Abstract
A global depletion of cellular copper as the result of a deficiency in high-affinity copper uptake was previously shown to affect the phenotype and life span of the filamentous fungus Podospora anserina. We report here the construction of a strain in which the delivery of copper to complex IV of the mitochondrial respiratory chain is affected. This strain, PaCox17::ble, is a PaCox17-null mutant that does not synthesize the molecular chaperone targeting copper to cytochrome c oxidase subunit II. PaCox17::ble is characterized by a decreased growth rate, a reduction in aerial hyphae formation, reduced female fertility, and a dramatic increase in life span. The mutant respires via a cyanide-resistant alternative pathway, displays superoxide dismutase (SOD) activity profiles significantly differing from those of the wild-type strain and is characterized by a stabilization of the mitochondrial DNA. Collectively, the presented data define individual components of a molecular network effective in life span modulation and copper as an element with a dual effect. As a cofactor of complex IV of the respiratory chain, it is indirectly involved in the generation of reactive oxygen species (ROS) and thereby plays a life span-limiting role. In contrast, Cu/Zn SOD as a ROS-scavenging enzyme lowers molecular damage and thus positively affects life span. Such considerations explain the reported differences in life span of independent mutants and spread more light on the delicate tuning of the molecular network influencing biological ageing.
The ascomycete Podospora anserina is a filamentous fungus characterized by a limited life span (54). After a strain-specific growth period, the growth of an individual culture ceases and finally the mycelium dies at the hyphal tips. Life span and ageing depend on environmental factors and genetic traits (for detailed reviews, see references 8, 22, 44, and 48). A variety of mutants characterized by different degrees of life span increase are available. Some of them are characterized by enormous differences in life span, even though the same molecular pathways are affected. Examples are mutants in which the respiration is switched from a cytochrome c oxidase (COX)-dependent to an alternative pathway (14, 19, 25, 37, 39, 46, 57). The observed differences make it clear that a variety of different factors contribute to the complex molecular network modulating life span. The details of the contribution of individual factors and their interactions are not known in any system.
Previously, we described the characteristics of the nuclear long-lived mutant grisea, a strain with a loss-of-function mutation in a gene coding for the copper-regulated transcription factor GRISEA. As a consequence, the target genes, including a gene encoding the high-affinity copper transporter PaCTR3, are not transcribed, and cellular copper levels are consequently low. Since copper is a cofactor of a variety of different enzymes (e.g., COX, tyrosinase, and laccase), different pathways are affected (9-13, 46). The contribution of every single pathway to the longevity phenotype is unclear.
In the present study, our aim was to address more carefully the effect of defined copper-dependent functions with respect to life span control. Specifically, we constructed a strain in which copper delivery to the COX was disrupted. Experimentally, this was possible via the generation of a transgenic strain in which a gene coding for the putative copper chaperone PaCOX17 was replaced by a selectable marker gene. Here we present the data demonstrating an important dual effect of copper on longevity.
MATERIALS AND METHODS
Strains and growth conditions.
The P. anserina wild-type strain s (21), the mitochondrial mutant strain ex1 (57), the nuclear mutant strains grisea (49) and PaCox17::ble (both derived from strain s; the present study) were used throughout the present study (Table 1). P. anserina strains were grown on standard cornmeal agar (BMM) at 27°C in the light (21). For the construction of the cDNA library, strains were grown with additional 33 μM BCS (bathocuproinedisulfonic acid) and 1 mM ascorbic acid to limit copper and to maximize the expression of genes that are repressed by this metal. For the preparation of RNA, native proteins, or mitochondria, cultures were subsequently grown in liquid complete medium (CM) for 3 or 7 days (14). To increase the copper concentration in the medium, filter-sterilized copper sulfate was added to a final concentration of 10 μM or 250 μM Cu2+. BMM containing 60 mM ammonium acetate was used as germination medium for ascospores.
TABLE 1.
Strains of P. anserina and S. cerevisiae used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| s | 21 | |
| grisea | grisea | 49 |
| ex1 | ex1 | 34 |
| T28 | heterokaryotic contining wild-type and PaCox17::ble nuclei | This study |
| PaCox17::ble-12 | PaCox17::ble | This studya |
| PaCox17::ble-37312 | PaCox17::ble | This studyb |
| PaCox17::ble-47690 | PaCox17::ble | This studyb |
| PaCox17::ble-47692 | PaCox17::ble | This studyb |
| W303 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | -c |
| W303ΔCOX17 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox17::TRP1 | 28 |
| W303ΔCOX17-pAD4 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox17::TRP1 pAD4 | This study |
| W303ΔCOX17-PaCox17 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 ΔCox17::TRP1 PaCox17 | This study |
Derived from a homokaryotic spheroplast of strain T28, which bears the PaCox17::ble mutation.
Derived from the progeny (monokaryotic ascospore) of a cross between PaCox17::ble-12 (mat−) and wild-type strain s (mat+).
Rodney Rothstein, Department of Human Genetics, Columbia University.
The Saccharomyces cerevisiae strains (W303, W303ΔCOX17, W303ΔCOX17-pAD4, and W303ΔCOX17-PaCox17) used in the present study (Table 1) were grown on yeast extract-peptone-dextrose (YPD) or YPG (YPD in which the dextrose is replaced by 3% [vol/vol] glycerol) medium at 30°C (59). The ability to grow on a nonfermentable carbon source was tested on YPG. All yeast mutants are in the genetic background of the strain W303ΔCOX17 (28), which was kindly provided by A. Tzagoloff (New York, N.Y.).
Preparation of a cDNA library and transformation of yeast.
P. anserina wild-type strain s was grown on BMM and subsequently in liquid CM containing 33 μM BCS (a Cu+ chelator) and 1 mM ascorbic acid (reduces Cu2+ to Cu+). RNA was isolated as described previously (14). Synthesis of cDNA was performed by using the Superscript II kit (Gibco). The cDNA fragments were ligated to HindIII and XhoI adapters and cloned in the yeast expression vector pAD4 cut with HindIII and SalI, whereby SalI and XhoI generate compatible ends (pAD4 was kindly provided by S. M. Jazwinski, New Orleans, La.). The cDNA library was transformed into Escherichia coli MRF′ cells (Stratagene) and grown on standard Luria-Bertani (LB) plates supplemented with 50 mg of ampicillin/liter (LBA). Colonies were washed from the surface of agar plates in a minimal volume of LBA. For the isolation of plasmids, 2 ml of suspension was used to inoculate 500 ml of LBA liquid medium, and the culture was grown overnight at 37°C.
Yeast transformation was performed by using the lithium acetate method (56). To identify plasmids carrying a cDNA complementing the mutant strain W303ΔCOX17, colonies were washed from the sorbitol-dextrose (Leu−) plates with sterile water and plated onto YPG plates.
Isolation and sequencing of PaCox17.
The cDNA on plasmid pAD4-PaCox17 selected from a yeast clone complemented to respiration competence was sequenced on both strands, starting from the ADH-promoter and from the ADH-terminator, by using oligonucleotides AD4 (5′-TAA TCT TTT GTT TCC TCG-3′) and AD4-ter (5′-TTA GAA GTG TCA ACA ACG-3), respectively (accession number AJ578462). Further sequencing was performed by primer walking. The PaCox17-cDNA clone was used to screen a genomic cosmid library of wild-type strain s. An 8-kbp subfragment (cut with HindIII and XhoI) containing the open reading frame (ORF) of PaCox17 and flanking regions was cloned into pBluescript SK II(+) (Stratagene), resulting in plasmid pPaCox17g-1 (accession number AJ578463; Fig. 3).
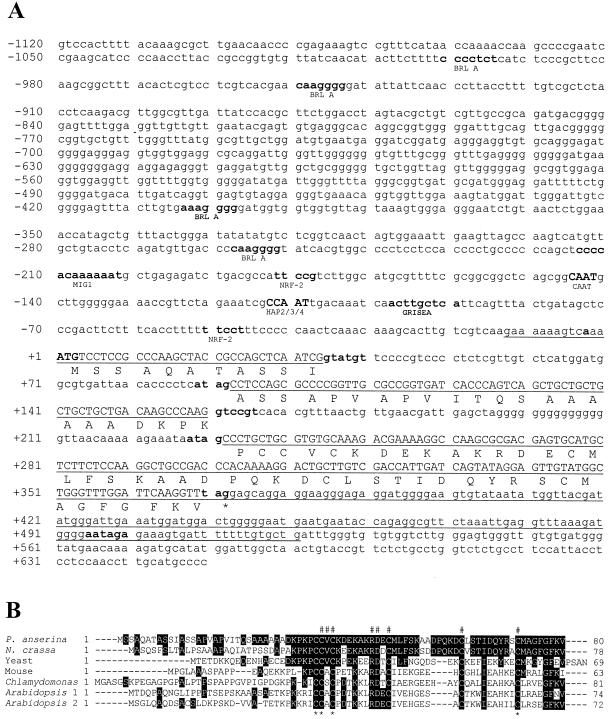
FIG. 3.
(A) Sequence of the PaCox17 locus and of the derived amino acid sequence. The coding sequence is shown in capital letters. Two introns (58 and 71 bp, respectively) disrupt the ORF. The 5′ splice sites GTATGT and GTCCGT resemble the consensus (GTANGT) for filamentous fungi, and the two 3′ splice sites ATAG match with the consensus (A/C)(C/T)AG. A putative polyadenylation site AATAGA (consensus, AATAAA) is found at position 495 (4). Protein-binding sites are described in the text. PaCox17 codes for a copper chaperone of 80 amino acids. The sequence of the cDNA is underlined. Putative promoter binding sites are shown in boldface, with the name of the binding protein below. (B) Amino acid comparison between PaCOX17 and COX17 from other species (Neurospora crassa; S. cerevisiae, accession number NP_013092; Mus musculus, accession number BAB32486; Chlamydomonas reinhardtii, accession number AAF82382; and Arabidopsis thaliana, accession numbers AAK73496 and AAK73497). Identical amino acids are shown by inverted colors. In the first lane, positions which were recently shown to be critical for restoring the respiratory competence of a Cox17-null mutant are indicated by “#” (53). The four essential cysteinyl residues are marked by asterisks.
The sequence of both strands of all constructs was determined by automated sequencing. BLASTN and BLASTX were used to search the databases (2).
The Tfsitescan program of the Institute for Transcriptional Informatics (IFTI [http://www.ifti.org/]), which has access to the object-oriented transcription factor database ooTFD, was used to analyze putative transcription factor binding sites of the PaCox17 5′ region (26). Sequence information provided by (3, 18, 27) was utilized to analyze the DNA sequence at the PaCox17 locus.
Plasmid construction.
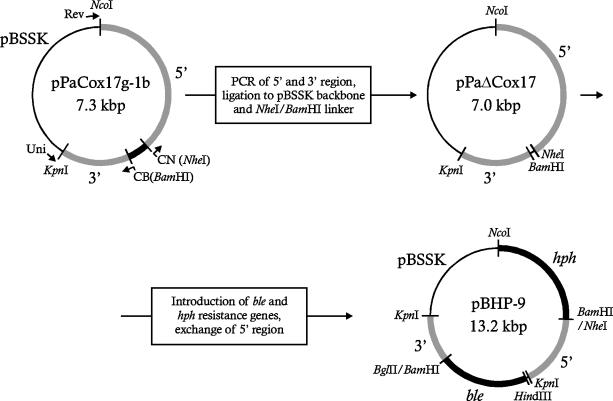
Plasmid pPaCox17g-1 containing a genomic fragment with the PaCox17 ORF and flanking regions was utilized for the construction of the PaCox17 gene replacement plasmid pBHP-9 (Fig. 1). First, the two flanking regions upstream of position −138 and downstream of position +429 of the PaCox17 locus were amplified with two sets of primers. In each PCR, one primer was either the universal (5′-GTA AAA CGA CGG CCA GT-3′) or the reverse (5′-GGA AAC AGC TAT GAC CAT G-3′) primer of pBluescript SK II(+), and the second primer was complementary to the sequence of the PaCox17 locus upstream (CB [5′-TAT GGA TCC GAT ATG GGA TTG-3′) or downstream (CN [5′-CCC GCT AGC ATT GCC GCT GAG CCG-3′) of the PaCox17 ORF (Fig. 1). The latter primers were designed to result in the introduction of specific restriction sites. After digestion with the appropriate enzymes, these two fragments, the KpnI/NcoI-digested pBSSK and a short BamHI/NheI linker, were ligated. The resulting hybrid plasmid, pPaΔCox17, contains the flanking regions of the PaCox17 ORF upstream of position −138 and downstream of position +429. In a series of subsequent subcloning experiments the bleocin resistance cassette (BRC) and the hygromycin B resistance gene were introduced into pPaΔCox17. In addition, the BRC cassette located on a HindIII/BglII fragment of pUT703 (15) was used to replace the BamHI/NheI linker fragment in pPaΔCox17. This cloning step led to plasmid pPaCox17::ble-hph (not shown). Subsequently, the hygromycin B resistance gene on a HindIII/BglII fragment of plasmid pAN7-1 (51) was introduced, resulting in plasmid pBHP-9 (Fig. 1 and 4A). In this plasmid the BRC is flanked by a continuous 2.5-kbp 5′ and a continuous 1.6-kbp 3′ region of the genomic PaCox17 locus.
FIG. 1.
Construction of knockout plasmid pBHP-9. In the first step, parts of the plasmid upstream and downstream of the PaCox17 ORF were amplified by PCR and ligated into pBSSK in the presence of a short BamHI/NheI linker, resulting in plasmid pPaΔCox17. The genomic sequence located upstream of the PaCox17 ORF which turned out to be discontinuous in pPaΔCox17 was replaced by a continuous genomic fragment. The BamHI/NheI linker was replaced by a ble resistance cassette, and then a hygromycin B resistance cassette (hph) was integrated in the bacterial backbone. The resulting plasmid is pBHP-9.
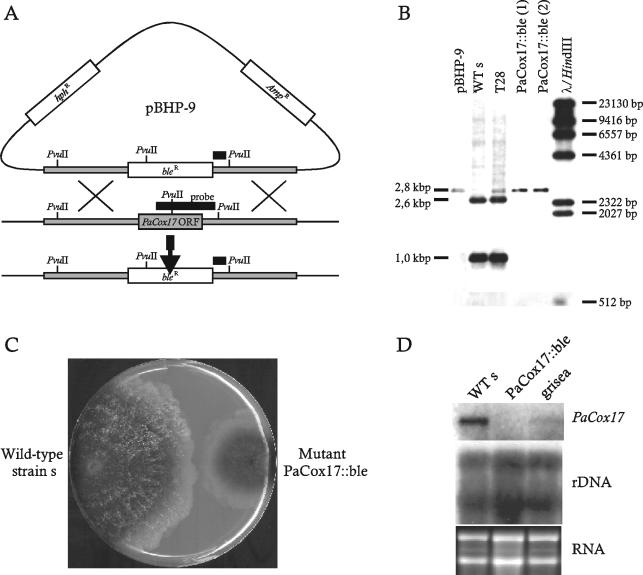
FIG. 4.
(A) Plasmid pBHP-9 containing the PaCox17 locus in which the PaCox17 ORF is replaced by a bleocin resistance cassette (bleR). Further, the backbone of pBHP-9 contains a hygromycin resistance cassette (hphR) as a second marker. Crossing over on both sides of the genomic PaCox17 ORF lead to its replacement by the bleocin resistance cassette of plasmid pBHP-9, resulting in the formation of transgenic strain PaCox17::ble. Distances in the schematic drawings are not drawn to scale. (B) Southern analysis of wild-type strain s and mutant strains PaCox17::ble-12 (PaCox17::ble, lane 1) and PaCox17::ble-37312 (PaCox17::ble, lane 2). Isolated genomic DNA was cut with PvuII (positions shown in panel A) and hybridized to the DIG-labeled PaCox17 S1*S2 probe, which encompasses the second half of the PaCox17-ORF and a 122-bp 3′ region (black bars in panel A). The first two lanes contain DNA of knockout plasmid pBHP-9 and DNA from wild-type strain s− (monokaryotic) as controls; the third lane contains DNA of the heterokaryotic primary transformant T28. The fourth and fifth lanes contain DNA from the two examples of progeny (mutants PaCox17::ble) derived from monokaryotic ascospores of the primary transformant and wild-type strain s, respectively. DIG-labeled DNA of phage lambda was cut with HindIII and used as length standard (sixth lane at the right). (C) Phenotype of wild-type and mutant PaCox17::ble. Wild-type strain s is shown on the left; the mutant PaCox17::ble-37312 strain is shown on the right side on one agar plate. In contrast to the wild type, the mutant shows slower growth and mycelia lacking aerial hyphae. (D) Northern analysis of wild-type strain s (lane 1), mutant PaCox17::ble-37312 (lane 2), and mutant grisea (lane 3). An rDNA probe (plasmid pMY60 [67] containing a HindIII fragment of the rDNA unit of S. carlsbergensis) and an ethidium bromide-stained gel were used as loading controls.
Plasmid pSP17 containing two copies of the first intron (pl-intron) of the mitochondrial gene PaCoxI was used for the isolation of the plDNA probe (63). The probe was labeled by using the digoxigenin (DIG) system.
Production, regeneration, and transformation of spheroplasts.
P. anserina strain s was grown 3 days on five BMM plates and subsequently for 2 days in a total volume of 1 liter of CM. Then, 30 g of mycelium was washed with TPS (5 mM Na2HPO4 · 2H2O, 45 mM KH2PO4, 0.8 M sucrose), chopped in a Waring blender (twice for 5 s each time [slowly], twice for 5 s each time [quickly]), and incubated at 35°C for 2 h with 20 mg of filter-sterilized Glucanex (Novo Nordisk Ferment AG)/ml. Subsequently, the suspension was first filtered through gauze and then three to four times through glass wool. Spheroplasts were pelleted by centrifugation for 10 min at 4,000 rpm. Subsequently, the pellet was washed with TPS three times.
In order to regenerate the spheroplasts, the pellet was recovered in TPS and different dilutions were plated onto plates containing regeneration agar (70 mM NH4Cl, 2 g of tryptone/liter, 1 g of Casamino Acids/liter, 1 g of yeast extract/liter, 50 mM glucose, 1 M sucrose, 550 μM KH2PO4, 335 μM KCl, and 110 μM MgSO4, and MnSO4, FeSO4, CuSO4, and ZnSO4 [each at 50 μg/liter]) with 10 μg of bleocin/ml. After 7 days of growth, mycelia of developing cultures were transferred to BMM. DNA transformation of spheroplasts of P. anserina strain s (mat−) was performed as previously described (47).
Northern analysis.
RNA of strains s (wild type), grisea, and PaCox17::ble-37312 was prepared, blotted, and washed as described previously (14). To confirm equal loading of RNA samples, blots were reprobed with the ribosomal DNA (rDNA) probe (plasmid pMY60 [67], containing a HindIII fragment of the rDNA unit of Saccharomyces carlsbergensis).
Western blot analysis.
Mitochondrial proteins (strains s, grisea, ex1, and PaCox17::ble-37312) were isolated as described previously (14). From each sample, 60 μg of protein was loaded onto a sodium dodecyl sulfate-polyacrylamide gel and subsequently blotted onto nitrocellulose filters. The alternative oxidase of P. anserina (PaAOX) was detected by using a monoclonal mouse AOX antibody, generated against the alternative oxidase of Sauromatum guttatum (20) and the alkaline-phosphatase labeled goat anti-mouse secondary antibody. To confirm the equal loading of the mitochondrial protein probes, the blots were reprobed with an anti-βATPase V rabbit monoclonal antibody (42).
Oxygen consumption measurements.
Respiration of P. anserina strains s (wild type), grisea, and PaCox17::ble-37312 was determined with a Clark-type electrode (Rank Bros.) as described previously (14). To distinguish between COX- and AOX-dependent respiration, potassium cyanide (KCN; 2.2 mM) and salicylhydroxamic acid (SHAM; 4 mM) were used as inhibitors.
SOD activity assay.
Native proteins from wild-type strain s, mutants grisea, ex1, and PaCox17::ble-37312 were isolated as described previously (14). To detect SOD activity, 60 μg of each protein sample were separated on a 8.5% nondenaturing polyacrylamide gel. Purified SOD1 was used as a positive control. Gels were stained once in 2 g of nitroblue tetrazolium solution/liter and once in 2 mg of riboflavin solution/liter containing 0.1% N,N,N′,N′-tetramethylethylenediamine (TEMED) for 20 min in the dark (24). Gels were then developed on a light box (neon light) and scanned with a Hewlett-Packard Scanjet 4470c.
Mitochondrial DNA (mtDNA) analysis.
Total DNA of wild-type strain s and PaCox17::ble-47690 and -47692 strain mycelia of different ages was isolated as described elsewhere (35). Digestion with restriction enzymes, gel electrophoresis, blotting onto a nitrocellulose membrane, and hybridization with a DIG-labeled pl-intron probe (plasmid pSP17) were performed according to standard protocols (10, 55, 63).
Life span measurements.
Mononucleate ascospores were isolated from independent crosses between wild-type strain s and the long-lived mutant PaCox17::ble (isolate 37312). After germination for 3 days, the central piece of the culture was transferred to a race-tube containing BMM agar. Tubes were incubated at 27°C in the light. Growth was measured every day until the cultures reached a point where they stopped growing. The time elapsed to this point was taken as the life span of an individual culture.
Accession numbers.
The sequences of the cDNA and of the genomic copy of PaCox17 were submitted to the EMBL data bank. The accession numbers are AJ578462 and AJ578463, respectively.
RESULTS
Cloning of PaCox17, a gene coding for an ortholog of the COX17 copper chaperone of yeast.
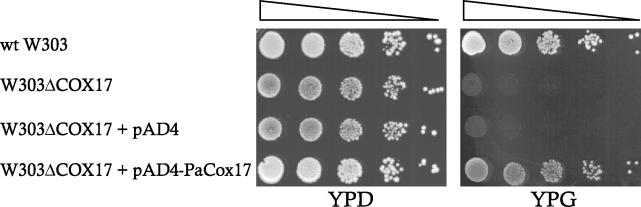
Previously, we reported that lowering the cellular copper levels affects the life span and ageing of P. anserina cultures significantly. This conclusion was derived from the characterization of the long-lived grisea mutant. The mutant is defective in high-affinity copper uptake. As a result, all cellular compartments are depleted of copper and various activities that depend on this metal are impaired (9, 10, 12, 14, 46). In order to define the functions that affect life span more precisely, we set out to construct a transgenic strain in which copper depletion is restricted to mitochondria. More specifically, our goal was to disrupt the pathway delivering copper to complex IV in the mitochondrial respiratory chain. To do this, we cloned PaCox17, a gene coding for a molecular chaperone targeting copper to subunit II of the COX. PaCox17 was isolated by heterologous complementation of the Cox17-deficient yeast strain W303ΔCOX17 (28) with a yeast expression cDNA library constructed from RNA of the P. anserina wild-type strain s grown on copper-depleted medium. Under these conditions maximal expression of PaCox17 was expected to occur. The cDNA library was used to transform the yeast strain W303ΔCOX17, a strain unable to respire and thus not growing on medium containing a nonfermentable carbon source such as glycerol (YPG) as sole carbon source. Successful complementation by a transforming plasmid led to yeast cells able to grow on YPG. From such transformants plasmids were isolated. One plasmid, later termed pAD4-PaCox17, containing a 0.4-kbp insert was further verified. Serial dilutions of the recipient strain transformed with this plasmid were plated onto YPD and in parallel onto YPG (Fig. 2). As controls, dilutions of the respiratory competent wild-type strain, the Cox17 deficiency recipient strain, and the same strain transformed with the yeast expression vector pAD4 containing no insert DNA were inoculated onto the same media. Clearly, plasmid pAD4-PaCox17 was found to reproducibly complement the deficiency of strain W303ΔCOX17 to grow on YPG.
FIG. 2.
Complementation of the yeast mutant strain W303ΔCOX17 by pAD4 carrying the P. anserina cDNA of PaCox17. The yeast strains are as follows (both plates): first line, wild-type W303; second line, respiratory-deficient Cox17 mutant strain W303ΔCOX17; third line, mutant W303ΔCOX17 transformed with empty pAD4; fourth line, mutant W303ΔCOX17 transformed with PaCox17 cDNA cloned in yeast expression plasmid pAD4. Strains were grown to an optical density at 600 nm of 1.0. Serial 10-fold dilutions were prepared, and 5 μl was spotted onto YPD or YPG plates. These strains were grown for 2 days (30°C) in YPG (right plate) or in YPD (left plate) medium.
Subsequently, the insert fragment of pAD4-PaCox17 was sequenced and analyzed. This fragment contains an ORF (Fig. 3A) able to code for a protein of 80 amino acids with a significant homology to the COX17 copper chaperone from different organisms. Importantly, the four cysteine residues that are essential for the function of COX17 are conserved in the different proteins (Fig. 3B) (32).
In the genomic sequence of PaCox17, which was isolated from a cosmid library of the P. anserina wild-type strain s by using the cDNA as a probe, two short introns were found to interrupt the PaCox17 ORF. In the 5′ upstream region a putative CCAAT (−113) and CAAT box (−145) (66), and putative binding sites for different regulatory proteins such as GRISEA (−98), NRF-2 (−51 and −182), MIG1 (−214), and BRL A (−258, −404, −950, and −1001) (1, 16, 38, 68) were localized. Downstream of the PaCox17 stop codon a putative polyadenylation site AATAGA (consensus, AATAAA) was identified in a region with an increased AT content (3). These data suggest that both regions are involved in controlling the expression of the PaCox17 ORF.
Finally, hybridization experiments revealed that PaCox17 is a single-copy gene (data not shown) corresponding to the situation in yeast and mice (28, 65). In contrast, two Cox17 copies are found in other organisms, such as Arabidopsis thaliana, and in humans (52, 69).
Construction of a PaCox17 deletion strain.
In order to generate a PaCox17-deficient strain, plasmid pBHP-9 (Fig. 1 and 4A) was constructed in which the sequence from −138 to +429 (containing the PaCox17 ORF) is replaced by the bleocin resistance cassette (BRC) of pUT703 (15). A 2.5-kbp region upstream and a 1.6-kbp region downstream of the PaCox17 ORF are retained in pBHP-9. These two regions allow cross over to occur on both sides of the BRC, a prerequisite for the exchange of the chromosomal PaCox17 copy by the selectable BRC of pBHP-9 (Fig. 4A). In order to discriminate between transformants arising from a double cross over and from the integration of the complete transforming plasmid via a single crossing over, pBHP-9 carries a second resistance marker (hygromycin B). Transformants of interest in which the PaCox17 gene is exchanged by the BRC should be resistant to bleocin but sensitive to hygromycin B. In order to select such transformants, we transformed spheroplasts of P. anserina wild-type strain s with pBHP-9 and isolated 649 independent bleocin-resistant primary transformants. Only 93 of them were sensitive to hygromycin B. Unfortunately, a Southern blot analysis revealed that none of the transformants was homokaryotic, containing exclusively nuclei with the deleted PaCox17 gene. However, among the selected transformants one transformant (T28) was identified as displaying a hybridization pattern characteristic for a heterokaryon containing predominantly nuclei with the PaCox17 wild-type gene but also a few nuclei with the PaCox17::ble mutation (Fig. 4B). This strain was chosen to select a genetically homogenous PaCox17::ble strain. In a first series of experiments, T28 was crossed with wild-type strain s and 22 monokaryotic ascospores were isolated and germinated. Unfortunately, no homokaryotic PaCox17-deficient transgenic strain could be selected. Therefore, as an alternative approach, mycelium of strain T28 was treated with the cell wall-degrading enzyme mixture Glucanex, and spheroplasts were subsequently selected on bleocin-containing media. Twelve transformants were verified by Southern blot analysis. Although the two PvuII fragments of 1.0 and 2.6 kbp containing the PaCox17 wild-type gene copy do not appear in all of these transformants (two are shown in Fig. 4B), a novel 2.8-kbp fragment hybridizes that results from the replacement of the PaCox17 ORF through the BRC of pBHP-9. These hybridization data clearly show that the selected strains are homokaryotic for the PaCox17::ble locus.
In order to further purify the PaCox17::ble strain, one of the selected primary transformants (PaCox17::ble-12) was crossed with wild-type strain s (mat+), and 42 monokaryotic ascospores were isolated. Three of these secondary transformants, PaCox17::ble-37312, -47690, and -47692, were used in subsequent experiments. PaCox17::ble-37312 was crossed with strain s, and 52 tetrads were isolated and analyzed. The germination rate of isolated ascospores from these tetrads that were derived from several crosses was between 90 and 93%. In these experiments, it turned out that ascospores giving rise to colonies with the mutant phenotype germinated only very poorly on germination medium (BMM containing 60 mM ammonium acetate), and the few developing hyphae stopped growing after about 1 to 5 mm of growth. However, it was possible to rescue these cultures by the transfer of hyphae to standard medium (BMM) without ammonium acetate. On this medium, the corresponding colonies are characterized by a strong reduction in the formation of aerial hyphae, a retarded pigmentation, strongly retarded fertility, and a growth rate (3.9 ± 0.2 mm/day) significantly lower than that of the wild-type strain s (6.5 ± 0.5 mm/day) (Fig. 4C). Perithecia of the mutant are formed after about 3 to 5 weeks in comparison to about 2 weeks in wild-type strains. In addition, they are only about two-thirds of the size of the wild-type strain and contain only few asci. A tetrad analysis revealed that the mutant phenotype (slow-growth, reduced aerial hyphae) was strictly linked to bleocin resistance. In addition, both phenotypes show a postreduction frequency (second strand division) of 79%. This is good initial evidence for a location of the bleocin gene at the chromosomal position at which PaCox17 is located in the wild-type genome.
The expression levels of PaCox17 in wild-type strain s, mutant PaCox17::ble, and mutant grisea were subsequently analyzed in Northern blot experiments. Although the grisea mutant displays lower PaCox17 transcript levels than the wild type, no transcript of this gene was detectable in mutant PaCox17::ble (Fig. 4D).
Finally, in order to experimentally demonstrate that the phenotype of PaCox17::ble-12 is the result of the deletion of the PaCox17 ORF and not due to other affected genes, transgenic strain PaCox17::ble-12 was transformed with plasmid pPaCox17g-2. This plasmid consists of pUC18 and a genomic fragment containing the wild-type copy PaCox17, including 2.5 kbp upstream and 0.3 kbp downstream of the ORF. Hygromycin B-resistant transformants were selected after cotransformation of PaCox17::ble-12 with pAN7-1 (51) and pPaCox17g-2. Southern blot analysis revealed that of 34 hygromycin B-resistant primary transformants, 11 had integrated pPaCox17g-2 at ectopic positions in the genome. These strains all displayed the wild-type phenotype (data not shown).
Collectively, the data from the molecular and the genetic analysis are all consistent and demonstrate the successful exchange of the PaCox17 gene by the selectable bleocin marker gene, resulting in a PaCox17-null mutant strain. The mutant phenotype of this strain is the result of the deletion of the PaCox17 ORF.
Induction of PaAOX in the PaCox17::ble transgenic strain.
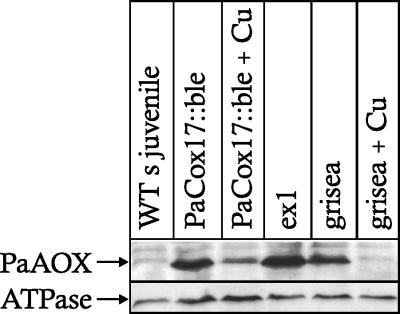
Since PaCox17 is not expressed in the PaCox17::ble transgenic strain, delivery of copper to the COX subunit II and consequently the assembly of complex IV of the respiratory chain were expected to be affected (6, 28, 33, 53, 60). Previous investigation revealed that in COX deficiency strains an alternative cyanide-resistant pathway is induced (12, 14, 19, 57). Consequently, we addressed this possibility. A Western blot analysis revealed high levels of PaAOX in the transgenic PaCox17::ble strain. PaAOX levels are higher than in the copper uptake mutant grisea and reach almost those levels found in the immortal ex1 mutant. Moreover, increasing copper in the growth medium lowers PaAOX levels (Fig. 5). This is in agreement with previous results demonstrating a copper-dependent reduction of transcription of PaAox (14).
FIG. 5.
Western blot analysis of AOX proteins in PaCox17::ble strains grown in rich medium or rich medium with 250 μM copper sulfate. The juvenile wild-type (WT) strains, mutant ex1, mutant grisea on rich medium, or on mutant grisea on medium containing 250 μM additional copper serve as controls. Mitochondria were isolated, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and blotted onto nitrocellulose. PaAOX was detected with monoclonal mouse antibodies against the AOX of Sauromatum guttatum strain Schott (20). An antibody against mitochondrial ATPase subunit β was used as loading control.
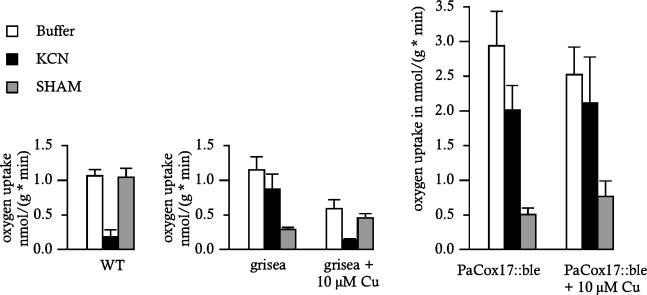
Consistent results were obtained in oxygen consumption experiments. In these experiments mycelial pellets from liquid cultures were incubated in a buffer saturated with oxygen and subsequently the consumption of oxygen was determined. The addition of KCN or SHAM allowed a discrimination between a PaCOX and a PaAOX-dependent respiration. The PaCox17::ble strain was clearly found to respire via a SHAM-sensitive and KCN-resistant alternative pathway. In contrast to the copper uptake mutant grisea, the molecular phenotype cannot be reverted by the addition of 10 μM CuSO4 (Fig. 6). Reversion was also not observed when cultures were grown in medium containing 250 μM CuSO4 (data not shown). Finally, it is remarkable that oxygen uptake by the PaCox17::ble strain is significantly higher than in the wild-type strain or in the grisea mutant. It may be speculated that, for yet unknown reasons, the uptake of oxygen by the mycelium of the Cox17 deficiency strain is enhanced. Thus, at the physiological level, mutants grisea and PaCox17::ble can clearly be differentiated corresponding to the fact that in both mutants different steps of the copper metabolism are impaired.
FIG. 6.
Oxygen uptake in the wild-type strain s, mutant grisea, and the transgenic PaCox17::ble strain. The type of respiration was discriminated by the addition of respiratory inhibitors (either 1 mM KCN or 4 mM SHAM). PaCox17::ble strains were compared to wild-type strain s and mutant grisea. Strains were grown in rich medium or in rich medium with an additional 10 μM copper sulfate. These experiments were each carried out at least five times with two or more independent isolates.
Cu/Zn SOD activities document the availability of copper in the cytoplasm of the PaCox17::ble strains.
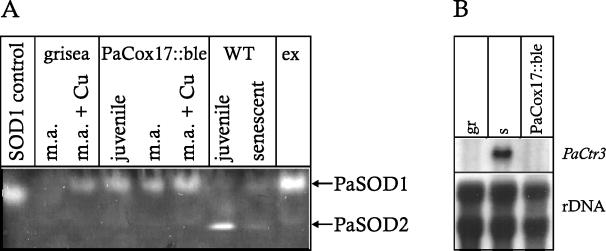
The aim of the present study was to construct a P. anserina strain in which copper depletion is restricted to the mitochondrial respiratory chain and not, as in mutant grisea, global in all cellular compartments. In the PaCox17::ble strain depletion of copper at the mitochondrial respiratory chain is suggested by the observed COX deficiency and the induction of the alternative respiratory pathway. Cytoplasmic copper levels were subsequently investigated indirectly via the determination of superoxide dismutase (SOD) activity. The activity of the copper depending SOD1 located in the cytoplasm and the mitochondrial intermembrane space depends on the availability of copper as a cofactor. Previously, we demonstrated that, due to cellular copper depletion, the copper uptake mutant grisea lacks a functional PaSOD1. This deficit can be overcome via the addition of extracellular copper (14). SOD activity analysis revealed that, in clear contrast to the wild-type strain s, PaSOD1 activities in PaCox17::ble are already high in juvenile cultures and remain at this level also in older age. In contrast to mutant grisea, in which the SOD1 activity can be induced by the addition of extracellular copper, such an induction is not or only very poorly observed in strain PaCox17::ble. Finally, in contrast to the juvenile wild-type strain, no PaSOD2 activity is detectable in juvenile cultures of PaCox17::ble strain (Fig. 7A). These data indicate that cytoplasmic copper levels are rather high in cultures of the new transgenic strain from juvenile to older age. This possibility was experimentally addressed by Northern blot analysis. As shown in Fig. 7B, transcripts of PaCtr3 encoding the high-affinity copper transporter of P. anserina are detected in RNA preparations of the wild-type strain s but not in mutant grisea and the transgenic strain PaCox17::ble. In mutant grisea the failure to transcribe PaCtr3 is the result of a null mutation in the gene encoding transcription factor GRISEA (10). In contrast, in transgenic strain PaCox17::ble increases in cytoplasmic copper levels seem to repress the transactivation activity of GRISEA and consequently the transcription of PaCtr3.
FIG. 7.
Activity of SODs and expression level of PaMt1. (A) Native proteins were isolated from PaCox17::ble strains that were juvenile (7 days) and middle aged (m.a.; 36 days), grown with or without 250 μM copper sulfate; wild-type strain s (juvenile [7 days] or senescent [36 days]); and mutant ex1 (ex) and mutant grisea (with or without 250 μM copper sulfate) as indicated. Subsequently, the proteins were separated by native polyacrylamide gel electrophoresis (see Materials and Methods). (B) Northern analysis of mutant grisea, wild-type strain s, and mutant PaCox17::ble-37312. PaCtr3 was used as probe; therefore, plasmid pAD4-PaCtr3 (12) was radioactively labeled. An rDNA probe (plasmid pMY60 [67] containing a HindIII fragment of the rDNA unit of S. carlsbergensis) was used as a loading control.
The mtDNA is stabilized in the PaCox17::ble strain.
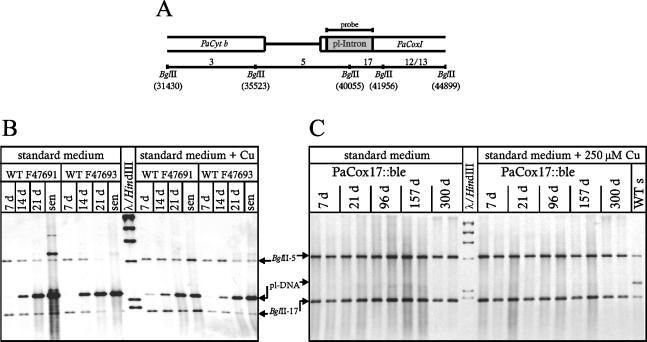
A hallmark of ageing of wild-type cultures of P. anserina is the extensive reorganization of the mtDNA. The most prominent age-related rearrangements occur in the region of the first PaCoxI intron, giving rise to the age-dependent amplification of the so-called plDNA (17, 45, 61, 62). This circular plasmid becomes amplified during ageing of the wild-type strain s on both standard and copper-supplemented growth media (Fig. 8B). In the long-lived copper-uptake mutant grisea grown on standard medium the mtDNA is stabilized. On medium supplemented with 250 μM CuSO4 copper grisea displays the wild-type specific instabilities (10). In sharp contrast, in the PaCox17::ble mutant the wild-type specific age-related mtDNA reorganizations are neither observed in cultures grown in standard growth medium nor in medium supplemented with 250 μM CuSO4. Under both conditions the mtDNA is stabilized.
FIG. 8.
Southern blot analysis of the mitochondrial PaCoxI region in two wild-type strains (B) and in two PaCox17::ble strains (C). (A) BglII restriction map of the PaCoxI and PaCyt b region of the mtDNA. The position of the hybridizing pl-intron and BglII fragments 17 and 5 are indicated. (B and C) Strains were grown on complex medium (left half of panels B and C) or with an additional 250 μM CuSO4 (right half of panels B and C). DNA samples were digested with BglII, separated on an 1% agarose gel, blotted, and hybridized to a plDNA specific probe. Signals corresponding to the mtDNA fragments 17 (1.9 kbp) and 5 (4.5 kbp) and the amplified plDNA are indicated by arrows. (B) DNA of two wild-type strains (7, 14, 21, and 28 days and senescent [ca. 36 days]) was used. (C) DNA of two independent PaCox17-null mutants was used (aged 7, 21, 96, 157, and 300 days).
PaCox17::ble displays a long-life phenotype.
Finally, we addressed the question whether the life span is affected in the newly generated PaCox17::ble strain. Initial measurements with four of the primary transformants indeed suggested a significant life span increase. These strains are now still growing after more than 1 year.
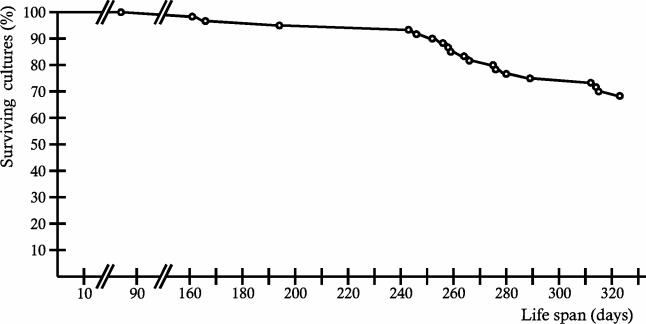
In order to raise statistical relevant life span data, we generated a larger number of independent cultures of mutant PaCox17::ble. These are derived from monokaryotic ascospores of a cross of the secondary transformant PaCox17::ble (strain 37312) mat+ with wild-type strain s (strain 690766) mat−. Sixty cultures (mat+ [n = 32]; mat− [n = 28]) with the mutant phenotype were selected, and life span measurements were performed in race tubes. Until now, after more than 300 days of cultivation on BMM, only 20 cultures expressed a senescent phenotype and stopped growing (Fig. 9). The remaining 40 cultures are still growing and do not display any symptoms of senescence.
FIG. 9.
Life span determination of transgenic strain PaCox17::ble. A total of 60 cultures derived from monokaryotic ascospores were grown on BMM agar in race tubes at 27°C in the light. After 320 days, 40 cultures were still alive, displaying no symptoms of aging.
DISCUSSION
In the filamentous ascomycete P. anserina a strong link between the type of respiration and life span has been demonstrated repeatedly. A switch from the standard COX-dependent respiration to an alternative respiration is accompanied by an increase in life span (14, 19, 37, 57). However, different COX-impaired mutants are characterized by extreme life span differences (e.g., 60% in the grisea mutant to immortality in the ex1 mutant), indicating that a variety of factors, in addition to the type of respiration, contribute to life span control. The systematic construction of specific transgenic strains and the comparative characterization of these strains in a more holistic sense is an important approach to define the relevant traits and their interactions.
Using this strategy, we focused on the relevance of cellular copper, which is essential for a COX-dependent respiration since copper is a cofactor of COX. The starting point for this analysis were previous data derived from the characterization of the long-lived grisea mutant (12, 13, 46). In comparison to other COX deficiency mutants, the grisea mutant is characterized by a rather moderate life span increase of ∼60%. In the present study, our specific interest was to define the molecular traits responsible for these differences and to investigate more thoroughly the impact of copper on ageing processes. Toward this goal, we cloned and characterized PaCox17 and demonstrated that it encodes an ortholog of the COX17 copper chaperone of yeast. COX17 recruits copper probably in the cytoplasm or directly from a copper importer by binding copper in a oligomeric polycopper complex. It is thought that COX17 delivers copper to SCO1, which is anchored in the inner mitochondrial membrane and contains a copper-binding site protruding in the mitochondrial intermembrane space. Subsequently, SCO1 passes copper to subunit II of COX located in the inner mitochondrial membrane (7, 29, 33, 43, 53, 58). Cloning of PaCox17 via complementation of a yeast Cox17-null mutant was possible because the amino acid sequence of the Podospora protein shares identical amino acids at positions that, in a recent mutagenesis analysis, were demonstrated to be important to restore wild-type respiration in a yeast Cox17-null mutant (53). In this study 11 mutations leading to a single amino acid exchange each at eight different positions were found to result in partial or complete failure to complement the Cox17-null mutant, demonstrating that these amino acids are of critical functional importance. Three amino acids were already known from an independent analysis to be of functional significance (32). Interestingly, in the P. anserina protein at all eight positions the same amino acid sequence was found as in the yeast protein, although only 28 of the 80 PaCOX17 amino acid residues were conserved in the protein of the two fungi.
In P. anserina, as in yeast, the deletion of Cox17 was found to be nonlethal. However, yeast and P. anserina strains survive for different reasons. Yeast Cox17 mutants are able to grow as “petite” colonies without the generation of ATP via respiration. In contrast, in the P. anserina Cox17-null mutant, as in the grisea mutant, an alternative respiratory pathway relying on the activity of the copper-independent AOX is induced. Whereas respiration and all other mutant characteristics of the grisea mutant can be reverted by growing this copper uptake mutant in medium containing increased amounts of copper, such a reversion is not possible in the PaCox17::ble strain. The two strains affected in copper metabolism are thus clearly distinguishable from each other.
The Cox17-null phenotype of yeast mutant strains can be reverted by growth in medium containing increased amounts of copper. Reversion is dependent on ScoI, a gene encoding a mitochondrial membrane protein accepting copper from COX17 and delivering it to COXII (28, 29). Although P. anserina also contains a nuclear gene coding for a putative SCO1 homolog (unpublished data), reversion of the PaCox17-null phenotype is not possible by simply growing the mutant in medium with increased amounts of copper. The reason for this is yet unknown. In contrast to the two fungal systems, in mice the deletion of Cox17 is lethal. During development of the homozygous Cox17 knockout strains died early during embryogenesis around the time of gastrulation (64). The lethality of Cox17 deletion in mice appears to be a consequence of the fact that mammals cannot meet their ATP demands by glycolysis and are not able to express an alternative respiratory chain. They rely on the generation of ATP via a COX-dependent respiration. These data demonstrate clearly that the molecular machinery involved in copper homeostasis, although conserved among species in many aspects, differs in details between species. Impairments of these machineries, depending on different characteristics of the individual system (e.g., ability to induce an alternative respiratory pathway or not), consequently lead to different phenotypes.
One of the most striking results obtained in the presented study is a clear correlation of the PaAOX levels in the different analyzed strains and the degree of life span increase. Importantly, in the PaCox17::ble strain, significant higher amounts of AOX are found than in the grisea mutant, reaching almost the level of the immortal ex1 mutant. However, it should be stressed at this point that the induction of the AOX in these strains is the result of impairments in the COX-dependent pathway and appears to be well regulated via yet-unknown mechanisms. The forced transgenic overexpression of the AOX in the wild-type background was found to have no impact on phenotypic characteristics. Even more, in the long-lived cox5::ble mutant background which, like the long-lived mutants investigated in the present study, respires via an alternative pathway, an increase of AOX levels above those normally found in this mutant leads to a reversion of the mutant phenotype (37). These data show that life span control depends on a delicate fine-tuning of the different types of respiration, as it naturally occurs in P. anserina. Modifying just one single component of this molecular network without adjusting other components may not lead to the desirable long-life phenotype.
From plants and P. anserina it is known that the respiration via the alternative pathway leads to a lower generation of reactive oxygen species (ROS) than in a COX-dependent respiration (19, 40). The increase in life span observed in the various COX deficiency mutants is in good agreement with the “mitochondrial free radical theory of ageing,” which postulates that ageing of biological systems is the result of a time-dependent accumulation of molecular damage, resulting from ROS mainly produced at the respiratory chain (reviewed in references 5, 23, 30, 31, and 41). In this context the level of ROS is of great significance. Lowering ROS levels can be achieved by interfering with the processes leading to their production (e.g., a switch from a COX-dependent pathway to an alternative pathway) or into the cellular ROS scavenging system. SODs are part of the latter system. In P. anserina we previously investigated two SODs, the Cu/Zn SOD (PaSOD1) and the MnSOD (PaSOD2) (12-14). In the wild-type strain s, PaSod1 was found to be constitutively expressed, but the activity of the encoded enzyme, which depends on the availability of copper, was found to increase during senescence. In accordance with other data, we concluded that during ageing the apoprotein is activated due to an increase of cytoplasmic copper level (12, 36, 50). In the long-lived copper uptake mutant grisea, no PaSOD1 activity was measured during the whole life time since copper is depleted in all cellular compartments. In addition, in this mutant the PaSod2 gene encoding the mitochondrial MnSOD is not transcribed due to the absence of the GRISEA transcription factor. In contrast, in the immortal mutant ex1 PaSOD2 activity and a very strong PaSOD1 activity were detected. In the present study, we report that the copper delivery mutant PaCox17::ble resembles the SOD activity profiles of the ex1 mutant (Fig. 7A). PaSOD1 activity was demonstrated to be rather high, although lower than in ex1, throughout the whole life span. These data and the results from the transcription analysis failing to detect transcripts of PaCtr3 clearly show that copper levels are increased in the cytoplasm of strain PaCox17::ble and that this mutant is well protected against cytoplasmic ROS and ROS generated in the mitochondrial intermembrane space.
Finally, a stabilization of the mtDNA was observed in the PaCox17::ble strain even after a very long period of growth. In contrast to mutant grisea, however, this holds true in strains grown on medium supplemented with copper salts. If in the transgenic strain the delivery of copper to the respiratory chain is specifically affected, this result clearly points to an indirect effect of copper on the mtDNA metabolism since in this case copper levels in the mitochondrial matrix should be normal. In contrast to the wild-type strain in which the mtDNA is efficiently rearranged, the mtDNA of mutant PaCox17::ble remains stable. This allows the expression of mtDNA encoded genes also in older cultures of the mutant and the remodeling of damaged mitochondrial proteins (e.g., proteins of the respiratory chain) and contributes to maintenance of functional mitochondria and to increased life span.
Collectively, the results of the present study demonstrate that very specific genetic modifications, as generated in the copper delivery mutant PaCox17::ble, may lead to multiple consequences. In the present study, the dual role of copper on life span control became obvious. On the one hand, copper indirectly leads to the age-related increase of mitochondrial oxidative stress and thus is active in limiting life span. On the other hand, as a cofactor of SOD1, it is part of a system lowering oxidative stress and as such life span increasing. This demonstrates that successful interference with the ageing process needs to be well designed. Transgenic strain PaCox17::ble is an example of an engineered strain with superior characteristics. However, still this strain has undesired attributes. Most importantly, due to the reduced ATP generation via the alternative pathway, PaCox17::ble is growing slower than the wild type and is affected in fertility. It will be interesting to see whether the observed impact on growth and life span can be dissected by specific manipulations.
Acknowledgments
We thank A. Tzagoloff (New York, N.Y.) for yeast strains, T. Elthon (Lincoln, Nebr.) for monoclonal AOX antibody, B. Ludwig (Frankfurt, Germany) for the βATPase V antibody (42), and S. M. Jazwinski (New Orleans, La.) for plasmid pAD4. The excellent technical assistance of A. Werner (Frankfurt) is gratefully acknowledged.
This study was supported by a grant of the Deutsche Forschungsgemeinschaft (Bonn, Germany) to H.D.O.
Footnotes
Dedicated to K. Esser on the occasion of his 80th birthday.
REFERENCES
- 1.Adams, T. H., H. Deising, and W. E. Timberlake. 1990. brlA requires both zinc fingers to induce development. Mol. Cell. Biol. 10:1815-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballance, D. J. 1986. Sequences important for gene expression in filamentous fungi. Yeast 2:229-236. [DOI] [PubMed] [Google Scholar]
- 4.Ballance, D. J., and G. Turner. 1986. Gene cloning in Aspergillus nidulans: isolation of the isocitrate lyase gene (acuD). Mol. Gen. Genet. 202:271-275. [DOI] [PubMed] [Google Scholar]
- 5.Beckman, K. B., and B. N. Ames. 1998. The free radical theory of aging matures. Physiol. Rev. 78:547-581. [DOI] [PubMed] [Google Scholar]
- 6.Beers, J., D. M. Glerum, and A. Tzagoloff. 1997. Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J. Biol. Chem. 272:33191-33196. [DOI] [PubMed] [Google Scholar]
- 7.Beers, J., D. M. Glerum, and A. Tzagoloff. 2002. Purification and characterization of yeast Sco1p, a mitochondrial copper protein. J. Biol. Chem. 277:22185-22190. [DOI] [PubMed] [Google Scholar]
- 8.Belcour, L., A. Sainsard-Chanet, C. Jamet-Vierny, and M. Picard. 1999. Stability of the mitochondrial genome of Podospora anserina and its genetic control, p. 209-228. In P. Lestienne (ed.), Mitochondrial diseases. Springer, Berlin, Germany.
- 9.Borghouts, C., S. Kerschner, and H. D. Osiewacz. 2000. Copper-dependence of mitochondrial DNA rearrangements in Podospora anserina. Curr. Genet. 37:268-275. [DOI] [PubMed] [Google Scholar]
- 10.Borghouts, C., E. Kimpel, and H. D. Osiewacz. 1997. Mitochondrial DNA rearrangements of Podospora anserina are under the control of the nuclear gene grisea. Proc. Natl. Acad. Sci. USA 94:10768-10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borghouts, C., and H. D. Osiewacz. 1998. GRISEA, a copper-modulated transcription factor from Podospora anserina involved in senescence and morphogenesis, is an ortholog of MAC1 in Saccharomyces cerevisiae. Mol. Gen. Genet. 260:492-502. [DOI] [PubMed] [Google Scholar]
- 12.Borghouts, C., C. Q. Scheckhuber, O. Stephan, and H. D. Osiewacz. 2002. Copper homeostasis and aging in the fungal model system Podospora anserina: differential expression of PaCtr3 encoding a copper transporter. Int. J. Biochem. Cell. Biol. 34:1355-1371. [DOI] [PubMed] [Google Scholar]
- 13.Borghouts, C., C. Q. Scheckhuber, A. Werner, and H. D. Osiewacz. 2002. Respiration, copper availability, and SOD activity in Podospora anserina strains with different lifespan. Biogerontology 3:143-153. [DOI] [PubMed] [Google Scholar]
- 14.Borghouts, C., A. Werner, T. Elthon, and H. D. Osiewacz. 2001. Copper-modulated gene expression and senescence in the filamentous fungus Podospora anserina. Mol. Cell. Biol. 21:390-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calmels, T., M. Parriche, H. Durand, and G. Tiraby. 1991. High efficiency transformation of Tolypocladium geodes conidiospores to phleomycin resistance. Curr. Genet. 20:309-314. [DOI] [PubMed] [Google Scholar]
- 16.Chang, Y. C., and W. E. Timberlake. 1993. Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics 133:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cummings, D. J., L. Belcour, and C. Grandchamp. 1979. Mitochondrial DNA from Podospora anserina. II. Properties of mutant DNA and multimeric circular DNA from senescent cultures. Mol. Gen. Genet. 171:239-250. [DOI] [PubMed] [Google Scholar]
- 18.Dhawale, S. S., and A. C. Lane. 1993. Compilation of sequence-specific DNA-binding proteins implicated in transcriptional control in fungi. Nucleic Acids Res. 21:5537-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dufour, E., J. Boulay, V. Rincheval, and A. Sainsard-Chanet. 2000. A causal link between respiration and senescence in Podospora anserina. Proc. Natl. Acad. Sci. USA 97:4138-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elthon, T. E., R. L. Nickels, and L. McIntosh. 1989. Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol. 89:1311-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esser, K. 1974. Podospora anserina, p. 531-551. In R. C. King (ed.), Handbook of genetics, vol. I. Plenum Press, Inc., New York, N.Y.
- 22.Esser, K., and P. Tudzynski. 1980. Senescence in fungi, p. 67-83. In K. V. Thimann (ed.), Senescence in plants. CRC Press, Boca Raton, Fla.
- 23.Finkel, T., and N. J. Holbrook. 2000. Oxidants, oxidative stress and the biology of ageing. Nature 408:239-247. [DOI] [PubMed] [Google Scholar]
- 24.Flohe, L., and F. Ötting. 1984. Superoxide dismutase assays. Methods Enzymol. 105:93-104. [DOI] [PubMed] [Google Scholar]
- 25.Frese, D., and U. Stahl. 1992. Oxidative stress and ageing in the fungus Podospora anserina. Mech. Ageing Dev. 65:277-288. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh, D. 1999. Object-oriented transcription factors database (ooTFD). Nucleic Acids Res. 27:315-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh, D. 2000. Object-oriented transcription factors database (ooTFD). Nucleic Acids Res. 28:308-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glerum, D. M., A. Shtanko, and A. Tzagoloff. 1996. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 271:14504-14509. [DOI] [PubMed] [Google Scholar]
- 29.Glerum, D. M., A. Shtanko, and A. Tzagoloff. 1996. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 271:20531-20535. [DOI] [PubMed] [Google Scholar]
- 30.Harman, D. 1998. Aging and oxidative stress. J. Int. Fed. Clin. Chem. 10:24-27. [PubMed] [Google Scholar]
- 31.Harman, D. 2001. Aging: overview. Ann. N. Y. Acad. Sci. 928:1-21. [DOI] [PubMed] [Google Scholar]
- 32.Heaton, D., T. Nittis, C. Srinivasan, and D. R. Winge. 2000. Mutational analysis of the mitochondrial copper metallochaperone Cox17. J. Biol. Chem. 275:37582-37587. [DOI] [PubMed] [Google Scholar]
- 33.Heaton, D. N., G. N. George, G. Garrison, and D. R. Winge. 2001. The mitochondrial copper metallochaperone Cox17 exists as an oligomeric, polycopper complex. Biochemistry 40:743-751. [DOI] [PubMed] [Google Scholar]
- 34.Kück, U., B. Kappelhoff, and K. Esser. 1985. Despite mtDNA polymorphism the mobile intron (plDNA) of the COI gene is present in ten different races of Podospora anserina. Curr. Genet. 10:59-67. [Google Scholar]
- 35.Lecellier, G., and P. Silar. 1994. Rapid methods for nucleic acids extraction from petri dish-grown mycelia. Curr. Genet. 25:122-123. [DOI] [PubMed] [Google Scholar]
- 36.Longo, V. D., E. Butler Gralla, and J. Selverstone Valentine. 1996. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. J. Biol. Chem. 271:12275-12280. [DOI] [PubMed] [Google Scholar]
- 37.Lorin, S., E. Dufour, J. Boulay, O. Begel, S. Marsy, and A. Sainsard-Chanet. 2001. Overexpression of the alternative oxidase restores senescence and fertility in a long-lived respiration-deficient mutant of Podospora anserina. Mol. Microbiol. 42:1259-1267. [DOI] [PubMed] [Google Scholar]
- 38.Lundin, M., J. O. Nehlin, and H. Ronne. 1994. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol. Cell. Biol. 14:1979-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marbach, K., J. Fernandez-Larrea, and U. Stahl. 1994. Reversion of a long-living, undifferentiated mutant of Podospora anserina by copper. Curr. Genet. 26:184-186. [DOI] [PubMed] [Google Scholar]
- 40.Maxwell, D. P., Y. Wang, and L. McIntosh. 1999. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 96:8271-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miquel, J., E. de Juan, and I. Sevila. 1992. Oxygen-induced mitochondrial damage and aging. EXS 62:47-57. [DOI] [PubMed] [Google Scholar]
- 42.Nelson, N., and G. Schatz. 1979. Energy-dependent processing of cytoplasmically made precursors to mitochondrial proteins. Proc. Natl. Acad. Sci. USA 76:4365-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nittis, T., G. N. George, and D. R. Winge. 2001. Yeast Sco1, a protein essential for cytochrome c oxidase function is a Cu(I)-binding protein. J. Biol. Chem. 276:42520-42526. [DOI] [PubMed] [Google Scholar]
- 44.Osiewacz, H. D. 2002. Genes, mitochondria, and aging in filamentous fungi. Ageing Res. Rev. 28:1-18. [DOI] [PubMed] [Google Scholar]
- 45.Osiewacz, H. D., and K. Esser. 1984. The mitochondrial plasmid of Podospora anserina: a mobile intron of a mitochondrial gene. Curr. Genet. 8:299-305. [DOI] [PubMed] [Google Scholar]
- 46.Osiewacz, H. D., and U. Nuber. 1996. GRISEA, a putative copper-activated transcription factor from Podospora anserina involved in differentiation and senescence. Mol. Gen. Genet. 252:115-124. [DOI] [PubMed] [Google Scholar]
- 47.Osiewacz, H. D., A. Skaletz, and K. Esser. 1991. Integrative transformation of the ascomycete Podospora anserina: identification of the mating-type locus on chromosome VII of electrophoretically separated chromosomes. Appl. Microbiol. Biotechnol. 35:38-45. [DOI] [PubMed] [Google Scholar]
- 48.Osiewacz, H. D., and S. W. Stumpferl. 2001. Metabolism and aging in the filamentous fungus Podospora anserina. Arch. Gerontol. Geriatr. 32:185-197. [DOI] [PubMed] [Google Scholar]
- 49.Prillinger, H., and K. Esser. 1977. The phenoloxidases of the ascomycete Podospora anserina. XIII. Action and interaction of genes controlling the formation of laccase. Mol. Gen. Genet. 156:333-345. [DOI] [PubMed] [Google Scholar]
- 50.Prinz, R., A. Schallies, and U. Weser. 1972. Superoxide dismutase from Saccharomyces cerevisiae. Hoppe-Seylers Z. Physiol. Chem. 353:1559-1560. [PubMed] [Google Scholar]
- 51.Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels, and C. A. van den Hondel. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56:117-124. [DOI] [PubMed] [Google Scholar]
- 52.Punter, F. A., D. L. Adams, and D. M. Glerum. 2000. Characterization and localization of human COX17, a gene involved in mitochondrial copper transport. Hum. Genet. 107:69-74. [DOI] [PubMed] [Google Scholar]
- 53.Punter, F. A., and D. M. Glerum. 2003. Mutagenesis reveals a specific role for Cox17p in copper transport to cytochrome oxidase. J. Biol. Chem. 278:30875-30880. [DOI] [PubMed]
- 54.Rizet, G. 1953. Sur la longevite des phenomen des souches de Podospora anserina. C. R. Acad. Sci. Paris 237:1106-1109. [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 57.Schulte, E., U. Kück, and K. Esser. 1988. Extrachromosomal mutants from Podospora anserina: permanent vegetative growth in spite of multiple recombination events in the mitochondrial genome. Mol. Gen. Genet. 211:342-349. [Google Scholar]
- 58.Schulze, M., and G. Rödel. 1989. Accumulation of the cytochrome c oxidase subunits I and II in yeast requires a mitochondrial membrane-associated protein, encoded by the nuclear SCO1 gene. Mol. Gen. Genet. 216:37-43. [DOI] [PubMed] [Google Scholar]
- 59.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 60.Srinivasan, C., M. C. Posewitz, G. N. George, and D. R. Winge. 1998. Characterization of the copper chaperone Cox17 of Saccharomyces cerevisiae. Biochemistry 37:7572-7577. [DOI] [PubMed] [Google Scholar]
- 61.Stahl, U., U. Kück, P. Tudzynski, and K. Esser. 1980. Characterization and cloning of plasmid-like DNA of the ascomycete Podospora anserina. Mol. Gen. Genet. 178:639-646. [DOI] [PubMed] [Google Scholar]
- 62.Stahl, U., P. A. Lemke, P. Tudzynski, U. Kück, and K. Esser. 1978. Evidence for plasmid-like DNA in a filamentous fungus, the ascomycete Podospora anserina. Mol. Gen. Genet. 162:341-343. [DOI] [PubMed] [Google Scholar]
- 63.Stahl, U., P. Tudzynski, U. Kück, and K. Esser. 1982. Replication and expression of a bacterial-mitochondrial hybrid plasmid in the fungus Podospora anserina. Proc. Natl. Acad. Sci. USA 79:3641-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takahashi, Y., K. Kako, S. Kashiwabara, A. Takehara, Y. Inada, H. Arai, K. Nakada, H. Kodama, J. Hayashi, T. Baba, and E. Munekata. 2002. Mammalian copper chaperone Cox17p has an essential role in activation of cytochrome c oxidase and embryonic development. Mol. Cell. Biol. 22:7614-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takahashi, Y., K. Kako, K. Ohmura, K. Tsumori, Y. Ohmasa, S. Kashiwabara, T. Baba, and E. Munekatat. 2001. Genomic structure of mouse copper chaperone, COX17. DNA Seq. 12:305-318. [DOI] [PubMed] [Google Scholar]
- 66.van Heeswijck, R., and M. J. Hynes. 1991. The amdR product and a CCAAT-binding factor bind to adjacent, possibly overlapping DNA sequences in the promoter region of the Aspergillus nidulans amdS gene. Nucleic Acids Res. 19:2655-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verbeet, M. P., J. Klootwijk, H. van Heerikhuizen, R. Fontijn, E. Vreugdenhil, and R. J. Planta. 1983. Molecular cloning of the rDNA of Saccharomyces rosei and comparison of its transcription initiation region with that of Saccharomyces carlsbergensis. Gene 23:53-63. [DOI] [PubMed] [Google Scholar]
- 68.Virbasius, J. V., and R. C. Scarpulla. 1991. Transcriptional activation through ETS domain binding sites in the cytochrome c oxidase subunit IV gene. Mol. Cell. Biol. 11:5631-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wintz, H., and C. Vulpe. 2002. Plant copper chaperones. Biochem. Soc. Trans. 30:732-735. [DOI] [PubMed] [Google Scholar]