manner of interfacing to cooperate with many other transcription factors [112]. In light of the close linkage of the clock to nuclear receptors it is of interest that FOXO interacts with numerous transcription factors that are nuclear receptors or associated elements (e.g. estrogen, androgen, progesterone, glucocorticoid receptors, constitutive androstane receptor (CAR), β-catenin, PGC-1α, PPAR-α, PPAR-γ, retinoic acid receptor (RAR), myocardin, thyroid hormone receptor, SMAD3 and SMAD4) (SMADS= mothers against decapentaplegic homolog). Vertebrate FOXOs contain a specific motif that mediates their interaction with nuclear receptors [112]. The progesterone receptor cooperates with FOXO to increase expression of IGFBP-1. FOXO- interactions with sex steroids such as the androgen receptor are implicated in development of cancers such as prostate and breast cancers and FOXO has tumor-suppressor impacts in such circumstances [112].
FOXO is also strongly associated with genes involved in energy metabolism (e.g., glucose 6 phosphatase (G6P), PCK-1, pyruvate dehydrogenase kinase-4 [PDK-4]). In many cases closely associated corticosteroid and FOXO response elements cooperate to regulate diverse promoters [96]. Possible cooperation between corticosteroids, the nuclear glucocorticoid receptor, other nuclear receptors and FOXO may represent critical processes dedicated to late sleep. Like IGFBP-1, PDK-4 expression is regulated by cooperation of GR, FOXO and p300/CREB binding protein. PDK4 has a glucose sparing function and is upregulated during fasting. Significantly, insulin suppresses corticosteroid mediation of PDK-4 expression by inhibiting FOXO [105]. PGC-1α and FOXO1 cooperate in the regulation of G6P and gluconeogenesis. This could reflect independent impacts on the G6P promoter or co-regulation by FOXO and PGC-1α [112]. FOXO1 synergism with C/EBPα (CCAAT/enhancer-binding protein α) regulates expression of adiponectin and reduced FOXO activity can impact insulin sensitivity, and glucose and lipid metabolism. SIRT1 deacetylates lysines in the FOXO1 region required for C/EBPα interaction [112]. Thus, activity of FOXO generally involves cooperation with other regulators and this can be modified by post-translational modifications.
Overall, the marshaled evidence is highly congruent with the reality of a dedicated window of IGF-1-TOR activity in early sleep followed by a window dominated by FOXO-mediated stress resistance and glucose metabolism in late sleep (Fig. 2) [10]. Although there are likely other periods of TOR and FOXO activity associated with ultradian insulin cycles in waking, these sleep windows are likely those most crucial to the regulation of aging rates and associated pathologies (Fig. 2). The fact that sleep may represent successive deployment of antagonistic and mutually exclusive functional suites represents a new conceptual framework for approaching health and aging. In particular, the idea that upregulation of FOXO must necessarily downregulate TOR predominates current theory where the temporal dimension is left out. The fact that TOR and FOXO occupy different temporal windows means they can be addressed with at least some independence.
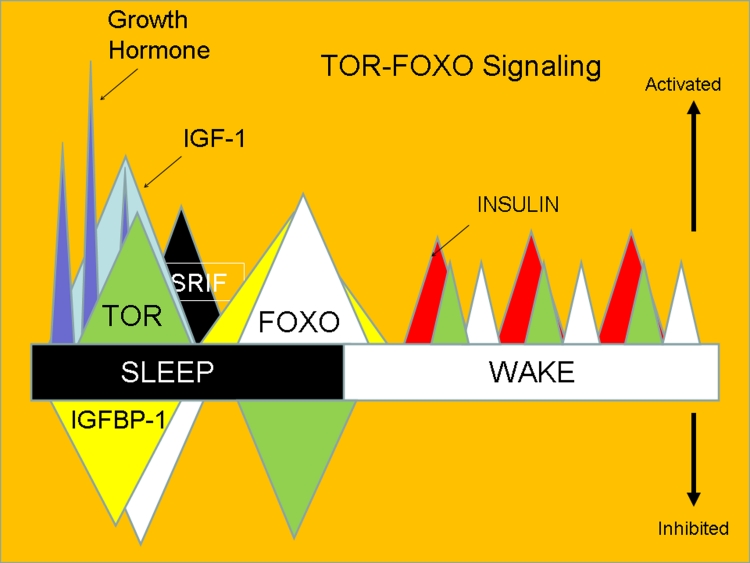
Figure 2.
A simplified illustration of the temporal distribution of the Target of rapamycin (TOR) and the Forkhead transcription factors (FOXO) across the circadian sleep-wake cycle. For humans, most daily growth hormone (GH) is secreted in large peaks shortly after initiation of sleep. GH stimulates insulin-like growth factor transcription (IGF-1) and suppresses IGF binding protein-1 (IGFBP-1), releasing plasma IGF-1 from IGFBP-3 to activate receptors and the MAPK/ERK and PI3K-Akt pathways. This imparts circadian rhythmicity to IGF-1 activity even though circulating levels do not cycle strongly. IGF-1 strongly activates TOR and mediates the synthetic and growth functions of the GH axis. TOR also downregulates IGF binding protein-1. Somatostatin (SRIF) then inhibits GH and stimulates IGFBP-1, thus shutting the TOR window via several mechanisms. Lack of insulin or IGF-1 signaling inhibits PI3K activity in late sleep, thus eliciting FOXO activation and translocation to the nucleus. FOXO and rising corticosteroid levels (not shown) also stimulate IGF binding protein-1. FOXO mediates numerous aspects of stress resistance that anticipate impending waking and these also may vary aging rates (as in dietary restriction). It is also likely that 3–4 h ultradian cycles associated with feeding and peaks of insulin also impact TOR and FOXO during waking.
INSULIN AND WAKING
If IGF-1 regulates TOR, and corticosteroids synergize FOXO, then insulin represents a predominant wake-associated hormone. Although intricately intermeshed to endogenous regulatory systems, insulin is mainly driven by food intake. Plasma insulin in humans normally shows three strong ultradian peaks (period of 3–4 h) corresponding to regular meals [113]. Such ultradian rhythmicity is of greater prominence in rodents, probably because their higher metabolic rates limit the length of tolerable fasts. Consequently, ultradian patterns may remain apparent even during the rodent rest-associated photophase [23]. Ultradian rhythms are also apparent in GH secretion, especially in rodents [23].
In addition to a significant TOR window associated with GH-IGF-1 signaling in early sleep, insulin is also likely to induce TOR activity associated with ultradian meal cycles. Insulin also downregulates IGFBP-1 so its ultradian signaling could be reinforced by IGF-1. Interestingly, rodent ultradian cycles can be identified by sequences of arousal, foraging, feeding and sleep [23]. Ultradian periods of sleep are also associated with GH secretion. Insulin expresses relatively low nadirs during inter-meal periods. It is interesting to speculate that reduced PI3K-Akt signaling at that time could generate ultradian SIRT/FOXO expression that would be highly adaptive for dealing with meal-associated influx of xenobiotics.
Differences in receptors and the presence of multiple PI3K and Akt isoforms suggests that insulin and IGF-1 signaling likely effect different levels of impact on common targets or may even traverse different pathways altogether (e.g., differential phosphorylation of Akt1 versus Akt2). Thus, insulin actions largely pertain to energy metabolism whereas GH-IGF-1 reigns over growth and cell proliferation [2]. It seems likely that the greatest activation of TOR relevant to aging and stress resistance is associated with the GH-IGF-1-TOR window in early sleep. However, increased insulin sensitivity in animals with downregulation of the GH axis may be an important factor in associated life extension [114].
Table 1 lists some important processes and mechanisms that are antagonistically regulated by TOR and FOXO. These include protein synthesis, cell proliferation, growth, proteasome and autophagy functions, apoptosis, redox status, immune function and stress resistance. These are not only relevant to aging, but also to fitness features such as athletic performance, resistance to disease, repair capacity and pathologies such as cancer. TOR-FOXO balance largely underlies predominant phenomena associated with altered aging rates - dietary restriction, reduced longevity of rapidly growing individuals intra-specifically and the extended longevity of species with prolonged development to larger body sizes [3, 115]. It is not possible to specifically examine all of these aspects here and redox aspects and stress resistance were thoroughly reviewed elsewhere [20]. Here I emphasize the implications of this triumvirate of circadian rhythmicity, particularly with respect to TOR-FOXO balance as well as a theoretical shift to recognize that the interplay of energy metabolism and redox may better explain aging rates than the free radical theory alone.
ADENOSINE MONOPHOSPHATE-ACTIVATED PROTEIN KINASE
An emphasis on energy highlights the importance of energy sensors and substrates. These are all closely intermeshed to TOR and FOXO signaling. ATP is the currency of cellular energy so many processes are highly sensitive to supply. Any modern rate of living theory must fulcrum on this factor. Regulatory signaling of energy status includes linkages to NAD(P)/NAD(P)H or AMP/ATP couples. A predominant signal of energy status is adenosine monophosphate-activated protein kinase (AMPK). AMPK is activated by a high ratio of AMP/ATP or Ca2+ elevations that reflect energy-ATP status. AMPK coordinates metabolic adjustments to energy shortfalls, including fasting, exercise and hypoxia [116, 117]. DR is regulated by sensors of both amino acids (TOR) and energy (AMPK) [118].
AMPK acts to restore energy balance by enhancing oxidative metabolism, lipid utilization, promoting mitochondrial biogenesis and by inhibiting energy consuming processes [117, 119]. An important mechanism of AMPK function is increasing levels of NAD+, which in turn promote SIRT1 activity. This is accomplished by a pathway promoting mitochondrial fatty acid oxidation [117]. Synthesis of NAD+ utilizes the AMP component of ATP which provides another possible linkage of NAD+ to AMPK [119]. Resveratrol activates AMPK which then supports expression of Nampt [121]. Resveratrol increased expression of genes involved in oxidative phosphorylation and mitochondrial biogenesis, reduced signaling by IGF-1 and increased AMPK and PGC-1α activity [122, 123].
AMPK-induced SIRT1 activity consequently deacetylates critical regulators of energy metabolism including PGC-1α, FOXO1 and FOXO3a. AMPK can also phosphorylate PGC-1α. Thus, AMPK and SIRT1 cooperate to regulate energy metabolism, and this likely extends to life extension under DR [117]. These results suggest that SIRT1 acts upstream of PGC-1α and FOXOs although the interaction of AMPK and SIRT1 may be reciprocal [117]. Note that these results particularly pertained to energy shortfalls induced by exercise, but a shift to lipid metabolism by the musculature during sleep is regarded as a mechanism that spares glucose for the brain during the sleep-associated fast.
Energy sensing by TOR also involves AMPK. AMPK downregulates expensive ATP-dependent processes and upregulates ATP generating mechanisms such as fatty acid oxidation. AMPK phosphorylates TSC2 (tuberous sclerosis factor 2), increasing its GTPase activity and inhibition of Rheb (Ras homolog enriched in brain). This then downregulates mTOR1/S6K [65, 124, 125]. AMPK also phosphorylates RAPTOR (regulatory associated protein of mTOR) inducing 14-3-3 binding and reducing TORC1 activity. AMPK phosphorylation of RAPTOR induced cell cycle arrest in growing cells [126]. REDD1 (regulated in development and DNA damage-1) stress signaling can independently inhibit TOR in response to energy stress [65].
AMPK is closely tied to clock function, particularly via interactions with SIRT1 [117]. AMPK also phosphorylates casein kinase Iε, increasing its activity and reducing PER2 stability [127]. AMPK also promotes phosphorylation and degradation of CRY1. This involves ubiquitination of CRY1 by the F box and leucine rich repeat protein, FBXL3. Nuclear localization of AMPK and CRY1 proteins showed inverse circadian phase [128]. Activation of AMPK caused phase advance in cell culture clock genes and in mice [129]. Mice with disrupted AMPKγ3 subunit express impaired clock induction of muscle genes and circadian shifts in energy metabolism [130].
Regulation of TOR, FOXO and the clock by AMPK highlights AMPK-mediated orchestration of TOR-FOXO balance critical to aging. Redox is also linked to this balance as growth is associated with free radical generation and stress resistance involves antioxidants and perhaps altered mitochondrial free radical generation. AMPK reduces intracellular ROS stress, in part by inducing FOXO translocation to the nucleus where it induced transcription of thioredoxin [131]. Thioredoxin contributes to ROS signaling networks by reducing oxidized cysteine residues on signaling proteins. The association of reducing conditions, fasting, FOXO and thioredoxin suggests that tyrosine phosphatase activity may be upregulated in late sleep. This would provide a barrier to insulin-IGF-1 signaling at that time. Such mechanisms may also bridge the linkage of redox and energy with respect to global signaling networks. Importantly, AMPK negatively regulates NOX activity in endothelial cells. Recall that NOX is required for MAPK-ERK and PI3K signaling to TOR in the early sleep window. Inhibition of AMPK results in increased NOX-mediated ROS generation, increased 26S proteasome activity, IĸBα degradation and NFĸB (nuclear factor kappa B) activation. Inhibition of the proteasome was ameliorating, presumably via blocking activation of NFĸB [132]. Given that the TOR window is associated with high NOX activity mediated by signaling of GH-IGF-1 (and other growth factors) via MAPK-ERK, PI3K-Akt, and JAK-STAT (janus kinase/signal transducer and activator of transcription), AMPK expression in the late fasting stage of sleep would further contribute to reducing conditions and suppression of the TOR window.
As noted above, AMPK linkage to the clock involves phosphorylation and destabilization of CRY1 [128]. Circadian rhythmicity of the AMPK regulatory subunit results in nuclear localization and activity of AMPK during the resting photophase period of the mouse. Mice deficient in the AMPK activator, liver kinase B1, showed elevation of CRY1 particularly in the resting photophase period [128]. This is consistent with activation of AMPK by the sleep-associated fast which would then be likely to activate SIRT1, PGC-1α and FOXOs. Thus, AMPK may be a critical contributor to the TOR-FOXO temporal transition.
Cyclic AMP activity and transcription regulated by cyclic AMP response elements (CRE) are also rhythmic and mutations in CREB2 alter the period of molecular and behavioral rhythms. Adenosine 3’,5’-monophosphate (cAMP) signaling impacts amplitude, period and phase of the circadian clock, and may contribute to continuity of one cycle to the next [133]. Interestingly, variation in cyclic AMP signaling modulates sleep independently of the clock [35].
FOXO3a binding and transcriptional activity was enhanced in dietary restriction by AMPK phosphorylation but AMPK did not affect FOXO localization [33,34]. Genes induced by nutrient deprivation that required AMPK phosphorylation of FOXO3 and other signals involved in stress and energy metabolism were also identified (including Pgc-1a, Ucp2, Gadd45a (growth arrest and DNA damage), and metallothioneins) [33]. Ucp3 and Pgc-1β showed circadian expression in skeletal muscle [136]. Overexpression of PGC-1α blocked induction of mitochondrial ROS production by hyperglycemia [119]. AMPK was associated with increases in MnSOD (manganese superoxide dismutase) that likely trace to FOXO [119, 137]. AMPK also phosphorylates and activates nitric oxide synthase (NOS) in endothelial cells [132].
AMPK likely fine-tunes transcriptional activity of FOXO3. AMPK was associated with Daf-16/FoxO in longevity extension of nematodes with an insulin-like receptor (Daf2) mutation [138] whereas longevity of Caenorhabditis elegans carrying mutated AMPK was reduced. Nematodes with mutated akk2 (AMPK) had reduced longevity and increased accumulation of lipofuscin-like materials [138]. AMPK was also involved in two methods of DR in C. elegans and was activated by resveratrol in cell cultures and in mice [118]. FOXO was dispensable for extension of longevity by resveratrol, even though AMPK otherwise required FOXO for life extension [118]. TOR pathways may be involved here. In flies, DR reduces amino acids which may particularly impact TOR and FoxA, whereas other mechanisms altering longevity may involve AMPK and FOXO [118].
Even acute DR strongly altered gene transcription profiles across tissues in mice. DR was associated with increased lipid catabolism and gluconeogenesis in liver and muscle as well as increased glucogenic amino acids in plasma. Expensive synthetic pathways and cell proliferation were reduced [139]. Lipid synthesis, steroid production and amino acid metabolism were all decreased. Genes downregulated in skeletal muscle by DR were associated with protein synthesis, cell growth, collagen synthesis, and blood vessel development whereas those associated with energy metabolism (carbohydrates, lipids) were elevated [139]. Overall such alterations are consistent with reduced TOR and upregulated AMPK-FOXO pathways in DR.
The AMP/ATP ratio increases with age in C. elegans, indicative of energy shortfalls and may provide a reliable biomarker of longevity. AMPK increases longevity in yeast, nematodes and flies, and a drug that upregulates AMPK (phenformin), extended mouse longevity by 20% [140]. AMPK mediated extension of longevity in C. elegans by heat shock (that increased the AMP/ATP ratio), low insulin signaling and glucose restriction [138, 141]. AMPK (but not sirtuin) was essential for lifespan extension by glucose restriction [141]. Remarkably, glucose restriction increased ROS generation (but also elevated catalase activity, oxidative stress resistance and longevity). Glucose or antioxidants inhibited induction of stress resistance and longevity in this circumstance [141].
CIRCADIAN GLUCONEOGENESIS
The clock regulates circadian cycles of fuel production, storage, and utilization and these pathways strongly modulating aging rates. Insulin, SIRT, FOXO are mediating elements and all are strongly implicated in aging and dietary restriction. Many regulatory neuropeptides and hormones provide feedback to the SCN, particularly via the arcuate nucleus which has access to information carried in ventricular fluid (e.g., levels of insulin, ghrelin, leptin, glucose) and strongly regulates feeding [36]. Many central regulators of metabolism, feeding and sleep show strong circadian rhythmicity (e.g., neuropeptide Y, insulin, glucagon, adiponectin, POMC (pro-opiomelanocortin), AgRP (agouti related peptide), leptin, ghrelin, and ATP) and may impact peripheral clocks [36, 142].
The pentose-phosphate pathway converts NAD(P) to NAD(P)H. This pathway is gated by glucose-6-phosphate 1-dehydrogenase. In Drosophila, this gene is maximally induced toward the end of the day. Opposing this activity and mediating gluconeogenesis is fructose biphosphatase which peaks near dawn. Thus, antagonistic pathways drive circadian rhythms of glucose and NAD(P)H metabolism [29, 143]. PCK-1 a key enzyme involved in gluconeogenesis peaked close to the light-dark transition and waking in the mouse [77].
FOXO activity is suppressed by feeding and insulin secretion but some aspects of energy metabolism are upregulated in sleep (e.g., carbohydrate anabolism and lipid catabolism). This is consistent with sleep as a fasting state but also indicates marshaling of enzymes and substrates to support impending waking [71]. Gluconeogenesis is largely a function of liver (and kidney) and is antagonistically regulated by insulin (inhibitory) and FOXO (stimulatory). FOXO was associated with changes in ∼28% of nutrient-sensitive genes in Drosophila highlighting it as a key regulator of nutritional metabolism [144].
Glucose-6-phosphatase is a key enzyme in hepatic gluconeogenesis inhibited by insulin/IGF-1 derived PI3K-Akt signaling. FOXO stimulates expression of glucose-6-phosphatase via interactions at an insulin response element [145]. FOXO also stimulated glycerol transport and amino acid catabolism [146]. FOXO binds and activates the promoters of Igfbp-1 (the primary regulator of IGF-1 supply) and Pck-1, whereas insulin is inhibitory [102, 146]. This may contribute to temporal segregation of TOR (early sleep) from FOXO (late sleep). FOXO upregulates IRS-2 (insulin receptor substrate 2) and receptors for leptin and adiponectin but inhibits glycolysis, the pentose-phosphate shunt, lipogenesis and steroid synthesis [146].
In rodents, plasma glucose increases across the photophase to peak in association with initiation of scotophase activity when glucose uptake and insulin sensitivity increase [147]. In skeletal muscle, insulin signaling induces translocation of GLUT4 to the cell membrane and import of glucose [65]. SIRT1, PGC-1α and FOXO are all linked to regulation of hepatic gluconeogenesis. Rising glucocorticoids in late sleep mediate gluconeogenesis as well. Dysregulation may explain abnormally high glucose (the dawn phenomenon) in diabetics [148]. In rodents, expression and activity of Pck-1 peaked in liver at ZT:13 (light-dark transition and waking). A nadir occurred at ∼ ZT:01 [77, 149–152].
Genes associated with glucose import and storage peak in the wake-feeding phase [152]. The human brain utilizes ∼20% of daily energy and the newborn brain consumes 50%, all fueled by glucose. The brain cannot store glycogen or perform gluconeogenesis and thus requires glucose for optimal function and survivial. Given that energy supply has recently been highlighted in aging, possible consequences of defects in gluconeogenic mechanism with age appear lacking (although the import of insulin resistance is widely recognized). Fasting glucose is derived via hepatic gluconeogenesis or from glycogen (glucogenolysis). Liver only stores ∼50% of glycogen needed for the brain [150].
Genes regulating redox and energy metabolism express circadian rhythmicity in the SCN [38]. Glucose homeostasis is disrupted in mice with SCN lesions or mutations in Bmal1, Clock, Per1 or Per2. Mice with loss of Bmal1 function are glucose intolerant whereas liver-specific deletion caused fasting-associated hypoglycemia and elevated glucose clearance [152–155]. This was associated with loss of Rev-Erbα expression and upregulation of Cry1 but no change in Per2. Bmal1 deficiency in liver suppressed expression of glucose-metabolic genes including glucose transporter 2, uncoupling protein 2, glucose-6-phosphate translocase, adenylate kinase and P450 oxidoreductase [152]. Rhythmicity of Pck-1 was altered in Bmal1 deficient mice such that normal fast-associated expression in liver was extended into the dark phase. Alternatively, expression of uncoupling protein 2 (Ucp2) normally peaking at ∼ZT:16 of the dark-activity phase was suppressed [152]. Glucose transporter 2 (Glut2) expression normally peaked in the mid-to-late fasting phase in liver but was suppressed in Bmal1 deficient mice [152]. Glucose export from liver during the fasting phase was deficient.
Gluconeogenesis and Pgc-1 are strongly upregulated by fasting, corticosteroids and cAMP [156]. This suggests association with late sleep when REMS and corticosteroids rise. Multiple clock genes possess glucocorticoid response elements including Per1 and Per2 (but not Clock) [157]. Glucocorticoids also confer rhythmicity to clock genes in cell cultures. Mice with Per1 mutations that ablated responsiveness to glucocorticoids showed no changes in glucose tolerance or insulin sensitivity during chronic glucocorticoid treatment but they developed glucocorticoid-mediated muscle wasting. Thus, interactions of glucocorticoids with the clock impact glucose homeostasis [157]. These authors suggest that this may involve leptin, which has a circadian rhythm out of phase to cortisol. Interestingly, leptin expresses circadian rhythmicity with peak expression and protein levels during sleep in humans [37].
Alterations in fuel utilization in DR are broadly similar to those seen in short-term fasting including that associated with sleep. This is consistent with the original formulation of the rate of living theory by Pearl (see below). PGC-1α, which impacts PPAR function, gluconeogenesis, lipolysis, β-oxidation and mitochondrial biogenesis is upregulated by DR in liver. Mitochondrial genes upregulated included ATP synthases, carnitine transporter proteins, cytochrome c oxidase (complex IV), succinate dehydrogenase (complex II) and the mitochondrial transcription regulator, Tfam [139].
CLOCKS AND LIPID METABOLISM
Besides gluconeogenesis, clock modulation of the key hormones regulating fat stores (including leptin and adiponectin) contribute to global lipid metabolism. Defective Clock or Per2 or insufficient sleep contribute to obesity [36, 57, 155]. Clock mutant mice show low expression of ghrelin, CART and orexins that regulate feeding. Decreased levels of POMC could account for increased photophase eating and hyperphagia [148, 155]. Melatonin may also contribute to glucose homeostasis and secretion is impaired in type II diabetes [148].
Glucose, lipid metabolism and the clock are tightly co-regulated, particularly in tissues like liver [48]. Sleep is tightly coupled to metabolic activity and utilization of substrates including glucose and lipids. Reduced glucose utilization by many tissues throughout the body, gluconeogenic impacts of GH, reduced insulin release and increased insulin resistance sequester plasma glucose for the brain during the sleep-associated fast [48]. Intact mice expressed rhythms of similar form for glucose and triglycerides but lipids were phase advanced compared to glucose by ∼ 4h. Peak corticosterone occurred around the nadir of triglycerides and peak adiponectin levels were associated with low glucose. REV-Erbα and BMAL1 are directly involved in adipose tissue differentiation and lipogenesis. Inactivation of Bmal1 and Clock suppressed diurnal variation in glucose and triglycerides and attenuated gluconeogenesis [154]. Bmal1 deficiency results in reduced differentiation of fibroblasts into adipocytes and lowers adipogenesis [47, 142].
Metabolic syndrome associated with obesity, cardiovascular disease and type II diabetes correlates with altered sleep and clock function [148]. Insufficient or poor quality sleep promotes obesity and type II diabetes in humans. Obesity and metabolic syndrome are associated with oxidative stress and major age-related human pathologies [122]. Homozygous Clock mutant mice (on an obesity-prone C57BL/6J background) expressed a metabolic syndrome with hyperphagia, obesity, hyperlipidemia, hyperglycemia and reduced insulin secretion. Locomotor rhythms were altered, energy expenditure was reduced and transcripts of Ghrelin, Orexins and Cart (cocaine and amphetamine regulated transcript) were attenuated in hypothalamus [155]. However, impacts of clock mutations vary radically with genetic background [48]. On an ICR mouse background clock mutants had reduced serum triglycerides and free fatty acids associated with poor fat absorption. Diet-induced obesity was also ameliorated [158].
The mouse plasminogen activator inhibitor-1 gene (PAI-1) is a risk factor for cardiovascular disease. PAI-1 levels increase with obesity and are strongly elevated in ob/ob mutant mice. PAI-1 contains an E-box element indicating clock control. Per2 antagonizes Clock:Bmal1 and associated activation of PAI-1 [159]. PAI-1 mRNA was normalized in most tissues and plasma (but not adipose tissue) by the clock mutation on an ob/ob background [158]. This was associated with exacerbated fat deposition in ob/ob mice carrying mutated Clock which induced adipocyte hypertrophy in both controls and ob/ob mice [158].
Integration of the clock to fat metabolism is reciprocal. High-fat diets altered locomotor rhythms, clock genes and clock controlled genes regulating fuel utilization [47]. Rhythms of glucose, insulin, corticosterone, leptin, neuropeptide Y, POMC and free fatty acids as well as orexin gene expression were all altered [47]. A high fat diet elevated the amplitude in insulin sensitivity and this was dependent on Clock. Clock regulation of insulin signaling suggests that reduced Clock activity could protect against diabetes [154]. Given the medical implications of obesity, metabolic syndrome and associated type II diabetes for aging and age-related human pathology, multiple and reciprocal linkages to clock function are potentially of great importance.
Adipogenesis and lipid deposition are upregulated by the TOR pathway. S6K1 blocks lipolysis and favors lipid deposition [65]. S6K-1 knockouts are 20% smaller at birth and are resistant to obesity due to elevated lypolysis and rapamycin reduces adipogenesis [65]. Alternatively, FOXO is associated with lipid mobilization and lipolysis. This makes sense in terms of the sleep-associated fast and circadian lipid metabolism. Perhaps the greatest period of fat deposition is associated with insulin signaling associated with meals. There are complications here as globally, insulin is associated with fat deposition, but GH is considered to promote lipid mobilization. It is possible that the TOR window in early sleep driven by GH and IGF-1 functions somewhat differently than meal-associated insulin signaling, or is the fat mobilization attributed to GH in fact engaging further downstream and outside the temporal realm of the TOR window?
TOR, FOXO AND PEARL’S RATE OF LIVING
Recent literature abounds with strong challenges to even venerated theories of aging, but new synthetic theory is slow to develop. The free radical theory of aging and its extension to mitochondria faces increasing challenges. In support of the free radical theory, oxidative stress increases with age and is reduced by DR. Mutations extending longevity often are associated with reduced oxidative stress or increased stress resistance [160]. Some recent studies, however, fail to find the expected correlations between longevity and levels of free radicals, oxidative damage, metabolic rate or antioxidants [161–165]. MnSOD+/− mice expressed oxidative damage to nuclear and mitochondrial DNA and doubled tumor incidence, but longevity and many biomarkers of aging were unaffected [166]. Of 18 genetic manipulations predicted to impact aging via altered free radical processes, some altered oxidative stress resistance but only deletion of the CuZnSOD gene had any effect on life span. This seriously questions the validity of the free radical theory of aging [160, 167].
Mitochondrial “mutator” mice express high rates of mutation accumulation resulting in severe electron chain defects and a suite of features resembling accelerated aging [168]. Mitochondrial mutations are expected to generate or be exacerbated by increased free radical generation, and indeed, mice with catalase targeted to mitochondria show lower levels of mitochondrial mutation than normal mice [169]. Mitochondrial mutator mice, however, showed no evidence of elevated oxidative stress, increased oxidative defenses and no evidence for ROS-induced apoptosis. Rather defects in the respiratory chain itself appear to account for accelerated aging [170]. This suggests that aging could trace to shortfalls in energy required for essential defense, repair and replacement processes.
Mutator mice were able to sustain a point-mutation load ∼500 fold greater than normal levels, suggesting that mitochondrial point mutations do not contribute to the aging of normal mice [169]. However, a subsequent examination pointed to mitochondrial DNA deletions associated with a recombinate repair process as a likely candidate contributing to aging in mice [170]. Energy-induced stress and apoptosis could be an important mechanism contributing to aging of mutator mice [171, 172]. It remains that repair processes are likely responding to some form of damage, perhaps generated by basal ROS. Would mutations accumulate faster in mutator mice that also express elevated ROS?
Oxygen consumption, ATP levels, ATP/AMP ratio, superoxide production capacity and reducing capacity all show steep age-related declines in nematodes [173, 174] and declining metabolic rate is a reliable biomarker of aging across phylogenies. In nematodes with mutation of the insulin/IGF-1 receptor, however, ATP was maintained at youthful levels and this was associated with maintenance of functions such as protein synthesis and detoxification capacity that otherwise decline steeply with age [173]. However, ATP does not necessarily correlate with longevity across longevity studies [173]. Some suggest that TOR signaling itself could explain aging [175]. A problem with simply invoking activity of TOR or FOXO as regulating aging is that an actual causal mechanism (energy limitation, some cause of molecular damage or something else) is missing. All other things being equal, even extended or high GH signaling of itself should not cause collagen to lose its elasticity, cataracts to form in old eyes or mutations causing cancer to arise in aging cells. Autophagy and the proteasome remove damaged cellular components and are regarded as critical to aging. What causes the damage?
Although gene arrays of the aging female brain reflect increasing immunological gene activation, men exhibit global decline in anabolic and catabolic capacity in association with a preponderance of reduced activity in genes contributing to energy production, protein synthesis and transport [176]. Restricting energy supply to neurons and in an Alzheimer’s mouse model caused elevated β-amyloid and plaque production via stress pathways impacting protein translational [177]. Alternatively it has been suggested that ATP deficiency could reduce free radical generation in the cytosol, thus increasing lifespan even in the face of mitochondrial oxidative stress [178]. ATP shortfalls, however, generally compromise most cellular functions including ubiquitin-proteasome and NAD-dependent functions. These include glutathione reductases, thioredoxin, P450 enzymes, PPAR, SIRT, and NOX. Energy shortfalls also engage stress pathways such as the unfolded protein response and hypoxia. Varying levels of energy may have complex impacts on systems that have evolved priorities in functional maintenance. Some variation among aspects of functional versus demographic aging may be rooted here.
Although energy (ATP) is singularly highlighted in the above examples, aging may better reflect a “complex interplay” among energy metabolism, oxidative damage, calcium overload and cell death [179]. Although discussed in the context of neurodegeneration, this perspective may generally apply to circumstances of high cellular stress. Interactions of ATP, ROS and calcium may particularly impact mitochondrial functions [179, 180]. Similar interactions are apparent in growth factor signaling where activation of membrane-bound NOX generates superoxide radical/hydrogen peroxide and associated calcium influx activates NOS. Interaction of superoxide and NO can generate peroxynitrite, elevate mitochondrial free radical generation and potentially reduce ATP production [181]. This likely extends to all growth factors including GH, IGF-1 and insulin. The key interactive players include NOX, NOS, mitochondria, endoplasmic reticulum, calcium, free radicals, ATP and longevity assurance systems (defense, repair, replacement). Hypoxia, ion channels (particularly ATP-sensitive ion channels and Ca2+) and free radical theories of aging can be unified within a temporal framework of metabolism and redox: the “electroplasmic cycle” [20, 25, 182].
A linkage of energy shortfalls to aging harkens to the long-standing, albeit rather creaky “Rate of Living Theory” (RLT) put forward by Pearl in 1928 [183]. The fact that it is still considered worthy of disproof attests to its power despite the need for many qualifications. Modern interpretations of the original RLT variously emphasize that lifespan reflects rates of respiration and energy metabolism [5, 184]. Pearl’s book surprisingly looks little like this. Rather, he viewed life as having an undefined vital capacity that was reflected equally well in longevity and starvation resistance. This was based on the fact that mortality curves associated with starvation and those for lifetime survivorship were of very similar form when viewed from a common scaling. Consequently, much of Pearl’s argument was based on starved flies and in particular, the growth rate and time to death of seedlings maintained without nutrients and in complete darkness. Given that FOXO manages the sleep-associated fast, dietary restriction, starvation responses and stress resistance, the idea that starvation resistance might provide a reliable biomarker of lifetime survivorship is actually quite interesting rather than improbable.
Shifts in resources between growth (TOR) versus stress resistance (FOXO) certainly modulate aging rates [15, 185, 186]. Pathways extending longevity at the expense of growth likely evolved as temporary interventions to protect against stress and ensure eventual reproductive success [186]. Thus, longevity assurance mechanisms are essentially stress response pathways, with energy and nutrient supply being key modulators and signals of favorable conditions for growth and reproduction. This emphasizes that the fitness value of “longevity assurance” mechanisms pertains to the success of youth capable of reproduction and that there has likely been little if any evolution of mechanisms actually functioning to ameliorate senescence or aging per se.
This circumstance may explain why we face the rather enormous and unexpected inconvenience of a regulatory structure that actually downregulates those pathways that might slow aging, as long as times are good for reproductive success. In other words, not only has there been little real selection for anti-aging adaptations, such potential may only be deployed in the context of growth and reproductive success in hard times. Pearl’s treatment of survival during normal aging and during starvation as being congruent reflections of the same thing [183] suggests that his intuition of the rate of living was deeper than previously appreciated. Further, given an emphasis here on growth associated with TOR, it is interesting that Pearl actually emphasized growth more than energy as a reliable biomarker of the rate of living [183]. Consequently, I suggest that a modified rate of living theory might be better construed as emphasizing growth/TOR as an exceptionally costly aspect of living (particularly because it suppresses stress resistance), whereas stress resistance/longevity assurance provided by FOXO represents opposing balance. Together they nicely constitute Pearl’s “vital essence.”
Despite required refinements, a metabolic basis of aging is supported by facts such as that the number of heartbeats for a mouse and an elephant really are roughly the same [187], that ectotherm longevity is strongly modulated by temperature [60] and that hibernating or diapausing animals live longer. Further variation may trace to how well lineages neutralize, repair or replace damage or even variation in constituents (e.g., saturation levels of fatty acids) that are differentially susceptible to ROS [188]. Recent findings suggest that in addition to damage and distorted signaling induced by ROS, declines in energy production with age may limit anabolic processes required to maintain defense, repair and replacement (e.g., accumulation of damaged proteins may reflect both ROS damage to the proteasome, and ATP limitations). Adding clock regulation of energy and redox adds further dimensionality, particularly since those key pathways regulating longevity assurance investments (TOR and FOXO) are highly sensitive to both redox and energy supply. Tropical birds expressed lower basal, field and maximal metabolic rates than temperate species and greater survivorship. Body temperature did not differ. Tropical birds had smaller cardio-pulmonary systems suggesting that they obtain a thermal subsidy from warmer climates. This was associated with slower growth rates of cells, and these cells had greater resistance to various stressors, including ROS [189]. These findings are consistent with a modified rate of living theory emphasizing the TOR-FOXO tradeoff.
The “Free Radical Theory of Aging” provided a concrete mechanism for the RLT involving processes that were directly and positively related to the degree of oxidative energy metabolism and identifying mitochondria as critical mediators. Assimilation of ROS as a mechanism for the RLT naturally derived a mitochondrial theory of aging centered on those organelles responsible for both energy metabolism and ROS production [190, 191]. This coherent vision required qualification, however, as lifetime energy utilization (metabolic scope) was found to vary remarkably among phylogenies (e.g., marsupials, lagomorphs, bats, primates, and birds). Thus, the RLT predicts that marsupials (with lower metabolic rates than eutherian mammals) should live longer, but the opposite is true. Similarly, birds have higher metabolic rates but live three-fold longer than mammals [5].
Generally, eutherian mammals expend ∼220–400 kcal/g in their lifetime [187] but primates average ∼450–1000 kcal/g. Birds expend ∼1000 kcal/g and some species even achieve 4,000 kcal/g [192, 193]. Cutler amassed evidence suggesting that longevity might be varied by “longevity assurance” investments [192, 193]. Although, many studies suggest that ROS generation predominates over stress resistance in determining aging rates, features associated with FOXO-mediated stress-resistance indeed alter aging rates. These include antioxidants, chaperones, metal chelators, DNA repair systems, proteasome function, autophagy and apoptosis, all enhanced by FOXO. Given their role in reversing oxidation in signaling processes, glutaredoxin and thioredoxin can be added to the repair category.
Relating metabolic rate to free radicals and longevity was further complicated by alterations in mitochondrial functioning that decrease ROS generation per unit of respiration [184, 194, 195]. Thus, extension of longevity by DR involves altered mitochondrial function that reduces ROS generation relative to respiratory rate. Similarly, the longer life and greater metabolic scope of birds compared to similar-sized mammals is associated with cleaner burning mitochondria [196–200]. If the mitochondria of birds are more uncoupled than mammals this could explain their longer lives. This is also consistent with the higher body temperatures of birds (∼44°C) compared to eutherian mammals (∼37°C).
For nematodes, however, insulin/IGF-1 receptor mutants (daf-2) appear to express higher rates of metabolism and ATP production but less heat per unit of oxygen consumed (suggesting increased coupling and greater efficiency of ATP synthesis). This seems contradictory as this feature might be expected to generate more ROS. This appears to indeed be the case, but increases in stress resistance elements (Cu/ZnSOD, heat shock proteins, glutathione S transferases, glutathione, metalothionien, catalase and xenobiotic capacity) offset ROS stress [173, 174]. Whereas functions like xenobiotic detoxification decline markedly with age in wild type nematodes, daf-2 mutants maintain this capacity (and ATP levels) into old age [173]. Daf-2 mutants also have upregulated gluconeogenesis and increased levels of ATP. Electron transport chain enzymes are not down regulated and the F1-ATPase inhibitor protein is upregulated [174].
Some expect that the RLT predicts that extended longevity should be associated with reduced ROS production [173]. Reconsidering this theory to reflect TOR-FOXO balance does not necessarily require that reduced metabolic rate or ROS generation determine longevity, but rather rates of damage and loss of function are the most important critical mechanisms (i.e., the rate of senescence as opposed to the rate of living). From this perspective aging rates may be varied by adjustments in longevity assurance systems (ie., defense, repair and replacement), reduced wake-associated ROS generation or lower TOR-associated anabolism. Thus, daf-2 mutants actually generated more ROS, but had reduced protein carbonyls in their aging mitochondria. Further, this was associated with slower rates of aging as reflected by delayed bradykinesis, lipofuscin deposition and sarcopenia [173].
Further challenge to the RLT comes from allometric analyses applying phylogenetic independent contrasts and correcting for body mass [184, 201, 202]. In all cases, larger species generally had lower mass-specific metabolic rates and lived longer but if body size and phylogeny were controlled for, little relationship between basal metabolic rate and longevity was evident. For this residual variation longevity and lifetime energy expenditure was associated with higher daily metabolic expenditure [184]. Interestingly, residual lifespan was negatively related to residual daily energy expenditure in mammals [184]. Patterns in mammals and birds were similar [184, 202].
The relationship of body mass to metabolic rate, however, is one of the strongest known allometric relationships. Thus, r2 values of at least 0.95 were obtained in one of these studies [201] and body mass accounts for nearly 97% of variation in metabolic rate across mammalian species [203]. A strong relationship of longevity to metabolic rate still holds then, given the caveat that this is largely associated with body size and growth processes. Treating body size as a “confounding” factor and studying residual variation when its contribution is removed is akin to studying the soap bubbles after removing the confounding baby and the bathwater. Rather, the remarkable association of growth rates with longevity suggests that some aspects of metabolism associated with growth (e.g., TOR-FOXO balance) are particularly important [3, 14, 115].
In terms of evolution, longevity assurance systems are considered to have a cost so a crucial modulator is the degree of extrinsic risk [5]. Species experiencing high mortality rates (like rats and rabbits) must mature quickly and reproduce sufficiently to offset losses (the balanced mortality hypothesis). Thus, the extrinsic mortality rate of birds can be inferred from the number of eggs in the nest. If animals are unlikely to live long due to extrinsic risk, resources invested in longevity assurance may be wasted. Better to allocate resources to assure reaching maturity and maximize early fecundity. Such ideas were formalized by Holliday and Kirkwood [204] in the “Disposable Soma Theory.”
The strong negative association of growth rate with longevity [115] and the parallel relationship to TOR-FOXO antagonism adds relevant dimensionality. Fitness under high risk requires rapid growth to ensure early maturation and still obtain the largest possible mature size as this is positively associated with fecundity (parental care and relative size of offspring aside). Besides stimulation of NAD(P)H oxidases by mitogens, rapid growth may benefit from high mitochondrial coupling that maximizes efficiency of ATP production at the expense of elevated ROS (see TOR section above). Thus, rapid growth and relatively greater ROS may be general consequences of extrinsic risk. This, and the highly conserved regulatory structure that downregulates FOXO-mediated stress resistance (=longevity assurance mechanisms) by TOR-driven synthetic and growth processes is likely sufficient to explain the strong association of the GH axis, PI3K signaling and aging rates [3, 115, 195].
The most recent challenge to conventional theory is the dissipation limit theory (DLT) put forward by Speakman and Krol [205, 206]. Prevailing views explaining the allometric relationship of body size, metabolic rates and longevity tend to highlight supply-side feeding, drinking, oxygen delivery, aerobic capacity and maintenance of body temperature [207–210]. Removal of waste (excretion, CO2) and dissipation of heat have always been appreciated, but the DLT argues that it is the inability to dissipate heat during periods of elevated metabolic rate that of itself determines levels of sustained performance (particularly reproduction) [205, 206]. The “Principle of Allocation” holds that (given limited ability to obtain and process energy) elevation in any one function must ultimately be traded off against others. This is a fundamental paradigm of evolutionary ecology and life history theory (“Risk” being the other) as well as the underlying framework for the disposable soma theory of aging. The DLT argues that energy is not limiting (even during reproduction), that functional tradeoffs are constrained to interactions bounded below dissipation limits and that the disposable soma theory should be discarded. I consider the DLT in some detail below as it is revolutionary and specifically attacks the disposable soma theory and the principle of allocation central to aging theory developed above.
The DLT is elegant, complex and sophisticated. I address it by emphasizing evidence against dissipation limits as global constraints. This is complicated because physics dictates that increasing body size associated with declining surface area to volume ratios indeed reduces dissipation and diffusion processes and ultimately could induce limits as suggested by the DLT. The question then becomes how closely species reside to such limits. One crucial and immediate point is that endothermy evolved to obtain a relatively stable thermal environment and increased rates of numerous fitness-related processes. This means that body temperature was selected for maintenance well above ambient so the crucial and prevailing problem was how to maintain body temperature in the face of dissipative loss across a relatively steep gradient rather than any dissipative limitation.
A central platform of conventional allometry has emphasized that selection on metabolic rates (endothermy is expensive) importantly reflects the need to maintain relatively similar temperatures despite variation in heat loss with changing body sizes. Thus, larger size can reduce thermoregulatory costs and selection against heat loss (dissipation) is evident in the nearly universal presence of hair, feathers or fat layers in birds, mammals, some extinct raptors and many warm-bodied insects. Naked mole rats don’t count because their heated niche and burrowing lifestyle allowed them to dispense with endothermy entirely. Speakman and Krol [205] count reduced insulation in warmer seasons as supporting their theory:
“If energy were a limited resource throughout the year, one wonders why they ever moult out of the winter coat into a summer coat that causes their energy demands to increase?”
Warmer ambient temperatures represent an energy subsidy that means that energy demands do not increase but rather the opposite. Further, nobody suggests that endothermy does not come with a price. Loss of winter insulation during summer would favor elevating exercise capacity in warmer weather, and reduce the cost of extra weight and maintenance of insulation. The fact that insulation is maintained still argues against a dissipation limit.
The near ubiquity of insulation of itself strongly argues against selection for increased dissipation. In fact, any animal that can save and redirect energy from thermogenesis to other fitness features may have a selective advantage. As heat generation increases the dissipative gradient increases, but problems certainly can arise at very high levels of performance or at high ambient temperatures. Lack of hair on a few large tropical animals like elephants and rhinos could well reflect an adaptation to allow sustained metabolic performance when required. Note however, that mastodons and mammoths (some larger than modern elephants) maintained robust insulation. Naked apes with whole-body sweating potential may reflect adaptations for heat dissipation during intense or prolonged exercise. Even humans however, dress for the climate. Other primates maintain insulation, including those living in tropical environments and some that are larger than us.
Bergmann’s rule argues that there is a trend for larger-size in organisms occupying colder environments as one adaptation to specifically reduce heat loss. Allen’s rule adds that bodies become more compact and extremities are reduced to lower surface area and dissipation in cold environments. Admittedly, such rules are weak and widely criticized [206] but there are no such rules in the opposite direction. Large size in cold environments could well serve to adaptively reduce dissipative heat loss and does not appear to impose any dissipative limit given the levels of activity expressed by animals like polar bears. Similarly, the relatively enormous size of cetaceans could be argued to represent release of growth by conductive transfer to cold water, but then why do cetaceans carry such enormous layers of fat? I walked upon a beached sperm whale that had been sliced by a propeller and marveled that it was basically a thickly fat-wrapped sausage (at least 30 cm of blubber). Increasing size of whales allows longer duration dives and better heat conservation.
Although reduced surface-area to volume ratios may serve to reduce heat loss, endotherms retain further adaptations designed to prevent heat dissipation. This includes the ability to alter thermal conductivity via vascular adjustments (vasoconstriction in your hand when placed in cold water) and evolution of counter-current heat exchange. The latter is especially pronounced in species inhabiting cold environments and warm-bodied fish like tuna can only remain elevated temperatures in cold water by employing sophisticated counter-current heat exchange. It is difficult to imagine any benefit of increased dissipation for such animals. The prevalence of adaptations to resist heat loss and conserve energy argues that energy and heat are precious and that dissipation limits are rare in the evolved niche. If heat dissipation is an important selective limitation, it could be easily offset by shifting to smaller sizes. This would impose a pattern of evolution where larger species tended to generate smaller ones whereas Cope’s rule suggests the opposite.
The DLT is unlikely to apply to ectotherms in any way similar to endotherms so it lacks generality. It would also be difficult to apply the DLT to the extremely rapid growth and early demise of cephalopods. This extends to inter- versus intra-specific trends as well. Intraspecifically the most rapidly growing and most fecund mammals tend to achieve larger mature sizes, diametrically opposite to predictions of the DLT. A modified rate of living theory is congruent with both intra- and inter-specific trends in growth rates, size and longevity [115]. Even within endothermic phylogenies eutherian mammals maintain higher body temperatures and metabolic rates than marsupials of similar size without meeting significant dissipation limitations. Placental mammals evolved efficient exchange between mother and fetus, increasing reproductive rates and allowing invasion of colder niches [203]. A significant dissipation limit could have constrained such a shift in warm climates. Birds have evolved even higher body temperatures and metabolic rates than eutherians, but still maintain insulative feathers in all climates. The argument that birds must retain feathers for flight does not apply to all feathers, mammalian fur or the original evolution of feathers for insulation in raptors. The residual variation in metabolic rate after correcting for body size has a positive relationship with body temperature that is the same in birds and mammals. Birds have a higher metabolic rate than equivalently sized mammals because they maintain a higher body temperature [211]. Remarkably, various lineages have been able to elevate their body temperatures substantially without meeting a dissipation limit.
An implied tenet of the DLT is that species (particularly large ones) have evolved to body sizes and metabolic rates that impinge on their dissipation limits, particularly during reproduction. It is highly unlikely that any species evolves to extremes that reduce global fitness. The DLT specifically predicts that the fecundity of large animals is limited by restricted dissipation. If so, any genetically smaller individuals could presumably increase their fecundity, placing them at a reproductive advantage that would eventually eliminate larger conspecifics. Body size would be selected away from the dissipation limit. The handicap principle aside, suggestions that species that evolve exaggerated features suffer reduced fitness and even possible extinction (like saber-toothed cats) invariably turn out to be wrong. Saber-toothed tigers were exquisitely suited to their niche (e.g., spearing thick-hided slow-moving ground sloths). Thus it seems more likely that the body size and metabolic rate of various species are selected to an optimum that must allow full expression of elevated rates and functions needed to maximize fitness. This would certainly include avoidance of any dissipation limit and especially if it was detrimental to a crucial function like reproduction. This extends to the argument by Speakman and Krol [205] that large species have low mortality rates because all those large (limited) species with high mortality rates went extinct. How would they ever get there in the first place?
With respect to reproduction the DLT holds that large animals have relatively low fecundity because their metabolic rates during reproduction are constrained. The increased production of offspring by animals experiencing high mortality is known as the “Balanced Mortality Hypothesis” and it has considerable empirical support (see above). In animals with no parental care, fecundity is in fact favored by achieving larger sizes. Large endotherms, however, generally allocate considerable resources to achieve larger, slower growing offspring that receive protracted parental care. This then limits the fecundity of the parents. The costs of parental care are high as evidenced by prolonged maintenance of elevated metabolism such as in foraging birds. In a world where persistence rather than maximization is the key fitness criteria, survival of a few large and well tended offspring is a life history strategy at least as viable as depositing 800 maggots. From an aging perspective, parental care may also select for longer persistence of parents rather than the common maximization strategy of reproducing once and then dropping dead. Thus, a dissipation limit is neither necessary nor sufficient to explain reduced fecundity in larger endotherms.
Speakman and Krol [205] suggest that rather than being limited, energy supplies in the environment are often unlimited, particularly when animals are breeding. Energy can never be unlimited because digestive systems have finite capacity and are of themselves expensive and constrained by functional integration into the morphological adaptive suite. In contrast, as evidence that energy supply is not limited Speakman and Krol [205] cite the near ubiquity of stomachs and crops.
“The universal existence of these structures” … [stomachs and crops] … “indicates that for almost all animals, the rate at which energy can be collected from the environment must at some phases of their lives be considerably greater than the rate at which it can be utilized”
Other than the time required to break up and break down food, or in ruminants, to allow symbiotic augmentation of nutrients, animals without crops or stomachs would need to engage in reduced but greatly protracted feeding rates. They would have no reserve for periods of even short-term circadian fasts other than fat. Considering that stomachs are like gas tanks or batteries highlights the point. The smaller the gas tank or charge, the more often refueling will be required. No gas tank – lots of service stations and liquid food franchises. Moreover, the presence of gas tanks does not preclude tradeoffs between propulsion and air conditioning, or variation in the price of gas. The size of the stomach or capacity of the digestive system would, however, impose an ultimate limit on uptake and processing of food and energy.
The DLT takes the argument of unlimited energy further to suggest that expensive longevity assurance investments need not be traded off against reproduction as the disposable soma infers, because “energy supply in the environment is not limited during critical phases of reproduction” [205]. To the extent that TOR contributes to the growth aspects of reproduction, the regulatory evidence suggests that the reduction of FOXO-associated longevity assurance mechanisms is not just a competitive trade-off, it is hard wired. The DLT suggests that it is unlikely that larger species could express high levels of longevity assurance investments because their metabolic rates are so low. However, just keeping investments relatively similar (FOXO side) as growth and metabolic rates decline (TOR) would provide exponentially increasing stress resistance likely to promote extended lifetimes. Stress resistance may well be allometric.
In terms of reproduction, many animals anticipate reproductive costs and accrue substantial fat reserves to sustain late pregnancy and subsequent lactation. Thus, to some extent, the costs of reproduction are partially paid earlier, and this reflects that energy demands during reproduction are likely to exceed the capacity of the system otherwise. Anticipatory storage would not be necessary if energy supply is otherwise unlimited during activities like migration, and reproduction. Nuptial gifts and variation in nutrition also vary fecundity, suggestive of resource limitations. Interestingly, pregnant women have little additional elevation in active metabolism over basal levels (1.6- to 1.8-fold) compared to other sedentary people. Physically active people maintained elevations of 2- to 3.4-fold [212]. Importantly, growing juveniles and pregnant and nursing mothers also tend to sleep more. Could this upregulate both TOR and FOXO?
Possible adjustments to cold include thicker fur, altered color, increased theromogenesis and altered feeding and digestive tracts. These are all indicators of energy limitation and heat conservation rather than dissipation limits. Reproduction of small animals like mice is much more constrained by cold temperatures because most of their available energy is diverted to heating and specific pregnancy block is engaged by such stress [213]. Speakman and Krol [205] argue convincingly that milk production by lactating females can be varied by altering temperature suggestive of dissipative regulation. It is worth noting in this context that eggs, juvenile birds and suckling mammals generally have poor thermogenesis, high surface area to volume ratios and incompletely developed insulation (other than those with precocial offspring). Heat subsides provided by parents in insulated nests (and huddling) are valuable adaptations allowing increased growth rates and growth efficiency of the developing young. Neither they nor their insulated parents appear to be at a dissipation limit. Many species (including bumblebees) utilize an un-insulated “brood patch” to specifically elevate and target dissipated heat to their young. Adaptively enhanced dissipative activity and maintenance of insulation during pregnancy, lactation and elevated foraging activity does not support a dissipation limit. Not only do mammals and birds maintain overall effective insulation, many build insulated nests where parents spend considerable time without overheating. It is not overly surprising that a lactating mouse increases both feeding and milk production when she is cold. She would also generate more heat. All of this would tune maternal function to altering needs of the young.
Debate regarding the mass exponent relating body mass to metabolic rate has centered around whether it conforms to the 2/3 rule predicted by simple Euclidean geometry versus the empirically derived ¾ power rule. The ecological theory of metabolism suggests that the geometry of nutrient supply is the key factor deriving the mass exponents and a value of 0.75 is generally recognized [207–210, 214]. Several studies suggest that field metabolic rates of mammals (but not birds) also are close to 0.75 [215]. However, empirical estimates of mass exponents for basal metabolic rate vary enormously [215].
Recent studies suggest that there is no single metabolic exponent [210, 211, 214, 215]. Differences were detected between ectotherms versus endotherms and between measures of rest, field and exercise metabolism [215]. Metabolic rate also varies with temperature, diet, reproduction, altitude, mode of locomotion, methods of calculation [203, 216] and phylogeny [203, 211, 214, 216]. For ectotherms the scaling exponent converged on a value >0.75. For endotherms, values fell between 0.67 and 0.75. Ectotherm values ranged from 0.80–0.87 [215]. A phylogenetic least-square model obtained global mass exponents for basal metabolism of 0.68–0.69, whereas an independent contrast approach yielded values between 0.71–0.74 [214]. Regardless, estimates differed from both 0.67 and 0.75. A mass exponent for basal metabolic rate based on 639 species obtained a value of 0.721 [203]. Across mammalian orders values varied between 0.54 and 1.05.
A recent breakthrough was the finding that metabolic mass exponents vary with body size such that small species approach values of ∼0.67 but values increase with body mass to an asymptote ∼0.75 [211]. This was also supported by an earlier comparison that obtained exponents of 0.69 for small species and 0.76 for large species [214]. This and other sources of variation go a very long way to explaining why empirical estimates vary so greatly. A mass exponent of 0.76 for larger species suggests that they do much better than the surface rule predicts. The value of 0.69 for small species may reflect that their tissues are less remote from atmospheric oxygen so they are more ruled by surface area dissipation loss. Small animals may scale as 0.67 because their tissues have relatively more direct access to oxygen and waste disposal. Large animals may show larger scaling exponents because they must improve over the basic surface area rule to maintain relatively high metabolic rates and temperatures, and still maintain sufficient scope to meet contingencies of reproduction and short term exertion.
A related discovery was that cell cultures from variously sized animals all converged on similar metabolic rates [217]. Comparing the difference between in vitro versus in vivo metabolic rates suggests that the in vivo metabolic rate of tissues in smaller animals more closely approached levels of in vitro cell cultures (i.e., they can better deliver oxygen to tissues throughout their small bodies). Large animals however, have progressively lower mass-specific metabolism compared to in vivo cell cultures, suggesting that their cells are adapted to relative hypoxia that could influence aging rates [25]. The ability of small animals to maintain oxygenation levels closer to ambient is supportive that they might be ruled more by the surface area rule whereas larger species facing greater limitations may have been more strongly selected for offsetting adaptations, including avoidance of dissipation limits.
The DLT predicts that daily energy expenditure should scale with a mass exponent of 0.63, rather than the value of 0.75 proposed by the metabolic theory of ecology. Using phylogenetically adjusted data, Speakman and Krol [205] found that field metabolic rates for mammals and birds had mass exponents of 0.679 and 0.576, respectively. The estimates of Speakman and Krol [205, 206] are exceptionally low compared to other recent literature [215, 216] and the 0.576 exponent for birds implies that they don’t even do as well as the surface area rule (0.67). This also applies to the theoretical exponent derived by the DLT of (0.63) [205]. The low exponents for field metabolic rates means that the relative degree of active metabolism declines as body size increases which is then considered support for the DLT [205].
Whether field metabolic rates are the best measure to use to test dissipation limits is questionable given that they include rest and sleep periods, often taken in insulated nests. Some studies of daily metabolism, for example, found lower exponents than for basal metabolic rates [216] (but see below). In that case a declining scope for field metabolic rates could actually reflect that the cost of locomotion is lower for large species [187] and their lower metabolic costs and thermal efficiency means they do not need to foraging as much or process as much food. Whereas shrews forage nearly continuously and sleep in short bouts, leopards may feed only once in several weeks and spend much of their time in leisure rest. Most footage of lions and cheetahs finds them laying about doing little. Going for the kill, however, requires very effective exercise capacity, and I will argue that this is most relevant to dissipation limits.
Allometry aside, for a given species field metabolic rates tend to be about 2–4 times greater than basal [209, 212] whereas maximal metabolic rates are usually about 10-fold higher (but can be elevated briefly by as much as 50-fold). Interestingly, human athletes can sustain metabolic elevations of 4- to 5.6-fold, exceeding elevations in field metabolisms of all but a few of other endotherm species examined [212]. Unlike many other aspects of metabolic allometry, the very high mass exponents for exercise metabolism are relatively consistent and reliable. Maximal metabolic scope (Max/Basal) itself has a positive mass exponent of 0.18, meaning that larger species are capable of increasingly greater metabolic elevation than small species [209, 218]. Whether field metabolic rates decline or increase with body mass are not as relevant to dissipation limits as the critical fitness aspect of exercise performance.
Slopes for both daily and field metabolic rates strongly vary. As discussed above, early studies support a lower slope for field metabolic rates than for basal rates suggesting that larger species have relatively reduced elevation of active metabolism in the field [219, 220]. More recent estimates found that field metabolism had substantially higher exponents than basal [216]. The overall mass exponent for field metabolic rates was ∼0.81. For eutherian mammals, birds, and reptiles (but not a small sample of marsupials) field metabolic rates were higher than basal. Large marsupials had exceptionally low metabolic rates with a scaling coefficient of 0.59 (i.e. for this sample they did worse than the surface area rule). For birds, however, basal metabolic rate scaled as ∼0.67, whereas for mammals it was ∼0.73 [216]. Regardless, the greater slope for field metabolism in these studies would suggest that larger animals have relatively greater elevation of their metabolic rate in the field, opposite to the DLT. Glazier [210] suggests mass exponents for birds and mammals of 0.66 and 0.69 for basal metabolism, and 0.70 and 0.75 for field metabolic rates, respectively. An exponent of 0.89 was given for running mammals. Another study obtained comparable estimates. These ranges between 0.74–0.78 for resting, 0.60–0.65 for daily metabolism (that would include sleep), 0.78–0.84 for field metabolism and 0.87–0.89 for exercise [215]. Given the great variation and evidence that there is no universal exponent, it is difficult to evaluate what any of these exponents really mean for dissipation limits other than with the reliably high exponents associated with exercise.
If dissipation limits do impinge on physiological function, they are most likely to impact physical (exercise) performance. Such limitations are strongly linked to crucial fitness aspects such as fight, flight and feeding. Running for your life, fighting for your wife or running for your dinner all require maximal performance where dissipation constraints would face strong negative selection pressure. If that is indeed the case, the idea that dissipation limits are important constraints on any lower levels of metabolic expenditures like lactation becomes unlikely, regardless of the duration of sustaining such activity.
The scaling exponents for maximal performance are far greater and consistent than for any other type of metabolism [209, 215, 218]. The mass exponent for maximal metabolic rate during exercise falls between 0.87–0.89 [209, 215]. For athletic species this was even higher (0.94) suggesting that additional factors than surface area are involved [209]. Such scaling may reflect the surface area of available mitochondria and systems managing resource delivery (O2, energy) and waste management for aerobic demands on cells. Interestingly, mitochondrial volume scaled almost identically to maximal metabolic scope (0.956 versus 0.962) and capillary volume also scaled as 0.984 [209]. Could a relative increase in mitochondrial number in larger species allow lower mitochondrial stress at normal levels of metabolism? Estimates of maximal metabolism have consistently obtained very high values, some approaching 1.00 (i.e. almost independent of the surface rule all together). Increased ventilation and directed blood flow to the musculature are two potential adjustments.
Most metabolic mass exponents are expected to range between 0.667 and 1.00 [210] but the fact is it would be extremely surprising if heat dissipation in animals, and especially large ones, conformed to the simple surface area to volume rule, and certainly not less. Animals are not simple shapes. Consider a cube with a hollow interior occupied by the additional surface area of convoluted lungs and a through-put digestive tract. Birds have the additional feature of large air sacs that allow oxygen to be extracted from the lungs during inhaling and exhaling. Surface area is increased by such features and invaginations on smaller scales without any change in volume. Now add a circulatory system ramifying blood throughout the body (and capable of regulating thermal conductance and specific targets for blood flow).
Dissipative processes are enormously increased by convection such as provided by breathing ventilation and fanning by large ears such as African elephants. The possibility that animals behave according to the surface rule becomes increasingly unlikely. Now add evaporative cooling, sweat, wallowing and the convection associated with movement itself. Evaporative cooling powerfully dissipates heat, thereby augmenting surface area. Movement-associated convective heat transfer is probably particularly important for flying animals that may also access cooler air at higher altitudes [206]. Interestingly, bumblebees and night flying moths that can fly at lower air temperatures are more insulated. Such animals must maintain a relatively high temperature to engage flight, and will “shiver” their flight muscles to reach functional temperatures. Dissipation limits associated with heat generated by sustained flight do not inhibit warm-bodied bumblebees even on hot days. The beating of their wings may actually contribute to convective heat loss at high levels of activity.
Speakman and Krol [206] carefully consider how animals like migrating birds can sustain highly elevated metabolisms because this challenges their theory. Such features, however, are exactly those explaining how dissipation limits can be avoided. Furthermore, migration and insulated hibernation are adaptations to avoid periods of energy stress and potential death associated with excessive heat dissipation in freezing air. The key point is that if birds or athletic animals face dissipation limits, they exploit their niche such as to avoid them. Size may also be a factor in locomotion because in addition to lower costs of transport, larger volumes may displace relatively more air over surfaces during movement.
Locomotion is also critical to warm-bodied tuna that can only extract the oxygen required for heat generation from water by engaging ram-jet water flow over the gills at high speeds. Recall that these animals also have remarkable counter-current heat exchange to maintain temperature against conductive heat loss into cold water. Dissipation is important – important to minimize. Although there can be little doubt that heat dissipation can be a problem, particularly in hot climates, thermoregulation evolved to elevate rates and functions above ambient and most adaptations serve to conserve this heat. It seems improbable that dissipation limits are the general and overriding determinate of mass-specific allometry and associated life history features. In particular the idea that animals have evolved to sizes that negatively impact reproduction, or that available energy for various functions is unlimited will likely prove difficult to support. For now at least, the principle of allocation and disposable soma theory might be considered as viable.
IMPLICATIONS
The fact that both pathways considered to most strongly regulate aging likely occur in sleep has pervasive ramifications (Fig. 2). That TOR and FOXO reside on opposite branches of the phosphorylated Akt bifurcation (Fig. 1) means that sleep consists of at least two antagonistic windows. In a temporal framework this can be considered as indicating that mutually synergistic aspects are marshaled together at a particular time and that separated windows are successively deployed to maximize global fitness. Order is also critical. One theoretical insight is that aging interventions that enhance one function to obtain life extension (e.g., FOXO upregulation by resveratrol) and/or suppress those functions seen as accelerating aging (e.g., rapamycin suppression of TOR) are somewhat misguided. Tradeoffs between functions may be avoided by appropriate management. The idea that changes in one pathway necessitate opposite alterations in the other does not hold if antagonistic or incompatible functions reside in separate temporal windows. Further, true youthful function must require robust activity in all waking and sleep windows. Each has evolved to maximize overall fitness so none can be neglected.
While robust and balanced rhythms are likely the necessary goal for healthy life extension, temporal features like sleep show age-related declines and fragmentation indicative of dysregulation, and waking functions like spontaneous activity markedly decline [195]. This suggests that aging may indeed represent distorted imbalance, in which case, interventions that differentially address one functional window may of themselves improve others. What is particularly frustrating from a perspective of life extension is that the hard-wired shifts that function to maintain the circadian march across compartmentalized functions (e.g., suppression of FOXO and associated stress resistance pathways by the TOR window) may emerge as formidable obstacles to our success. Aging has even been suggested to arise from continuation of TOR signaling beyond the growth period [221]. To the extent that clocks contribute to tissue-specific functions, dysfunction of clocks in old age could contribute to dysregulation of the nuclear receptors or tissue de-differentiation which has been proposed as a possible mechanism of aging.
One obvious source of imbalance is the increasingly oxidative conditions and greater levels of ROS with age. Elevated ROS could chronically elevate MAPK-ERK and PI3K-Akt-TOR activity [175, 222] even in the face of declining GH and IGF-1 signaling typical of aging. In fact, declines in GH axis signaling and elevations in the HPA axis could be compensatory. If so, reducing ROS could restore balance in these higher order regulatory systems [3].
Waking levels of energy and amino acid acquisition likely determine levels of subsequent TOR and FOXO activity, partly via sensors such as AMPK and S6K. Consequently, industrialized diets, particularly high in protein (e.g., meat) and excess energy likely elevate TOR. Dietary restriction, fasting or low-protein diets are more likely to favor FOXO. Although industrialized diets may benefit growth, health and fitness in youth, they may contribute to much of our avoidable aging-associated pathology. High protein diets that promote growth may specifically suppress those stress resistance pathways that would otherwise best serve us in old age, and counter-intuitively, could also contribute to obesity and metabolic syndrome [65]. The TOR target, S6K-1 is sensitive to insulin and amino acids and appears to reduce insulin signaling via inhibition of IRS-1 (Fig. 1). Thus, S6K-1 could induce insulin resistance under conditions of excess protein intake. S6K-1 knockouts are 20% smaller at birth and are resistant to obesity due to elevated lipolysis. Rapamycin also reduces adipogenesis [65].
Of potentially great practical importance, the temporal triumvirate suggests that dietary restriction utilizing alternate day feeding (ADF) likely represents an entirely different mechanism than chronic daily reduction of calories. Theoretically, TOR would be upregulated on feeding days and suppressed on fasting days. ADF could alternately stimulate the FOXO or TOR pathways, perhaps better maintaining benefits of both and offsetting age-related distortion. Whereas DR is very difficult for humans, ADF requires less discipline. Although the TOR and FOXO windows might be simultaneously manipulated via time-released drug delivery, ADF might be employed to alternately stimulate the TOR and FOXO pathways. This could circumvent the fact that FOXO normally resides behind the TOR window. A combination of ADF augmented by alternating pharmacological/ nutritional manipulation could prove particularly effective at ages where these pathways may become chronically distorted. Alternatively, could alternating high protein feeding days with low protein days (with somewhat reduced calories) achieve some level of impact?
The mechanisms mediating impacts of DR remain unresolved. Froy [36] suggested that ADF might engage stress resistance via alterations in the clock, and the framework described here suggests a more specific mechanism. Intermittent feeding yields similar benefits to chronic DR, but the reduction in calories may be smaller or even minimal [37]. Time restricted feeding and timing of ADF can reset and entrain the circadian clock. Some benefits of DR and ADF may arise from improved circadian coordination [37]. Disruption of circadian clocks is associated with hormonal imbalances, increased disease risks (including cardiovascular disease and cancer) and reduced longevity [37].
The present framework suggests that alterations in the clock, and especially sleep, could have enormous health and aging impacts via TOR and/or FOXO. A key factor in extending health and longevity may trace to robust cycling of the clock [37] and maintenance of balance among the three functional phases of the circadian rhythm (i.e., waking, anabolism, and defense). Thus, αMUPA mice have high amplitude clock rhythms associated with extended longevity, low IGF-1, smaller size, lower apoptotic thresholds and reduced cancer [37, 223]. The linkage of SIRT to the clock and its known involvement in DR strongly suggests a linkage of the clock to aging rates. Alternate day feeding may elevate stress resistance and DR downregulates TOR in mice [37]. Liver, muscle and colon showed increased activity in Per1, Per2, Per3 and downregulated Arnt1/Bmal1 (Arnt= aryl hydrocarbon receptor nuclear translocator) genes associated with circadian rhythmicity under DR. This suggests a role of the clock in alterations in energy metabolism and defense under resource shortfalls [139]. Linkage of ADF to the clock is highlighted by lack of hepatic clock rhythms in mice with feeding restricted to the photophase resting period [37].
Total duration of sleep declines but the duration of sleep episodes increases with increasing body size across mammalian species [224]. This may reflect lower growth demands or reduced stress that accompanies low metabolic rates and slow growth to larger sizes. Now the question becomes, what aspects of sleep are altered as species become larger? Is the TOR window reduced in period or intensity? Is the FOXO window expanded? The disposable soma theory would suggest that no naturally evolved species will reflect longevity assurance investments that could entirely eliminate aging. Can we extend longevity beyond our blindly evolved limitations by exaggeration of late sleep FOXO functions? A new and promising landscape stretches before us.
Acknowledgments
This work was supported by a grant to C. D. Rollo from the Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- [1].Tolkien JRR. The Lord of the Rings. Glascow: Harper Colins; 1954. [Google Scholar]
- [2].Spong A, Bartke A. Hormonal influences on aging and lifespan. In: Wolf NS, editor. Comparative biology of aging. Netherlands: Springer; 2010. pp. 43–68. [Google Scholar]
- [3].Rollo CD. Growth negatively impacts the life span of mammals. Evol Develop. 2002;4:55–61. doi: 10.1046/j.1525-142x.2002.01053.x. [DOI] [PubMed] [Google Scholar]
- [4].Rollo CD. Overview of research on giant transgenic mice with emphasis on the brain and aging. In: Samaras T, editor. Human body size and the laws of scaling. New York: Nova Scientific Publishers; 2007. pp. 235–60. [Google Scholar]
- [5].Austad SN. Animal size, metabolic rate, and survival, among and within species. In: Wolf NS, editor. The comparative biology of aging. Netherlands: Springer; 2010. pp. 27–41. [Google Scholar]
- [6].Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageingprocess. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- [7].Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–13. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- [8].Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Evens P, Cervera P, LeBouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–7. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- [9].Bonkowski MS, Rocha JS, Masternak MM, Al Regaiey KA, Bartke A. Targeted disruption of growth hormone receptor interferes with the beneficial actions of calorie restriction. Proc Natl Acad Sci USA. 2006;103:7901–5. doi: 10.1073/pnas.0600161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Taguchi A, Wartschow LM, White MF. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 2007;317:369–72. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- [11].Selman C, Lingard S, Choudhury AI, Batterham RL, Claret M, Clements M, Ramadani F, Okkenhaug K, Schuster E, Blanc E, Piper MD, Al-Qassab H, Speakman JR, Carmignac D, Robinson ICA, Thornton JM, Gems D, Partridge L, Withers DJ. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 2008;22:807–18. doi: 10.1096/fj.07-9261com. [DOI] [PubMed] [Google Scholar]
- [12].Steger RW, Bartke A, Cecim M. Premature aging in transgenic mice expressing different growth hormone genes. J Reprod Fert. 1993;46:61–75. [PubMed] [Google Scholar]
- [13].Wolf E, Kahnt E, Ehrlein J, Hermanns W, Brem G, Wanke R. Effects of long-term elevated serum levels of growth hormone on life expectancy of mice: Lessons from transgenic animal models. Mech Ageing Develop. 1993;68:71–87. doi: 10.1016/0047-6374(93)90141-d. [DOI] [PubMed] [Google Scholar]
- [14].Rollo CD, Carlson J, Sawada M. Accelerated aging of giant transgenic growth hormone mice is associated with elevated free radical processes. Can J Zool. 1996;74:606–20. [Google Scholar]
- [15].Rollo CD. Phenotypes: Their Epigenetics, Ecology and Evolution. London: Chapman and Hall; 1994. [Google Scholar]
- [16].Shapira M, Segal E, Botstein D. Disruption of yeast forkhead-associated cell cycle transcription by oxidative stress. Mol Biol Cell. 2004;5:5659–69. doi: 10.1091/mbc.E04-04-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kaeberlein M, Powers RW, III, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–6. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- [18].Castrillo JI, Zeef LA, Hoyle DC, Zhang N, Hayes A, Gardner DCJ, Cornell MJ, Petty J, Hakes L, Wardleworth L, Rash B, Brown M, Dunn WB, Broadhurst D, O’Donoghue K, Hester SS, Dunkley TPJ, Hart SR, Swainston N, Li P, Gaskell SJ, Paton NW, Lilley KS, Kell DB, Oliver SG. Growth control of the eukaryote cell: a systems biology study in yeast. J Biol. 2007;6:4. doi: 10.1186/jbiol54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rollo CD. Circadian Redox Regulation. In: Pantopoulos K, Schipper HM, editors. Principles of free radical biomedicine. New York: Nova Science Publishers; 2010. (In Press). [Google Scholar]
- [21].Salmon AB, Murakami S, Bartke A, Kopchick J, Yasumura K, Miller RA. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Physiol Endocrinol Metab. 2005;289:E23–E29. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- [22].Lachmansingh E, Rollo CD. Evidence for a trade-off between growth and behavioural activity in giant “Supermice” genetically engineered with extra copies of growth hormone genes. Can J Zool. 1994;72:2158–68. [Google Scholar]
- [23].Rollo CD, Foss J, Lachmansingh E, Singh R. Behavioural rhythmicity in transgenic growth hormone mice: trade-offs, energetics, and sleep-wake cycles. Can J Zool. 1997;75:1020–34. [Google Scholar]
- [24].Hajdu I, Obal F, Jr, Fang J, Krueger JM, Rollo CD. Sleep of transgenic mice producing excess rat growth hormone. Am J Physiol Regul Integr Comp Physiol. 2002;282:R70–R76. doi: 10.1152/ajpregu.00485.2001. [DOI] [PubMed] [Google Scholar]
- [25].Rollo CD. Dopamine and aging: intersecting facets. Neurochem Res. 2009;34:601–29. doi: 10.1007/s11064-008-9858-7. [DOI] [PubMed] [Google Scholar]
- [26].Murray DB, Engelen F, Lloyd D, Kuriyama H. Involvement of glutathione in the regulation of respiratory oscillation during a continuous culture of Saccharomyces cerevisiae. Microbiol. 1999;145:2739–45. doi: 10.1099/00221287-145-10-2739. [DOI] [PubMed] [Google Scholar]
- [27].Lloyd D, Eshantha L, Salgado J, Turner MP, Suller MTE, Murray D. Cycles of mitochondrial energization driven by the ultradian clock in a continuous culture of Saccharomyces cerevisiae. Microbiol. 2002;148:3715–24. doi: 10.1099/00221287-148-11-3715. [DOI] [PubMed] [Google Scholar]
- [28].Lloyd D, Murray DB. Redox rhythmicity: clocks at the core of temporal coherence. Bioessays. 2007;29:465–73. doi: 10.1002/bies.20575. [DOI] [PubMed] [Google Scholar]
- [29].Young MW. An ultradian clock shapes genome expression in yeast. Proc Natl Acad Sci USA. 2004;101:1118–9. doi: 10.1073/pnas.0400052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tu BP, McKnight SL. Metabolic cycles as an underlying basis of biological oscillations. Nature Rev Mol Cell Biol. 2006;7:696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- [31].Tu BP, McKnight SL. The yeast metabolic cycle: insights into the life of a eukaryotic cell. Cold Spring Harb Symp Quant Biol. 2007;72:339–43. doi: 10.1101/sqb.2007.72.019. [DOI] [PubMed] [Google Scholar]
- [32].Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–8. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- [33].Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci USA. 2007;104:16886–91. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Ann Rev Gen Hum Genet. 2004;5:407–41. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Allada C, Chung BY. Circadian organization of behavior and physiology in Drosophila. Ann Rev Physiol. 2010;72:26.2–26.20. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Froy O. Metabolism and circadian rhythms- implications for obesity. Endocr Rev. 2010;31:1–24. doi: 10.1210/er.2009-0014. [DOI] [PubMed] [Google Scholar]
- [37].Froy O, Miskin R. Effect of feeding regimens on circadian rhythms: Implications for aging and longevity. Aging. 2010;2:7–27. doi: 10.18632/aging.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- [39].McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–99. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–10. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- [42].Yang X. A wheel of time: the circadian clock, nuclear receptors, and physiology. Genes Dev. 2010;24:741–747. doi: 10.1101/gad.1920710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- [44].Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Ann Rev Genet. 2006;40:409–48. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- [45].Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–9. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- [46].Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Ann Rev Biochem. 2001;71:307–31. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- [47].Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–21. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- [48].Laposky AD, Turek FW. Physiologic and health consequences of circadian disruption in animal models. Sleep Med Clin. 2009;4:127–42. [Google Scholar]
- [49].Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–57. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Duez H, Staels B. Rev-erb-α: an integrator of circadian rhythms and metabolism. J Appl Physiol. 2009;107:1972–80. doi: 10.1152/japplphysiol.00570.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yuan X, Ta TC, Lin M, Evans JR, Dong Y, Bolotin E, Sherman MA, Forman BM, Sladek FM. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS ONE. 2009;4(5):e5609. doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Burris TP. Nuclear hormone receptors for heme: REV-ERBα and REV-ERBβ are ligand-regulated components of the mammalian clock. Mol Endocrinol. 2008;22:1509–20. doi: 10.1210/me.2007-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kumar N, Liu D, Wang H, Robidoux J, Collins S. Orphan nuclear receptor NOR-1 enhances cAMP-dependent uncoupling protein-1 gene transcription. Mol Endocrinol. 2008;22:1057–64. doi: 10.1210/me.2007-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Pearen MA, Myers SA, Raichur S, Ryall JG, Lynch GS, Muscat GEO. The orphan nuclear receptor, NOR-1, a target of β-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinol. 2008;149:2853–65. doi: 10.1210/en.2007-1202. [DOI] [PubMed] [Google Scholar]
- [55].Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms. 2006;21:350–61. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
- [56].Son GH, Chung S, Choe HK, Kim HD, Baik SM, Lee H, Lee HW, Choi S, Sun W, Kim H, Cho S, Lee KH, Kim K. Adrenal peripheral clock controls the autonomous circadian rhythm of glucocorticoid by causing rhythmic steroid production. Proc Natl Acad Sci USA. 2008;105:20970–5. doi: 10.1073/pnas.0806962106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yang S, Liu A, Weidenhammer A, Cooksey RC, McClain D, Kim MK, Aguilera G, Abel ED, Chung JH. The role of mPer2 clock gene in glucocorticoid and feeding rhythms. Endocrinol. 2009;150:2153–60. doi: 10.1210/en.2008-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Leslie NR. The redox regulation of PI3-Kinase–dependent signaling. Antioxid Redox Signal. 2006;8:1765–74. doi: 10.1089/ars.2006.8.1765. [DOI] [PubMed] [Google Scholar]
- [59].Vellai T. Autophagy genes and ageing. Cell Death Different. 2009;16:94–102. doi: 10.1038/cdd.2008.126. [DOI] [PubMed] [Google Scholar]
- [60].Partridge L. The new biology of ageing. Phil Trans R Soc B. 2010;365:147–54. doi: 10.1098/rstb.2009.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Selman C, Tullet JMA, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, Woods A, Robinson ICA, Schuster E, Batterham RL, Kozma SC, Thomas G, Carling D, Okkenhaug K, Thornton JM, Partridge L, Gems D, Withers DJ. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–4. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB, III, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKBβ) Science. 2001;292:1728–31. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- [63].Cho H, Thorvaldsen J, Chu Q, Feng F, Birnbaum M. Akt1/PKBα is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001;276:38349–52. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- [64].Garofalo RS, Orena SJ, Rafidi K, Torchia AJ, Stock JL, Hildebrandt AL, Coskran T, Black SC, Brees DJ, Wicks JR, McNeish JD, Coleman KG. Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKBβ. J Clin Invest. 2003;112:197–208. doi: 10.1172/JCI16885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Um SH, D’Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- [66].Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, Johnston ED, Tchao BN, Pak DN, Welton KL, Promislow DEL, Thomas JH, Kaeberlein M, Kennedy BK. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18:564–70. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle1 CM, Flurkey K, Nadon NC, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kaeberlein M, Shamieh LS. The role of TOR signaling in aging. In: Wolf NS, editor. Comparative biology of aging. Netherlands: Springer; 2010. pp. 147–61. [Google Scholar]
- [69].Sharp ZD, Strong R. The role of mTOR signaling in controlling mammalian life span: What a fungicide teaches us about longevity. J Gerontol A Biol Sci Med Sci. 2010;65A:580–9. doi: 10.1093/gerona/glp212. [DOI] [PubMed] [Google Scholar]
- [70].Baserga R, Hongo A, Rubini M, Prisco M, Valentinis B. The IGF-I receptor in cell growth, transformation and apoptosis. Biochim Biophys Acta Rev Cancer. 1997;1332:F105–F126. doi: 10.1016/s0304-419x(97)00007-3. [DOI] [PubMed] [Google Scholar]
- [71].Mackiewicz M, Shockley KR, Romer MA, Galante RJ, Zimmerman JE, Naidoo N, Baldwin DA, Jensen ST, Churchill GA, Pack AI. Macromolecule biosynthesis: a key function of sleep. Physiol Genomics. 2007;31:441–57. doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- [72].Eckel-Mahan KL, Phan T, Han S, Wang H, Chan GCK, Scheiner ZS, Storm DR. Circadian oscillation of hippocampal MAPK activity and cAMP: implications for memory persistence. Nature Neurosci. 2008;11:1074–82. doi: 10.1038/nn.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Cao R, Lee B, Cho HY, Saklayen S, Obrietan K. Photic regulation of the mTOR signaling pathway in the suprachiasmatic circadian clock. Mol Cell Neurosci. 2008;38:312–24. doi: 10.1016/j.mcn.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cao R, Li A, Cho HY, Lee B, Obrietan K. Mammalian target of rapamycin signaling modulates photic entrainment of the suprachiasmatic circadian clock. J Neurosci. 2010;30:6302–14. doi: 10.1523/JNEUROSCI.5482-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Conover CA, Bale LK. Loss of pregnancy-associated plasma protein A extends lifespan in mice. Aging Cell. 2007;6:727–29. doi: 10.1111/j.1474-9726.2007.00328.x. [DOI] [PubMed] [Google Scholar]
- [76].Lawrence JB, Oxvig C, Overgaard MT, Sottrup-Jensen L, Gleich GJ, Hays LG, Yates JR, III, Conover CA. The insulin-like growth factor (IGF)-dependent IGF binding protein-4 protease secreted by human fibroblasts is pregnancy-associated plasma protein-A. Proc Natl Acad Sci USA. 1999;96:3149–53. doi: 10.1073/pnas.96.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–50. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- [78].Norrelund H, Fisker S, Vahl N, Børglum J, Richelsen B, Christiansen JS, Jørgensen JOL. Evidence supporting a direct suppressive effect of growth hormone on serum IGFBP-1levels. Experimental studies in normal, obese and GH-deficient adults. Growth Horm IGF Res. 1999;9:52–60. doi: 10.1054/ghir.1998.0087. [DOI] [PubMed] [Google Scholar]
- [79].Levine AJ, Feng Z, Mak TM, You H, Jin S. Coordination and communication between the p53 and IGF-1–AKT–TOR signal transduction pathways. Genes Dev. 2006;20:267–75. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- [80].Sachdev D, Yee D. The IGF system and breast cancer. Endocr Rel Cancer. 2001;8:197–209. doi: 10.1677/erc.0.0080197. [DOI] [PubMed] [Google Scholar]
- [81].Averous J, Maurin AC, Bruhat A, Jousse C, Arliguie C, Fafournoux P. Induction of IGFBP-1 expression by amino acid deprivation of HepG2 human hepatoma cells involves both a transcriptional activation and an mRNA stabilization due to its 3′UTR. FEBS Lettr. 2005;579:2609–2614. doi: 10.1016/j.febslet.2005.03.077. [DOI] [PubMed] [Google Scholar]
- [82].Finlay D, Ruiz-Alcaraz AJ, Lipina C, Perrier S, Sutherland C. A temporal switch in the insulin-signalling pathway that regulates hepatic IGF-binding protein-1 gene expression. J Mol Endocrinol. 2006;37:227–37. doi: 10.1677/jme.1.02084. [DOI] [PubMed] [Google Scholar]
- [83].Sharp ZD, Bartke A. Evidence for down-regulation of phosphoinositide 3-kinase/Akt/ mammalian target of rapamycin (PI3K/Akt/mTOR)-dependent translation regulatory signaling pathways in Ames dwarf mice. J Gerontol Ser A Biol Sci Med Sci. 2005;60:293–300. doi: 10.1093/gerona/60.3.293. [DOI] [PubMed] [Google Scholar]
- [84].Miquet JG, Gonzalez L, Matos MN, Hansen CE, Louis A, Bartke A, Turyn D, Sotelo AI. Transgenic mice overexpressing GH exhibit hepatic upregulation of GH-signaling mediators involved in cell proliferation. J Endocrinol. 2008;198:317–30. doi: 10.1677/JOE-08-0002. [DOI] [PubMed] [Google Scholar]
- [85].Hayashi AA, Proud CG. The rapid activation of protein synthesis by growth hormone requires signaling through mTOR. Am J Physiol Endocrinol Metab. 2007;292:E1647–E1655. doi: 10.1152/ajpendo.00674.2006. [DOI] [PubMed] [Google Scholar]
- [86].Ramm P, Smith CT. Rates of cerebral protein synthesis are linked to slow wave sleep in the rat. Physiol Behav. 1990;48:749–53. doi: 10.1016/0031-9384(90)90220-x. [DOI] [PubMed] [Google Scholar]
- [87].Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- [88].Cirelli C. The genetic and molecular regulation of sleep: from fruit flies to humans. Nature Rev Neurosci. 2009;10:549–60. doi: 10.1038/nrn2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Methippara MM, Bashir T, Kumar S, Alam DN, Szymusiak R, McGinty D. Salubrinal, an inhibitor of protein synthesis, promotes deep slow wave sleep. Am J Physiol Regul Integr Comp Physiol. 2009;296:R178–R184. doi: 10.1152/ajpregu.90765.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Boldt HB, Conover CA. Pregnancy-associated plasma protein-A (PAPP-A): A local regulator of IGF bioavailability through cleavage of IGFBPs. Growth Horm IGF Res. 2007;17:10–18. doi: 10.1016/j.ghir.2006.11.003. [DOI] [PubMed] [Google Scholar]
- [91].Conover CA, Bale LK, Mader JR, Mason MA, Keenan KP, Marler RJ. Longevity and age-related pathology of mice deficient in pregnancy-associated plasma protein-A. J Gerontol A Biol Sci Med Sci. 2010;65A:590–599. doi: 10.1093/gerona/glq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Ren SG, Ezzat S, Melmed S, Braunstein GD. Somatostatin analog induces insulin- like growth factor binding protein-1 (IGFBP-1) expression in human hepatoma cells. Endocrinol. 1992;131:2479–81. doi: 10.1210/endo.131.5.1385103. [DOI] [PubMed] [Google Scholar]
- [93].Yan L, Lavin VA, Moser LR, Cui Q, Kanies C, Yang E. PP2A regulates the pro-apoptotic activity of FOXO1. J Biol Chem. 2008;283:7411–20. doi: 10.1074/jbc.M708083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zhao W, Qin W, Pan J, Wu Y, Bauman WA, Cardozo C. Dependence of dexamethasone induced Akt/FOXO1signaling, upregulation of MAFbx, and protein catabolism upon the glucocorticoid receptor. Biochem Biophys Res Comm. 2009;378:668–72. doi: 10.1016/j.bbrc.2008.11.123. [DOI] [PubMed] [Google Scholar]
- [95].Cho JE, Fournier M, Da X, Lewis MI. Time course expression of Foxo transcription factors in skeletal muscle following corticosteroid administration. J Appl Physiol. 2010;108:137–45. doi: 10.1152/japplphysiol.00704.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Waddell DS, Baehr LM, van den Brandt J, Johnsen SA, Reichardt HM, Furlow JD, Bodine SC. The glucocorticoid receptor and FOXO1 synergistically activate the skeletalmuscle atrophy-associated MuRF1 gene. Am J Physiol Endocrinol Metab. 2008;295:E785–E797. doi: 10.1152/ajpendo.00646.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Suwanichkul A, Allander SV, Morris SL, Powell DR. Glucocorticoids and insulin regulate expression of the human gene for insulin-like growth factor-binding protein-1 through proximal promoter elements. J Biol Chem. 1994;269:30835–41. [PubMed] [Google Scholar]
- [98].Holden JP, Butzow TL, Laughlin GA, Ho M, Morales AJ, Yen SC. Regulation of insulin-like growth factor binding protein-I during the 24-hour metabolic clock and in response to hypoinsulinemia induced by fasting and sandostatin in normal women. J Soc Gynecol Inv. 1995;2:38–44. [PubMed] [Google Scholar]
- [99].Martinelli CE, Jr, Yateman ME, Cotterill AM, Moreira AC, Camacho-Hubner C. Correlation between cortisol and insulin-like growth factor-binding proteins (IGFBPs) under physiological conditions in children. Clin Endocrinol. 1999;50:767–74. doi: 10.1046/j.1365-2265.1999.00724.x. [DOI] [PubMed] [Google Scholar]
- [100].Gillies G. Somatostatin: the neuroendocrine story. Trends Pharmacol Sci. 1997;18:87–95. doi: 10.1016/s0165-6147(96)01032-2. [DOI] [PubMed] [Google Scholar]
- [101].Vaccarino FJ, Sovran P, Baird JP, Ralph MR. Growth hormone-releasing hormone mediates feeding-specific feedback to the suprachiasmatic circadian clock. Peptides. 1995;16:595–8. doi: 10.1016/0196-9781(95)00018-f. [DOI] [PubMed] [Google Scholar]
- [102].Hall RK, Yamasaki T, Kucera T, Waltner-Law M, O’Brien R, Granner DK. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin: the role of winged helix/forkhead proteins. J Biol Chem. 2000;275:30169–75. doi: 10.1074/jbc.M004898200. [DOI] [PubMed] [Google Scholar]
- [103].Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–23. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Conover CA, Bale LK, Harrington SC, Resch ZT, Overgaard MT, Oxvig C. Cytokine stimulation of pregnancy-associated plasma protein A expression in human coronary artery smooth muscle cells: inhibition by resveratrol. Am J Physiol Cell Physiol. 2006;290:C183–C188. doi: 10.1152/ajpcell.00199.2005. [DOI] [PubMed] [Google Scholar]
- [105].Kwon HS, Huang B, Unterman TG, Harris RA. Protein kinase B-α inhibits human pyruvate dehydrogenase kinase-4 gene induction by dexamethasone through inactivation of FOXO transcription factors. Diabetes. 2004;53:899–910. doi: 10.2337/diabetes.53.4.899. [DOI] [PubMed] [Google Scholar]
- [106].van der Heide LP, Hoekman MFM, Smidt MP. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem J. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Gan L, Pan H, Unterman TG. Insulin response sequence-dependent and -independent mechanisms mediate effects of insulin on glucocorticoid-stimulated insulin-like growth factor binding protein-1 promoter activity. Endocrinol. 2005;146:4274–80. doi: 10.1210/en.2005-0224. [DOI] [PubMed] [Google Scholar]
- [108].Tsuchiya T, Dhahbi JM, Cui X, Mote PL, Bartke A, Spindler SR. Additive regulation of hepatic gene expression by dwarfism and caloric restriction. Physiol Genomics. 2004;17:307–15. doi: 10.1152/physiolgenomics.00039.2004. [DOI] [PubMed] [Google Scholar]
- [109].Wu N, Yin L, Hanniman EA, Joshi S, Lazar MA. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbα. Genes Dev. 2009;23:2201–9. doi: 10.1101/gad.1825809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Handschin C, Lin J, Rhee J, Peyer AK, Chin S, Wu PH, Meyer UA, Spiegelman BM. Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1α. Cell. 2005;122:505–15. doi: 10.1016/j.cell.2005.06.040. [DOI] [PubMed] [Google Scholar]
- [111].Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- [112].Van der Vos KE, Coffer PJ. FOXO-binding partners: it takes two to tango. Oncogene. 2008;27:2289–99. doi: 10.1038/onc.2008.22. [DOI] [PubMed] [Google Scholar]
- [113].Polonski KS, Given BD, Van Cauter E. Twenty-four-hour profiles and pulsatile patterns of insulin secretion in normal and obese subjects. J Clin Invest. 1998;81:442–8. doi: 10.1172/JCI113339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Masternak MM, Panici JA, Bonkowski MS, Hughes LF, Bartke A. Insulin sensitivity as a key mediator of growth hormone actions on longevity. J Gerontol A Biol Sci Med Sci. 2009;64A:516–21. doi: 10.1093/gerona/glp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Rollo CD. The evolutionary ecology of body size with special reference to allometry and survivorship. In: Samaras T, editor. Human body size and the laws of scaling. New York: Nova Science Publishers; 2007. pp. 213–34. [Google Scholar]
- [116].Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–31. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–60. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–27. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Kukidome D, Nishikawa T, Sonoda K, Imoto K, Fujisawa K, Yano M, Motoshima H, Taguchi T, Matsumura T, Araki E. Activation of AMP-activated protein kinase reduces hyperglycemia-induced mitochondrial reactive oxygen species production and promotes mitochondrial biogenesis in human umbilical vein endothelial cells. Diabetes. 2006;55:120–127. [PubMed] [Google Scholar]
- [120].Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–57. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Bechtold DA. Energy responsive timekeeping. Genetics. 2009;87:447–58. doi: 10.1007/s12041-008-0067-6. [DOI] [PubMed] [Google Scholar]
- [122].Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- [124].Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- [125].Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nature Rev Mol Cell Biol. 2009;10:307–18. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- [126].Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–26. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH. Activation of 5′-AMP-activated kinase with diabetes drug metformin induces casein kinase Iɛ (CKIɛ)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282:20794–8. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- [128].Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, Thompson CB, Evans RM. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–40. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Vieira E, Nilsson EC, Nerstedt A, Ormestad M, Long YC, Garcia-Roves PM, Zierath JR, Mahlapuu M. Relationship between AMPK and the transcriptional balance of clock-related genes in skeletal muscle. Am J Physiol Endocrinol Metab. 2008;295:E1032–E1037. doi: 10.1152/ajpendo.90510.2008. [DOI] [PubMed] [Google Scholar]
- [131].Li XN, Song J, Zhang L, LeMaire SA, Hou X, Zhang C, Coselli JS, Chen L, Wang XL, Zhang Y, Coselli JS, Chen L, Wang XL, Zhang Y, Shen YH. Activation of the AMPK-FOXO3 pathway reduces fatty acid-induced increase in intracellular reactive oxygen species by upregulating thioredoxin. Diabetes. 2009;58:2246–57. doi: 10.2337/db08-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Wang S, Zhang M, Liang B, Xu J, Xie Z, Liu C, Viollet B, Yan D, Zou MH. AMPKα2 deletion causes aberrant expression and activation of AD(P)H oxidase and consequent endothelial dysfunction in vivo: role of 26S proteasomes. Cir Res. 2010;106:1117–28. doi: 10.1161/CIRCRESAHA.109.212530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].O’Neill JS, Maywood ES, Chesha JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–53. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].MPMP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–19. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- [135].Burhans WC, Heintz NH. The cell cycle is a redox cycle: Linking phase-specific targets to cell fate. Free Radical Biol Med. 2009;47:1282–1293. doi: 10.1016/j.freeradbiomed.2009.05.026. [DOI] [PubMed] [Google Scholar]
- [136].Zhang X, Dube TJ, Esser KA. Working around the clock: circadian rhythms and skeletal muscle. J Appl Physiol. 2009;107:1647–54. doi: 10.1152/japplphysiol.00725.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–21. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- [138].Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Selman C, Kerrison ND, Cooray A, Piper MDW, Lingard SJ, Barton RH, Schuster EF, Blanc E, Gems D, Nicholson JK, Thornton JM, Partridge L, Withers DJ. Coordinated multitissue transcriptional and plasma metabonomic profiles following acute caloric restriction in mice. Physiol Genomics. 2006;27:187–200. doi: 10.1152/physiolgenomics.00084.2006. [DOI] [PubMed] [Google Scholar]
- [140].Curtis R, Geesaman BJ, DiStefano PS. Ageing and metabolism: drug discovery opportunities. Nature Rev Drug Discov. 2005;4:569–80. doi: 10.1038/nrd1777. [DOI] [PubMed] [Google Scholar]
- [141].Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–93. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- [142].Froy O. Cytochrome P450 and the biological clock in mammals. Curr Drug Metab. 2009;10:104–15. doi: 10.2174/138920009787522179. [DOI] [PubMed] [Google Scholar]
- [143].Claridge-Chang A, Wijinen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- [144].Gershman B, Puig O, Hang L, Peitzsch RM, Tatar M, Garofalo RS. High-resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiol Genomics. 2007;29:24–34. doi: 10.1152/physiolgenomics.00061.2006. [DOI] [PubMed] [Google Scholar]
- [145].Schmoll D, Walker KS, Alessi DR, Grempler R, Burchell A, Guo S, Walther R, Unterman TG. Regulation of glucose-6-phosphatase gene expression by protein kinase Bα and the forkhead transcription factor FKHR: Evidence for insulin response unit-dependent and –independent effects of insulin on promoter activity. J Biol Chem. 2007;275:36324–33. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- [146].Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, Heidenreich KA, Sajan MP, Farese RV, Stolz DB, Tso P, Koo SH, Montminy M, Unterman TG. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic and lipogenic gene expression. J Biol Chem. 2006;281:10105–7. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- [147].Kalsbeek A, Palm IF, La Fleur SE, Scheer FAJL, Perreau-Lenz S, Ruiter M, Kreier F, Cailotto C, Buijs RM. SCN outputs and the hypothalamic balance of life. J Biol Rhythms. 2006;21:458–69. doi: 10.1177/0748730406293854. [DOI] [PubMed] [Google Scholar]
- [148].Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ Res. 2010;106:447–62. doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Kida K, Nishio T, Yokozawa T, Nagai K, Matsuda H, Nakagawa H. The circadian change of gluconeogenesis in the liver in vivo in fed rats. J Biochem. 1980;88:1009–13. doi: 10.1093/oxfordjournals.jbchem.a133051. [DOI] [PubMed] [Google Scholar]
- [150].Nagai K, Nakagawa H, editors. Central Regulation of Energy Metabolism With Special Reference to Circadian Rhythm. CRC Press. Boca Raton, FL: CRC Press; 1992. [Google Scholar]
- [151].Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–81. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- [152].Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–7. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].La Fleur SE, Kalsbeek A, Wortel J, Fekkes ML, Buijs RM. A daily rhythm in glucose tolerance: a role for the suprachiasmatic nucleus. Diabetes. 2001;50:1237–43. doi: 10.2337/diabetes.50.6.1237. [DOI] [PubMed] [Google Scholar]
- [154].Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, FitzGerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2(11):e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–5. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–8. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- [157].So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc Natl Acad Sci USA. 2009;106:17582–7. doi: 10.1073/pnas.0909733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Oishi K, Atsumi G, Sugiyama S, Kodomari I, Kasamatsu M, Machida K, Ishida N. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lettr. 2006;580:127–30. doi: 10.1016/j.febslet.2005.11.063. [DOI] [PubMed] [Google Scholar]
- [159].Oishi K, Miyazaki K, Uchida D, Ohkura N, Wakabayashi M, Doi R, Matsuda J, Ishida N. PERIOD2 is a circadian negative regulator of PAI-1 gene expression in mice. J Mol Cellul Cardiol. 2009;46:545–52. doi: 10.1016/j.yjmcc.2009.01.001. [DOI] [PubMed] [Google Scholar]
- [160].Perez VI, Bokov A, Van Remmen H, Mele J, Ran Q, Ikeno Y, Richardson A. Is the oxidative stress theory of aging dead? Biochim Biophys Acta. 2009;1790:1005–14. doi: 10.1016/j.bbagen.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Miwa S, Riyahi K, Partridge L, Brand MD. Lack of correlation between mitochondrial reactive oxygen species production and life span in Drosophila. Ann NY Acad Sci. 2004;1019:388–91. doi: 10.1196/annals.1297.069. [DOI] [PubMed] [Google Scholar]
- [162].Buffenstein R, Edrey YH, Yang T, Mele J. The oxidative stress theory of aging: embattled or invincible? Insights from non-traditional model organisms. AGE. 2008;30:99–109. doi: 10.1007/s11357-008-9058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Doonan R, McElwee JJ, Matthijssens F, Walker GA, Houthoofd K, Back P, Matscheski A, Vanfleteren JR, Gems D. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008;22:3236–3324. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Gems D, Doonan R. Oxidative stress and aging in the nematode Caenorhabditis elegans. In: Miwa S, Beckman KB, Muller FL, editors. Oxidative stress in aging: From model systems to human diseases. Totowa NJ: Humana Press; 2008. pp. 81–110. [Google Scholar]
- [165].Vijg J, Campisi J. Puzzles, promises and a cure for ageing. Nature. 2008;454:1065–71. doi: 10.1038/nature07216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang TT, Nelson J, Strong R, Richardson A. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- [167].Salmon AB, Perez VI, Bokov A, Jernigan A, Kim G, Zhao H, Levine RL, Richardson A. Lack of methionine sulfoxide reductase A in mice increases sensitivity to oxidative stress but does not diminish life span. FASEB J. 2009;23:3601–8. doi: 10.1096/fj.08-127415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–23. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- [169].Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nature Genet. 2007;39:540–3. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- [170].Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, Spelbrink JN, Wibom R, Jacobs HT, Larsson NG. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc Natl Acad Sci USA. 2005;102:17993–8. doi: 10.1073/pnas.0508886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, Loeb LA. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nature Genet 2008. 2008;40:392–4. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- [172].Trifunovic A, Larsson NG. Mitochondrial dysfunction as a cause of ageing. J Intern Med. 2008;263:167–78. doi: 10.1111/j.1365-2796.2007.01905.x. [DOI] [PubMed] [Google Scholar]
- [173].Brys K, Vanfleteren JR, Braeckman BP. Testing the rate-of-living/oxidative damage theory of aging in the nematode model Caenorhabditis elegans. Exp Gerontol. 2007;42:845–851. doi: 10.1016/j.exger.2007.02.004. [DOI] [PubMed] [Google Scholar]
- [174].Braeckman BP, Houthoofd K, Vanfleteren JR. Intermediary metabolism. In: Kuwabara P, editor. The Wormbook. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Blagosklonny MV. Aging: ROS or TOR. Cell Cycle. 2008;7:3344–3354. doi: 10.4161/cc.7.21.6965. [DOI] [PubMed] [Google Scholar]
- [176].Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, Beach T, Miller C, Troncoso J, Trojanowski JQ, Zielke HR, Cotman CW. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci USA. 2008;105:15605–10. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].O’Connor T, Sadleir KR, Maus E, Velliquette RA, Zhao J, Cole SL, Eimer WA, Leslie BH, Bembinster LA, Lammich S, Lichtenthaler SF, Henert SS, De Strooper B, Haass C, Bennet DA, Vassar R. Phosphorylation of the translation initiation factor eIF2 increases BACE1 levels and promotes amyloidogenesis. Neuron. 2008;60:988–1009. doi: 10.1016/j.neuron.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Lapointe J, Hekimi S. Early mitochondrial dysfunction in long-lived Mclk1+/− mice. J Biol Chem. 2008;283:26217–27. doi: 10.1074/jbc.M803287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Beal MF. Aging, energy, and oxidative stress in neurodegenerative diseases. Ann Neurol. 1995;38:357–66. doi: 10.1002/ana.410380304. [DOI] [PubMed] [Google Scholar]
- [180].Brookes PS, Yoon Y, Robotham JL, Anders MW, Sheu SS. Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am J Physiol Cell Physiol. 2004;287:C817–C833. doi: 10.1152/ajpcell.00139.2004. [DOI] [PubMed] [Google Scholar]
- [181].Ricci C, Pastukh V, Leonard J, Turrens J, Wilson G, Schaffer D, Schaffer SW. Mitochondrial DNA damage triggers mitochondrial-superoxide generation and apoptosis. Am J Physiol Cell Physiol. 2008;294:C413–C422. doi: 10.1152/ajpcell.00362.2007. [DOI] [PubMed] [Google Scholar]
- [182].Rollo CD. Multidisciplinary aspects of regulatory systems relevant to multiple stressors: aging, xenobiotics and radiation. In: Mothersill C, editor. Multiple stressors: A challenge for the future. New York: Springer; 2007. pp. 185–224. [Google Scholar]
- [183].Pearl R. The Rate of Living. London: University of London Press; 1928. [Google Scholar]
- [184].Speakman JR. Body size, metabolism and lifespan. J Exp Biol. 2005;208:1717–30. doi: 10.1242/jeb.01556. [DOI] [PubMed] [Google Scholar]
- [185].Bauer M, Hamm AC, Bonaus M, Jacob A, Jaekel J, Schorle H, Pankratz MJ, Katzenberger JD. Starvation response in mouse liver shows strong correlation with life-span-prolonging processes. Physiol Genomics. 2004;17:230–244. doi: 10.1152/physiolgenomics.00203.2003. [DOI] [PubMed] [Google Scholar]
- [186].Schumacher B, Garinis GA, Hoeijmakers JHJ. Age to survive: DNA damage and aging. Trends Genet. 2008;24:77–85. doi: 10.1016/j.tig.2007.11.004. [DOI] [PubMed] [Google Scholar]
- [187].Calder WA. Size, Function, and Life History. Cambridge: Harvard University Press; 1984. [Google Scholar]
- [188].Hulbert AJ. Metabolism and longevity: Is there a role for membrane fatty acids? Integr Comp Biol. 2010 doi: 10.1093/icb/icq007. [DOI] [PubMed] [Google Scholar]
- [189].Williams JB, Miller RA, Harper JM, Wiersma P. Functional linkages for the pace of life, life-history, and environment in birds. Integr Comp Biol. 2010 doi: 10.1093/icb/icq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- [191].Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–7. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- [192].Cutler RG. Antioxidants, aging and longevity. In: Pryor WA, editor. Free radicals in biology. VI. New York: Academic Press; 1984. pp. 371–428. [Google Scholar]
- [193].Cutler RG. Evolutionary biology of aging and longevity in mammalian species. In: Johnson JE, editor. Aging and cell function. New York: Plenum Press; 1984. pp. 1–147. [Google Scholar]
- [194].Speakman JR, Talbot DA, Selman C, Snart S, McLaren JS, Redman P, Krol E, Jackson DM, Johnson MS, Brand MD. Uncoupled and surviving: individual mice with high metabolism have greater mitochondrial uncoupling and live longer. Aging Cell. 2004;3:87–95. doi: 10.1111/j.1474-9728.2004.00097.x. [DOI] [PubMed] [Google Scholar]
- [195].Aksenov V, Long J, Lokuge S, Foster JA, Liu J, Rollo CD. A dietary supplement ameliorates locomotor, neurotransmitter and mitochondrial aging. Exp Biol Med. 2010;335:66–76. doi: 10.1258/ebm.2009.009219. [DOI] [PubMed] [Google Scholar]
- [196].Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–81. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- [197].Barja G. Endogenous oxidative stress: relationship to aging, longevity and caloric restriction. Ageing Res Rev. 2002;1:397–411. doi: 10.1016/s1568-1637(02)00008-9. [DOI] [PubMed] [Google Scholar]
- [198].Barja G. Rate of generation of oxidative stress related damage to animal longevity. Free Radical Biol Med. 2002;33:1167–1172. doi: 10.1016/s0891-5849(02)00910-3. [DOI] [PubMed] [Google Scholar]
- [199].Barja G. Free radicals and aging. Trends Neurosci. 2004;27:595–600. doi: 10.1016/j.tins.2004.07.005. [DOI] [PubMed] [Google Scholar]
- [200].Brand MD, Affourtit C, Esteves TC, Green K, Lambert AL, Miwa S, Pakay JL, Parker N. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radical Biol Med. 2004;37:755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- [201].de Magalhaes JP, Costa J, Church GM. An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J Gerontol A Biol Sci Med Sci. 2007;62:149–160. doi: 10.1093/gerona/62.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Furness LJ, Speakman JR. Energetics and longevity of birds. AGE. 2008;30:75–87. doi: 10.1007/s11357-008-9054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].McNab BK. An analysis of the factors that influence the level and scaling of mammalian BMR. Comp Biochem Physiol A. 2008;151:5–28. doi: 10.1016/j.cbpa.2008.05.008. [DOI] [PubMed] [Google Scholar]
- [204].Kirkwood TBL. A systematic look at an old problem. Nature. 2008;451:644–7. doi: 10.1038/451644a. [DOI] [PubMed] [Google Scholar]
- [205].Speakman JR, Krol E. The heat dissipation limit theory and evolution of life histories in endotherms - time to dispose of the disposable soma theory? Integ Comp Biol. 2010 doi: 10.1093/icb/icq049. [DOI] [PubMed] [Google Scholar]
- [206].Speakman JR, Krol E. Maximal heat dissipation capacity and hyperthermia risk: neglected key factors in the ecology of endotherms. J Anim Ecol. 2010;79:726–46. doi: 10.1111/j.1365-2656.2010.01689.x. [DOI] [PubMed] [Google Scholar]
- [207].Brown JH, Gillooly JF, Allen AP, Savage VM, West GB. Toward a metabolic theory of ecology. Ecology. 2004;85:1771–89. [Google Scholar]
- [208].Savage VM, Gillooly JF, Woodruff WH, West GB, Allen AP, Enquist BJ, Brown JH. The predominance of quarter-power scaling in biology. Funct Ecol. 2004;18:257–282. [Google Scholar]
- [209].Weibel ER, Hoppeler H. Exercise-induced maximal metabolic rate scales with muscle aerobic capacity. J Exp Biol. 2005;208:1635–44. doi: 10.1242/jeb.01548. [DOI] [PubMed] [Google Scholar]
- [210].Glazier DS. A unifying explanation for diverse metabolic scaling in animals and plants. Biol Rev. 2010;85:111–38. doi: 10.1111/j.1469-185X.2009.00095.x. [DOI] [PubMed] [Google Scholar]
- [211].Clarke A, Rothery P, Isaac NJB. Scaling of basal metabolic rate with body mass and temperature in mammals. J Anim Ecol. 2010;79:610–619. doi: 10.1111/j.1365-2656.2010.01672.x. [DOI] [PubMed] [Google Scholar]
- [212].Peterson CC, Nagy KA, Diamond J. Sustained metabolic scope. Proc Natl Acad Sci USA. 1990;87:2324–28. doi: 10.1073/pnas.87.6.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Rollo CD, Rintoul J, Kajiura LJ. Lifetime reproduction of giant transgenic mice: the energy stress paradigm. Can J Zool. 1997;75:1336–45. [Google Scholar]
- [214].White CR, Blackburn TM, Seymour RS. Phylogenetically informed analysis of the allometry of mammalian basal metabolic rate supports neither geometric nor quarter-power scaling. Evolution. 2009;63:2658–67. doi: 10.1111/j.1558-5646.2009.00747.x. [DOI] [PubMed] [Google Scholar]
- [215].White CR, Cassey P, Blackburn TM. Allometric exponents do not support a universal metabolic allometry. Ecology. 2007;88:315–23. doi: 10.1890/05-1883. [DOI] [PubMed] [Google Scholar]
- [216].Nagy KA. Field metabolic rate and body size. J Exp Biol. 2005;208:1621–25. doi: 10.1242/jeb.01553. [DOI] [PubMed] [Google Scholar]
- [217].Brown MF, Gratton TP, Stuart JA. Metabolic rate does not scale with body mass in cultured mammalian cells. Am J Physiol Regul Integr Comp Physiol. 2007;292:R2115–R2121. doi: 10.1152/ajpregu.00568.2006. [DOI] [PubMed] [Google Scholar]
- [218].Weibel ER, Bacigalupe LD, Schmitt B, Hoppeler H. Allometric scaling of maximal metabolic rate in mammals: muscle aerobic capacity as determinant factor. Respir Physiol Neurobiol. 2004;140:115–32. doi: 10.1016/j.resp.2004.01.006. [DOI] [PubMed] [Google Scholar]
- [219].Koteja P. On the relation between basal and field metabolic rates in birds and mammals. Funct Ecol. 1991;5:56–64. [Google Scholar]
- [220].Degen AA, Kam M. Scaling of field metabolic rate to basal metabolic rate ratio in homeotherms. Ecoscience. 1995;2:48–54. [Google Scholar]
- [221].Blagosklonny MV, Hall MN. Growth and aging: a common molecular mechanism. Aging. 2009;1:357–62. doi: 10.18632/aging.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Droge W, Schipper HM. Oxidative stress and aberrant signaling in aging and cognitive decline. Aging Cell. 2007;6:361–70. doi: 10.1111/j.1474-9726.2007.00294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Froy O, Chapnik N, Miskin R. Long-lived αMUPA transgenic mice exhibit pronounced circadian rhythms. Am J Physiol Endocrinol Metab. 2006;291:E1017–E1024. doi: 10.1152/ajpendo.00140.2006. [DOI] [PubMed] [Google Scholar]
- [224].Zeplin H, Rechtschaffen A. Mammalian sleep, longevity, and energy metabolism. Brain Behav Evol. 1974;10:425–70. doi: 10.1159/000124330. [DOI] [PubMed] [Google Scholar]