Abstract
The long-term goal of our study is to identify chronic obstructive pulmonary disease (COPD)-related bronchoalveolar lavage fluid (BALF) nitroproteins to clarify COPD pathological mechanisms and to discover biomarkers of COPD. The goal of the present study was to detect the presence of, and potential roles of, nitroproteins in, human ex-smoker (without COPD) BALF samples. Nitroproteins were immunoprecipitated from two separate BALF samples, and digested with trypsin; and tryptic peptides were analyzed with matrix-assisted laser desorption/ionization (MALDI)-tandem mass spectrometry (MS/MS). Each MS/MS spectrum was composed of accumulated scans (n = 50–100). The MS/MS data were searched with BioWorks 2.0 TuboSequest in the SwissProt database to generate the amino acid sequence, which was evaluated manually. Eleven nitrotyrosine sites were identified in eight proteins, including progestin and adipoQ receptor family member III, zinc finger protein 432, proteasome subunit alpha type 2, NADH-ubiquinone oxidoreductase B14, slit homolog 1 protein, lysozyme, aldose 1-epimerase, and PTS system lactose-specific EIICB component. Each nitrotyrosine site was located within a specific protein domain and motif. Those identified nitrated proteins could be involved in multiple functional metabolic systems, including transcriptional regulation, mitochondrial complex, immune system, and energy metabolism.
Keywords: Protein tyrosine nitration, Nitroproteomics, Chronic obstructive pulmonary disease, Bioinformatics, Bronchoalveolar lavage fluid, Tandem mass spectrometry
Cigarette smoke, a major cause of inflammatory-related lung diseases such as chronic obstructive pulmonary disease (COPD), contains a variety of oxidants and nitrosants such as nitric oxide (NO) [1] that could initiate the inflammatory response to airway and lung insult. Alveolar macrophages, neutrophils, CD8+ lymphocytes, other inflammatory cells, and airway epithelial cells generate and release reactive oxygen species (ROS) such as the superoxide anion (O2.−), hydrogen peroxide (H2O2), and the hydroxyl radical (OH·) through an NADPH-oxidase-dependent mechanism; and the nitric oxide (NO·) free radical through a nitric oxide synthase (NOS)-dependent mechanism. Inducible (iNOS) can generate large amounts of NO for an extended period of time during an inflammatory response compared to neuronal (nNOS) and endothelial NOS (eNOS). Most of the cellular toxic effects of NO result from its rapid reaction with O2.− to form the strong nitrosant peroxynitrite anion (ONOO−)[2, 3]. Peroxynitrite causes the nitration (addition of –NO2) of a protein tyrosine residue to provoke inhibition of mitochondria respiration, protein dysfunction, and damage to cell membranes [4, 5] to involve the pathophysiological processes of inflammatory-related lung diseases. Tyrosine nitration is also possibly generated through heme peroxidase (such as myeloperoxidase, MPO; and xanthine oxydase, XO)-dependent mechanisms [4].
Studies have shown that oxidants and nitrosants such as peroxynitrite in the gas phase of cigarette smoke can pass through the pulmonary alveolar wall into blood to induce systemic oxidative stress and to increase the serum level of 3-nitrotyrosine [6]. The level of 3-nitrotyrosine in a smoker is significantly higher than in a non-smoker [7]. Tobacco smoke impairs the normal process of NO generation to alter the function of peripheral muscle [8]. Tobacco nicotine significantly increased superoxide anions and 3-nitrotyrosine-positive proteins, and exogenous peroxynitrite mimicked the effects of nicotine on the AMP-activated protein kinase (AMPK) that acts as a major energy sensor and regulator in adipose tissues [9], and the tyrosine nitration of sarcoplasmic-endoplasmic reticulum Ca2+ adenosine triphosphatase 2 (SERCA2) in COPD patients could contribute to their low body mass index (BMI) [10]. Chronic smoking causes the decreased exhaled NO and an increased level of nitrotyrosine [11]. Cigarette smoke-mediated oxidative stress induced an inflammatory response in the lungs by stimulating the release of proinflammatory cytokines such as interleukin 8 (IL-8) via the activation of NF-kB [12]. Smoking can cause nitration of a protein tyrosine residue [13]. Antioxidants such as superoxide dismutase can protect smoke-induced inflammation and conteract the proteolytic cascade that leads to emphysema formation [14].
Protein nitration adds 45 mass units (NO2 – H) to a tyrosine residue [15]. Tandem mass spectrometry (MS/MS) enables one to monitor that mass shift, obtain an amino acid sequence of a nitropeptide, and identify the low-abundance nitrotyrosine sites (∼ 1 in 106 tyrosines [16, 17]) after a nitrotyrosine antibody-based immunoaffinity preferential enrichment of endogenous nitrotyrosine-containing proteins [18, 19].
The goal of the present study was to detect the presence of, and the potential roles of, nitroproteins in human ex-smoker (without COPD) BALF samples. Nitroproteins in BALF were preferentially selected with immunoprecipitation (IP). Nitrotyrosine sites were identified with tandem mass spectrometry. Each nitrotyrosine site was located within a specific protein domain and motif. Those nitroproteins could be involved in multiple functional metabolic systems, including transcriptional regulation, mitochondrial complex, immune system, and energy metabolism; which provides new insights into the mechanisms of oxidative/nitrative stress in cigarette smoke-related lung diseases and new clues to biomarker discovery.
MATERIALS AND METHODS
Human BALF collection
The protocol of collecting human BALF was approved by the Institutional Review Board at Chiesi Pharmaceutical Company (Parma, Italy). Written informed consent was obtained from each subject to enroll in the study. Briefly, BAL was performed by professional clinicians. Five separate 30 ml aliquots of 0.9% sterile saline were instilled into the right middle lobe or lingula. BALF samples were collected, aliquoted into polypropylene tubes, and stored at −80 °C prior to further analyses. A 5-ml aliquot of BALF on dry-ice was shipped by air to the University of Tennessee Health Science Center in Memphis for nitroproteomics analysis.
BALF protein preparation
Two separate 5-ml BALF samples (male, ex-smoker without COPD, 56 and 66 year-old) were vacuum-dried (0°C), and the residues were redissolved in 180μl of Pierce M-PER protein extraction buffer; the solutions were centrifuged (15,000 x g; 30 min). The supernatants of the two samples were combined (∼ 350 μl) for nitrotyrosine immunoaffinity analysis.
Nitroprotein immunoprecipitation
Nitrotyrosine immunoprecipitations were performed as described previously [19]. Briefly, rabbit anti-nitrotyrosine polyclonal antibody (92 μg, Chemicon International, Temecula, CA, USA) was coupled with immunopure immobilized protein G beads (400 μl, Pierce) for 2h with gentle shaking in binding buffer (0.14M NaCl, 0.008M sodium phosphate, 0.002M potassium phosphate, and 0.01M KCl; pH 7.4), and washed to remove any unbound antibody (Pierce Immunoprecipitation Kit, product 45225). The bound antibody was covalently crosslinked to protein G with gentle shaking (40 min) in disuccinimidyl suberate (final concentration 0.0025%); the beads were washed briefly with a low pH buffer (pH 2.8) to remove non-crosslinked antibody, and re-equilibrated into the above binding buffer. The immobilized anti-nitrotyrosine antibody (∼92 μg) was incubated (4 °C, overnight) with the prepared BALF protein samples to capture the nitrotyrosine-containing proteins; the column was washed (3x) with the binding buffer (400 μl), and nitroproteins were eluted with Pierce Elution buffer into a volume of ∼250 μL.
Tryptic peptide preparation
An aliquot (20 μL) of IP products was treated with 1 μL of 1 M Tris (pH 9.5). Sequencing grade-modified trypsin (20 μg, Promega, Madison, WI, USA) was dissolved in a volume (100 μl, pH 8.2) of solution that consisted of 5 μl trypsin resuspension buffer that contained 50 mM acetic acid (pH 2.8) and 95 μl of 200 mM NH4HCO3 (pH 8.2). The enzyme digestion reaction system (100 μl, final concentration of NH4HCO3 = 50 mM, pH8.1), which consisted of 21 μl neutralized IP products, 25 μl trypsin solution, and 54 μl ddH2O, was incubated (overnight; 37 °C). An aliquot (20 μL) of the tryptic peptide mixture was purified with a Millipore ZipTip C18 column according to the manufacturer’s instructions (Millipore, Billerica, MA, USA). The purified tryptic peptides were eluted directly from the microcolumn onto a vMALDI plate with 2 μL of an α-cyano-4-hydroxycinnamic acid (CHCA) solution (2.5 mg/ml CHCA in 50 % v/v acetonitrile/0.1 % v/v trifluoroacetic acid); that solution was dried in ambient air for mass spectrometry (MS) analysis.
Protein identification with MALDI-tandem mass spectrometry
The tryptic peptides were analyzed with a vMALDI-LTQ tandem mass spectrometer in the “Nth-order double play” data-dependent experimental mode to obtain the amino acid sequence of each peptide [19]. Briefly, the crystal-positioning system (CPS) and auto spectrum filter (ASF; threshold = 500 counts for an MS scan and 250 counts for an MS2 scan) were enabled for the vMALDI source. The automatic gain control (AGC) was enabled to automatically adjust the number of laser shots to maintain the quality of the spectra. For an MS scan, high-mass range (m/z 600–4000), normal scan rate, full scan, polarity, profile data type, and five microscans of each experiment were used. For an MS2 scan, the 50 most-intense peaks in the full MS spectrum, high-mass range (m/z 50–4000), normal scan rate, polarity, centriod data type, isolation width 3.0 Th, normalized collision energy 40 (arbitrary units), default charge state 1, minimal signal threshold 100 counts, activation Q value 0.25, activation time 30 ms, and five microscans of each experiment were used. Instrument operation and data acquisition utilized the Xcalibur software package (ThermoFinnigan). Initial protein identifications from MS/MS data utilized the Bioworks TurboSequest software search engine (ThermoFinnigan, version 3.2) and the Swiss-Prot protein database. The Swiss-Prot database search parameters included 2 allowed missed tryptic cleavage sites, precursor-ion mass tolerance = 2 Da, fragmention mass tolerance = 1.0 Da, and protein modifications for Tyr nitration (+ 45 mass unit), Asn and Gln deamidation (+ 1 mass unit), and Met oxidation (+ 16 mass unit). Each nitrotyrosine-containing peptide was examined manually, as described previously [19]. The residue that preceded the N-terminus or C-terminus must be K or R with singly charged b-, y-, and a-ion series for a nitrotyrosine-positive search result. An accumulated MS2 spectrum (n = 50–100 scans) of each nitrotyrosine-positive search result was acquired on the vMALDI-LTQ Tune page to improve the signal-to-noise (S/N) ratio, and was used to search the Swiss-Prot database with Bioworks 3.2 to corroborate each positive search result.
Furthermore, most of the prominent peaks within the MS spectra were selected to directly acquire the accumulated (n = 50) MS2 spectrum whether it was a nitropeptide or not. The MS/MS data were used to identify the protein and nitrotyrosine-site as described above. NCBI BLASTP (version 2.2.17; http://ca.expasy.org/cgi-bin/blast.pl) was used to determine the identity of each nitropetide that was aligned to a protein in the UniProtKB Database (6,489K sequences that consist of Swiss-Prot and TrEMBL databases; July 1, 2008).
Protein domain and pathway analyses
Protein domain and motif analyses were carried out with ScanProsite (http://us.expasy.org/tools/scanprosite/), and Motifscan (http://myhits.isb-sib.ch/cgi-bin/motif_scan). Each nitrotyrosine-site was located within the corresponding protein domain and motif. The pathway networks that could involve the nitroproteins were analyzed with Ingenuity Pathways Analysis software (Ingenuity® Systems, www.ingenuity.com).
RESULTS
Nitroproteins identified in BALF
The tryptic peptides were subjected to vMALDI MS/MS analysis, after nitroprotein immunoaffinity enrichment and digestion with trypsin. MS/MS data provided the amino acid sequences of nine nitrotyrosine-containing peptides from human ex-smoker BALF (Table 1; Supplemental Figures A–H). Each peptide amino acid sequence and nitrotyrosine-site was confirmed with a manual analysis of the MS/MS spectrum, including spectral quality and the interpretation of the b- and y-ion series. Furthermore, each nitropeptide was also confirmed with accumulated MS/MS spectra (n = 50–100). The amino acid sequence of each nitropeptide was exactly matched to a protein with Bioworks TuboSequest analysis in the Swiss-Prot protein database: progestin and adipoQ receptor family member III, zinc finger protein 432, proteasome subunit alpha type 2, NADH-ubiquinone oxidoreductase B14, slit homolog 1 protein, lysozyme, aldose 1-epimerase, and PTS system lactose-specific EIICB component. Table 1 contains those nitroproteins: protein name, species, Swiss-Prot database accession number, nitropeptide amino acid sequence, nitrotyrosine site, singly-charged precursor ion ([M + H]+), and TuboSequest Score.
Table 1.
Nitroproteins Identified in Human Ex-smoker BALF
| Protein Name (Species) | Swiss-Prot No. | Nitrotyrosine-containing peptide | Nitrotyrosine site | [M +H]+ | Sequest Score | E-value (NCBI 6489K) | Protein domain and motif containing nitrotyrosine | Protein Function |
|---|---|---|---|---|---|---|---|---|
| Progestin and adipoQ receptor family member III (Human) | Q6TCH7 | R.LYTY*EQIPGSLKD NPYITDGYRAYLPSR. L |
Y33 | 3338.6 | 3.53 | 3.0E-19 | Cytoplasmic | Multi-pass membrane protein |
| Zinc finger protein 432 (Human) | O94892 | K.DLYRDVMLEIY*SN LLSMGYQVSKPDALS K.L |
Y41 | 3393.7 | 3.04 | 7.0E-20 | KRAB domain | Transcriptional regulation |
| Proteasome subunit alpha type 2 (Human) | P25787 | K.DY*LAAIA.- | Y229 | 781.4 | 1.61 | 4.6E+00 | Functional chain | Multicatalytic proteinase complex with an ATP-dependent proteolytic activity |
| NADH-ubiquinone oxidoreductase B14 subunit (Human) | P56556 | R.ELY*RAWY*REVP NTVHQFQ#LDITVK.M |
Y36, Y40 | 3096.6 | 2.91 | 5.0E-16 | Functional chain, component of mitochondrial Complex I | Accesory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase |
| Slit homolog 1 protein (Rat) | O88279 | K.HDCVN#GGVCVDG IGNYTCQCPLQY*TG R.A |
Y1035 | 2918.2 | 2.74 | 1.0E-18 | EGF-like domain 3 | Secreted; Act as molecular guidance cue in cellular migration. |
| Lysozyme (ANOGA) | Q17005 | K.NKNGSTDYGIFQIN NKY*WCDSGYGSN#D CK.I |
Y79 | 3337.4 | 2.08 | 9.0E-21 | Tyrosine Kinase phosphorylation motif | Have primarily a bacteriolytic function; associated with enhance the activity of immunoagents. |
| K.NKNGSTDYGIFQIN N#KYWCDSGY*GSND CK.I |
Y85 | 3337.4 | 2.08 | 9.0E-21 | ||||
| Aldose 1-epimerase (ACICA) | P05149 | K.FSLDGKTY*NLEKN NGPN#SLHSGN#PGFD K.R |
Y118 | 3097.5 | 3.77 | 6.0E-18 | Tyrosine kinase phosphorylation motif | Periplasma, involved glucose metablism |
| PTS system lactose-specific EIICB component (STAAU) | P11162 | K.EY*QLIILAPQVAS NY*EDIKQ#DTDR.L |
Y515, Y528 | 2913.4 | 3.26 | 5.0E-15 | Casein kinase II phosphorylation motif | Multi-pass membrane protein; involved in lactose transport |
Y* = nitrotyrosine.
N# = deamidated Asn.
Q# = Deamidated Gln
Protein domain versus nitrotyrosine site
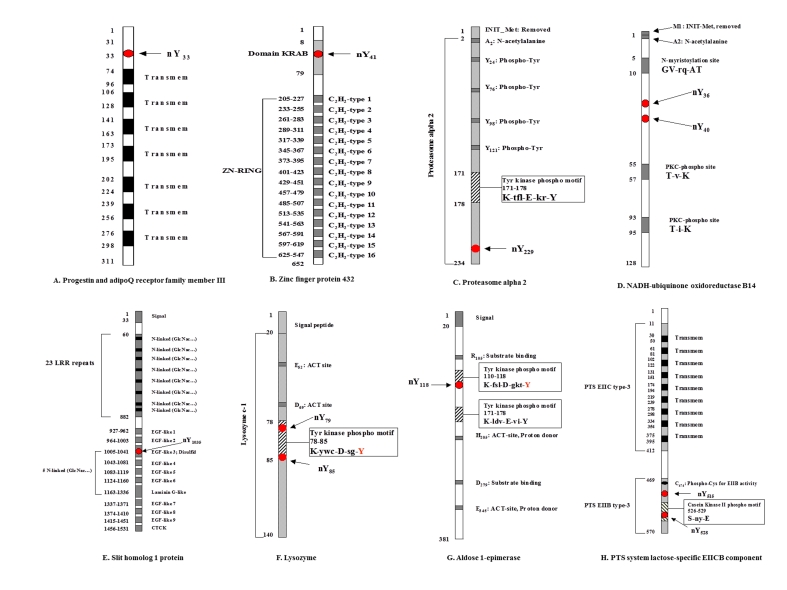
Protein-domain analysis demonstrated that each nitrotyrosine site was located within a specific protein domain and motif (Figure 1 and Table 1). Two of the identified nitrotyrosine sites also appear to be the targets of tyrosine phosphorylation, including lysozyme (nY85 was located at the tyrosine kinase phosphorylation motif K-ywc-D-sg-Y85; Figure 1F) and aldose 1-epimerase (nY118 was located at the tyrosine kinase phosphorylation motif K-fsl-D-gkt-Y118; Figure 1G). Nitrotyrosine (nY79) was also detected in the tyrosine kinase phosphorylation motif (K-y79wc-D-sg-Y) of lysozyme (Figure 1F). Nitrotyrosines (nY515 and nY528) appear in the PTS EIIB type-3 domain of PTS system lactose-specific EIICB component (Figure 1H), and nY528 occurred in the casein kinase II phosphorylation motif (S-ny-E). The other identified nitrotyrosine sites were detected in the cytoplasmic region (nY33) of multi-pass membrane protein progestin and adipoQ receptor family member III (Figure 1A), the KRAB domain (nY41) of zinc finger protein 432 (Figure 1B), the functional chain (nY229) of proteasome alpha 2 (Figure 1C), the mitochondrial complex I functional chain (nY36 and nY40) of NADH-ubiquinone oxidoreductase B14 (Figure 1D), and the EGF-like domain 3 (nY1035) of slit homolog 1 protein (Figure 1E). The relationship analyses of each nitrotyrosine site and protein domain suggest that each nitrotyrosine site resides within an important domain and motif of each protein; that correlation suggests the physiological significance of tyrosine nitration in each individual protein functional alteration.
Figure 1.
Nitrotyrosine was located within specific protein domains and motifs. Two nitrotyrosine sites (nY79 and nY85) were present in two individual peptides from lysozyme, respectively.
Metabolic networks that involve nitroproteins
Metabolic network systems that involve nitroproteins could reveal the significance of tyrosine nitration in human ex-smoker lung physiology. Pathway analysis suggested that those nitroproteins could be involved in multiple functional metabolic systems, including transcriptional regulation, mitochondrial complex, immune system, energy metabolism, protein metabolism, etc. (Table 1). Proteasome subunit alpha type 2 (PSMA2), which is nitrated, and is an important component of the proteasome complex, was identified (Figure 2A); the proteasome complex is involved in protein synthesis, cancer formation, and lung fibrosis disease; and the ubiquitin-proteasome pathway is involved in the mechanisms of reduced contractile protein content in the diaphragm of patients with mild to moderate COPD [20]. NADH-ubiquinone oxidoreductase B14, which is nitrated, is involved in the mitochondrial membrane respiratory chain NADH dehydrogenase system (Figure 2B), and is a component of the mitochondrial complex I that is involved in the apoptosis signaling pathways in lung diseases [21]; recent studies found that a cigarette smoke-induced blockade of the mitochondrial respiratory chain was involved in the mechanisms of increased lung cell apoptosis and necrosis in COPD patients [22]. Therefore, the nitration of that component of the mitochondrial respiratory chain could involve this process.
Figure 2.
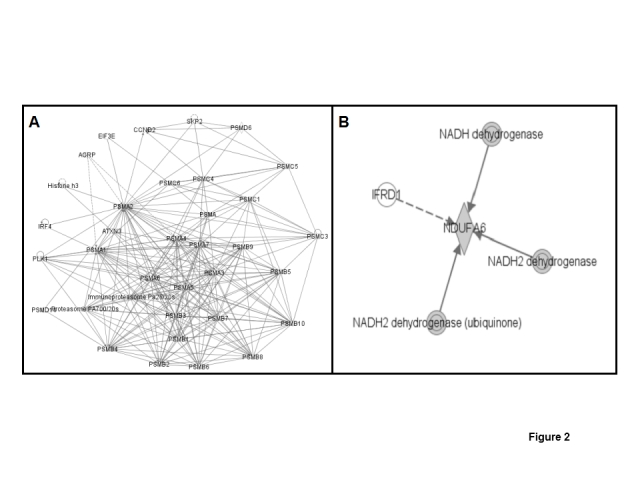
Multiple metabolic networks that were involved in nitroproteins. A. Nitrated proteasome alpha 2-involved network. B. Nitrated NADH-ubiquinone oxidoreductase B14-involved network.
DISCUSSION
Oxidative/nitrative stress plays important roles in cigarette smoke-related inflammatory airway diseases, including COPD [3], such as an amplification of inflammation via the activation of transcription factors such as NF-kB and AP-1, reduction of antiproteases to accelerate the breakdown of elastin, increase of apoptosis and death in endothelial and epithelial cells, and even increased resistance of anti-inflammation processes to corticosteroids. Nitrotyrosine, as a biomarker of oxidative/nitrative stress, significantly increases in smoke-related inflammatory lung diseases [5]. The goal of this present study was to identify the targets of nitrotyrosine in human ex-smoker BALF to further clarify the mechanism of cigarette smoke-related inflammatory airway disease and to provide an optimized experimental approach and control reference for a COPD nitroprotein study.
Nitrotyrosine immunoaffinity enrichment was used to preferentially isolate nitroproteins from human ex-smoker BALF, and to improve the sensitivity to detect nitrotyrosine sites. The accumulated (n = 50) MS2 spectrum of each tryptic peptide from the IP product was acquired with a vMALDI-LTQ to optimize the S/N and to improve the quality of each MS2 spectrum to accurately identify nitrotyrosine sites. To select for low-abundance nitrotyrosine-containing peptides, Bioworks TuboSequest database searches, in combination with a manual inspection and interpretation of each MS/MS spectrum, identified nine nitropeptides (Table 1), with support of reliable amino acid sequence information (Supplemental Figures A–H). BLAST homolog analysis provided the statistically significant identification of nitrotyrosine sites in seven human BLAF proteins, with an NCBI BLAST E-value range from 5E-15 to 9E-21 (Table 1), except for proteasome subunit alpha type 2 (E-value = 4.9), which, however, was supported by the reliable amino acid sequence (Supplemental Figure C).
Bioinformatics analyses of protein domains and metabolic networks clarify the biological significance of identified endogenous BALF nitroproteins. Each nitrotyrosine site was located in an important functional and structural domain or motif of each protein (Figure 1), and nitroproteins are involved in multiple metabolic networks (Figure 2).
The biological significance of each identified nitrotyrosine-containing protein was rationalized into pathophysiological processes of cigarette smoke-related inflammatory pulmonary disease such as COPD (Figure 3). Cigarette smoke contains a variety of oxidants and nitrosants [1] that activates the alveolar macrophage, neutrophil, CD8+ lymphocyte, epithelial cells, and other inflammatory cells to further produce endogenous oxidants such as O2.−, ·OH, H2O2, and nitrosants such as NO·. Peroxynitrite (ONOO−), which is derived from the rapid reaction between NO· and O2.−, is a toxic oxidant and nitrosant, and is a primary source to nitrate a protein tyrosine residue. Our results showed that this nitration often occurred at specific protein domains or important motifs that could alter the protein functions (Figure 1) and could affect the corresponding functional networks. The nitrated proteasome subunit alpha type 2 (PSMA2) is involved in the ATP-dependent ubiquitin-proteasome pathway, which is an important intracellular nonlysosomal proteolytic pathway [23, 24], and which is also experimentally found to be involved in the processes of reduced contractile protein content in the diaphragm of COPD patients [20]. The nitrated NADH-ubiquinone oxidoreductase B14 is a component of the mitochondrial complex I and of the mitochondrial membrane respiratory chain NADH dehydrogenase system, which is involved in the mechanisms of increased lung cell apoptosis and necrosis in COPD patients [22]. The nitrated aldose-1-epimerase (mutarotase) is involved in the interconversion of alpha- and beta-hexoses, which are essential for normal carbohydrate metabolism and the production of complex oligosaccharides, and could affect energy metabolism and synthesis of cell-surface glycoproteins and glycolipids [25]. The nitrated PTS system lactose-specific EIICB component catalyzes the intake and phosphorylation of lactose, which is involved in sugar and energy metabolism [26, 27]. Lysozyme is induced by bacterial infection, is expressed in salivary glands and tubules, and has primarily a bacteriolytic function [28]; in tissues and body fluids, lysozyme is associated with the moncyte-macrophage system and enhances the activity of immunoagents, and contributes to the antimicrobial defense of the alveolar lining layer [29] and respiratory system defense [30–32]. Tyrosine nitration (nY79 and nY85), which occurred within the tyrosine kinase phosphorylation motif (K-y79wc-D-sg-Y85) of lysozyme (Figure 1F), could interfere with its bacteriolytic function and inhibit the activity of immunoagents, which could promote bronchoalveolar infection and inflammation. Slit homolog 1 protein, which was nitrated, is secreted and functions as a ligand for the Roundabout (Robo) receptor-signaling pathway, which is implicated in neurogenesis, angiogenesis, and the immune response [33]. For zinc finger 432, which is involved in the transcription regulation of a broad range of genes, nitration (nY41) occurred within the kruppel-associated box (KRAB) domain (Figure 1B) that functions as a transcriptional suppressor [34, 35]; this nitration might impair transcriptional suppression of proinflammatory genes. Progestin and adipoQ receptor III (PAQR3) is a multiple-pass membrane protein [36], and interacts with ligand progesterone. Nitration (nY33) occurred within the extra-membrane region (Figure 1A), and could interfere with receptor-ligand binding and the signal transduction of progesterone. Hormone-replacement therapy, such as the use of estrogen and progesterone, has been used in chronic airway diseases with protean effects [37–39]. Therefore, the nitrated PAQR could interfere with effects of hormone replacement therapy (such as progesterone) in chronic airway inflammatory diseases.
Figure 3.
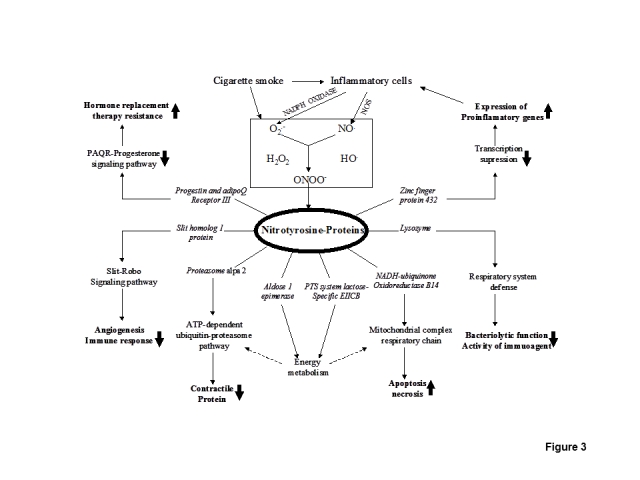
Biological significance of tyrosine nitration in cigarette smoke-related inflammatory pulmonary disease.
In summary, this present study is the first report to discover the presence of, and the potential biological significance of, nitroproteins in human ex-smoker BALF. Nitrotyrosine sites occurred within specific protein domains and motifs that could alter protein functions, and that are involved in multiple metabolic networks, including transcriptional regulation, mitochondrial complexes, immune system, and energy metabolism. This present study clarifies contribution of oxidative and nitrative stress to cigarette smoke-related airway inflammatory diseases. Further investigation is required to determine the biological consequences of these tyrosine nitration events and the relevance to the pathophysiological mechanisms of chronic airway inflammatory diseases.
Supplemental Figures
A representative MS/MS spectrum (Supplemental Figure A) and the data interpretation used to identify the nitrotyrosine - containing 28 amino acid peptide from progestin and adipoQ receptor family member III is described here in detail. A singly charged ion ([M + H]+, m/z = 3340.6) in the MS spectrum was selected to accumulate MS/MS spectra (n = 100) with a normalized collision energy = 35 arbitrary units, followed by a Bioworks TuboSequest analysis in Swiss-Prot Database. The singly charged b - and y - product ions (y9, y11, y12, y14, y15, y16, y18, y19, y21, y22, y26, y26, y26, b9, b12, b18, b21, b23, b25, b26, and b27) are labeled in the MS/MS spectrum with the corresponding amino acid sequence LYTY*EQIPGSLKDNPYITDGYRAYLPSR shown, where Y* = nitrotyrosine. Furthermore, the singly charged a - productions (a7, a9, a11, a12, a13, a14, a16, a19, and a24) were detected in the MS/MS spectrum. Three ion - series of singly charged b -, y -, and a -ions determined, with a high level of confidence, this nitropeptide amino acid sequence and the assignment of the nitrotyrosine site. This peptide sequence matched exactly to residues 30 – 57 of the protein, and tyrosine nitration was assigned to Y 33 (Table 1). Similar approaches were used to interpret the eight other MS/MS spectra from human ex - smoker BALF shown in Supplemental Figures B–H.
Progestin and adipoQ receptor family member III
Zinc finger protein 432
Proteasome alpha 2
NADH-ubiquinone oxidoreductase B14
Slit homolog 1 protein
Lysozyme
Aldose 1- epimerase
PTS system lactose - specific EIICB component
Acknowledgments
The authors acknowledge the financial support from Chiesi Pharmaceuticals (Parma, Italy).
Footnotes
Conflict of Interest
Each nitroprotein identified in this article has been protected with patent (Application number: 20100183578).
REFERENCES
- [1].Norman V, keith CH. Nitrogen oxides in tobacco smoke. Nature. 1965;205:915–6. [Google Scholar]
- [2].Beckman JS, Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and the ugly. Am J Physiol. 1996;271:C1431–7. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- [3].Barnes PJ. Oxidative stress in COPD. In: Hansel TT, Barnes PJ, editors. Recent Advances in pathophysiology of COPD. Birkhauser; 2004. pp. 61–75. [Google Scholar]
- [4].Van der Vliet A, Eiserich JP, Shigenaga MK, Cross CE. Reactive nitrogen species and tyrosine nitration in the respiratory tract: epiphenomena or a pathobiologic mechanism of disease? Am J Respir Crit Care Med. 1999;160:1–9. doi: 10.1164/ajrccm.160.1.9807044. [DOI] [PubMed] [Google Scholar]
- [5].Ricciardolo FLM, Caramori G, Ito K, Capelli A, Brun P, Abatangelo G, Papi A, Chung KF, Adcock I, Barnes PJ, Bonner CF, Rossi A, Stefano AD. Nitrosative stress in the bronchial mucosa of severe chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2005;116:1028–35. doi: 10.1016/j.jaci.2005.06.034. [DOI] [PubMed] [Google Scholar]
- [6].Yamaguchi Y, Nasu F, Harada A, Kunitomo M. Oxidants in the gas pahse of cigarette smoke pass through the lung alveolar wall and raise systemic oxidative stress. J Pharmacol Sci. 2007;103:275–82. doi: 10.1254/jphs.fp0061055. [DOI] [PubMed] [Google Scholar]
- [7].Zhang WZ, Lang C, Kaye DM. Determination of plasma free 3-nitrotyrosine and tyrosine by reversed-phase liquid chromatography with 4-fluoro-7-nitrobenzofurazan derivatization. Biomed Chromatogr. 2007;21:273–8. doi: 10.1002/bmc.750. [DOI] [PubMed] [Google Scholar]
- [8].Montes de Oca M, Loeb E, Torres SH, De Sanctis J, Hernandez N, Talamo C. Peripheral muscle alterations in non-COPD smokers. Chest. 2008;133:13–8. doi: 10.1378/chest.07-1592. [DOI] [PubMed] [Google Scholar]
- [9].An Z, Wang H, Song P, Zhang M, Geng X, Zou MH. Nicotine-induced activation of AMP-activated protein kinase inhibits fatty acid synthase in 3T3L1 adipocytes: a role for oxidant stress. J Biol Chem. 2007;282:26793–801. doi: 10.1074/jbc.M703701200. [DOI] [PubMed] [Google Scholar]
- [10].Morla M, Iglesias A, Sauleda J, Cosio B, Agusti A, Busquets X. Reduced expression of the sarcoplasmic calcium pump SERCA2 in skeletal muscle from patients with chronic obstructive pulmonary disease and low body weight. Arch Bronconeumol. 2007;43:4–8. doi: 10.1016/s1579-2129(07)60013-5. [DOI] [PubMed] [Google Scholar]
- [11].Rytila P, Rehn T, Ilumets H, Rouhos A, Sovijarvi A, Myllarniemi M, Kinnula VL. Increased oxidative stress in asymptomatic current chronic smokers and GOLD stage 0 COPD. Respir Res. 2006;7:69. doi: 10.1186/1465-9921-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yang SR, Chida AS, Bauter MR, Shafiq N, Seweryniak K, Maggirwar SB, Kilty I, Rahman I. Cigarette smoke induces proinflammatory cytokine release by activation of NF-kappa B and posttranslational modifications of histone deacetylase in macrophages. Am J Physiol Lung Cell Mol Physiol. 2006;291:L46–57. doi: 10.1152/ajplung.00241.2005. [DOI] [PubMed] [Google Scholar]
- [13].Puhakka AR, Harju TH, Paakko PK, Soini YM, Kinnula VL. Nitric oxide synthases are associated with bronchial dysplasia. Lung Cancer. 2006;51:275–82. doi: 10.1016/j.lungcan.2005.11.005. [DOI] [PubMed] [Google Scholar]
- [14].Foronjy RF, Mirochnitchenko O, Propokenko O, Lemaitre V, Jia Y, Inouye M, Okada Y, D’Armiento JM. Superoxide dismutase expression attenuates cigarette smoke- or elastase-generated emphysema in mice. Am J Respir Crit Care Med. 2006;173:623–31. doi: 10.1164/rccm.200506-850OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhan X, Desiderio DM. The human pituitary nitroproteome: detection of nitrotyrosyl-proteins with two-dimensional Western blotting, and amino acid sequence determination with mass spectrometry. Biochem Biophys Res Commun. 2004;325:1180–6. doi: 10.1016/j.bbrc.2004.10.169. [DOI] [PubMed] [Google Scholar]
- [16].Haddad IY, Pataki G, Hu P, Galliani C, Beckman JS, Matalon S. Quantitation of nitrotyrosine levels in lung sections of patients and animals with acute lung injury. J Clin Invest. 1994;94:2407–13. doi: 10.1172/JCI117607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shigenaga MK, Lee HH, Blount BC, Christen S, Shigeno ET, Yip H, Ames BN. Inflammation and NO(X)-induced nitration: assay for 3-nitrotyrosine by HPLC with electrochemical detection. Proc Natl Acad Sci USA. 1997;94:3211–6. doi: 10.1073/pnas.94.7.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhan X, Du Y, Crabb JS, Gu X, kern TS, Crabb JW. Targets of tyrosine nitration in diabetic rat retina. Mol Cell Proteomics. 2008;7:864–74. doi: 10.1074/mcp.M700417-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [19].Zhan X, Desiderio DM. Nitroproteins from a human pituitary adenoma tissue discovered with a nitrotyrosine affinity column and tandem mass spectrometry. Anal Biochem. 2006;354:279–89. doi: 10.1016/j.ab.2006.05.024. [DOI] [PubMed] [Google Scholar]
- [20].Ottenheijm CA, Heunks LM, Li YP, Jin B, Minnaard R, van Hees HW, Dekhuijzen PN. Activation of the ubiquitin-proteasome pathway in the diaphragm in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;174:997–1002. doi: 10.1164/rccm.200605-721OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kuwano K, Yoshimi M, Maeyama T, Hamada N, Yamada M, Nakanishi Y. Apoptosis signaling pathways in lung diseases. Med Chem. 2005;1:49–56. doi: 10.2174/1573406053402497. [DOI] [PubMed] [Google Scholar]
- [22].Van der Toorn M, Slebos DJ, de Bruin HG, Leuvenink HG, Bakker SJ, Gans RO, Koeter GH, van Oosterhout AJ, Kauffman HF. Cigarette smoke-induced blockade of the mitochondrial respiratory chain switches lung epithelial cell apoptosis into necrosis. Am J Physiol Lung Cell Mol Physiol. 2007;292:L1211–8. doi: 10.1152/ajplung.00291.2006. [DOI] [PubMed] [Google Scholar]
- [23].Tamura T, Lee DH, Osaka F, Fujiwara T, Shin S, Chung CH, Tanaka K, Ichihara A. Molecular cloning and sequence analysis of cDNAs for five major subunits of human proteasomes (multi-catalytic proteinases complexes) Biochem Biophys Acta. 1991;1089:95–102. doi: 10.1016/0167-4781(91)90090-9. [DOI] [PubMed] [Google Scholar]
- [24].Kristensen P, Johnsen AH, Uerkvitz W, Tanaka K, Hendil KB. Human proteasome subunits from 2-dimensional gel identified by partial sequencing. Biochem Biophys Res Commun. 1994;205:1785–9. doi: 10.1006/bbrc.1994.2876. [DOI] [PubMed] [Google Scholar]
- [25].Pai T, Chen Q, Zhang Y, Zolfaghari R, Ross AC. Galactomutarotase and other galactose-related genes are rapidly induced by retinoic acid in human myeloid cells. Biochemistry. 2007;46:15198–207. doi: 10.1021/bi701891t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Breidt Jr F, Hengstenberg W, Finkeldei U, Stewart GC. Identification of the genes for the lactose-specific components of the phosphotransferase system in the lac operon of Staphylococcus aureu. J Biol Chem. 1987;262:16444–9. [PubMed] [Google Scholar]
- [27].Peters D, Frank R, Hengstenberg W. Lactose-specific enzyme II of the phosphoenolpyruvate-dependent phosphotransferase system of Staphylococcus aureus. Purification of the histidine-tagged transmembrane component IICBLac and its hydrophilic IIB domain by metal-affinity chromatography, and functional characterization. Eur J Biochem. 1995;228:798–804. doi: 10.1111/j.1432-1033.1995.0798m.x. [DOI] [PubMed] [Google Scholar]
- [28].Li B, Calvo E, Marinotti O, James AA, Paskewitz SM. Characterization of the c-type lysozyme gene family in Anopheles gambiae. Gene. 2005;360:131–9. doi: 10.1016/j.gene.2005.07.001. [DOI] [PubMed] [Google Scholar]
- [29].Shelley SA, Paciga JE, Balis JU. Lysozyme is an ozone-sensitive component of alveolar type II cell lamellar bodies. Biochim Biophys Acta. 1991;1096:338–44. doi: 10.1016/0925-4439(91)90070-p. [DOI] [PubMed] [Google Scholar]
- [30].Dajani R, Zhang Y, Taft PJ, Travis SM, Starner TD, Olsen A, Zabner J, Welsh MJ, Engelhardt JF. Lysozyme secretion by submucosal glands protects the airway from bacterial infection. Am J Respir Cell Mol Biol. 2005;32:548–52. doi: 10.1165/rcmb.2005-0059OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Skerrett SJ. Lysozyme in pulmonary host defense: new tricks for an old dog. Am J Respir Crit Care Med. 2004;169:435–6. doi: 10.1164/rccm.2312018. [DOI] [PubMed] [Google Scholar]
- [32].Taylor DC, Cripps AW, Clancy RL. A possible role for lysozyme in determining acute exacerbation in chronic bronchitis. Clin Exp Immunol. 1995;102:406–16. doi: 10.1111/j.1365-2249.1995.tb03798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Katoh Y, Katoh M. Comparative genomics on SLIT1, SLIT2, and SLIT3 orthologs. Oncol Rep. 2005;14:1351–5. [PubMed] [Google Scholar]
- [34].Margolin JF, Friedman JR, Meyer WKH, Vissing H, Thiesen HJ, Rauscher FJ., III Kruppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci USA. 1994;91:4509–13. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Witzgall R, O’Leary E, Leaf A, Oenaldi D, Bonventre JV. The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc Natl Acad Sci USA. 1994;91:4514–8. doi: 10.1073/pnas.91.10.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tang YT, Hu T, Arterburn M, Boyle B, Bright JM, Emtage PC, Funk WD. PAQR proteins: a novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J Mol Evol. 2005;61:372–80. doi: 10.1007/s00239-004-0375-2. [DOI] [PubMed] [Google Scholar]
- [37].Barr RG, Camargo CA., Jr Hormone replacement therapy and obstructive airway diseases. Treat Respir Med. 2004;3:1–7. doi: 10.2165/00151829-200403010-00001. [DOI] [PubMed] [Google Scholar]
- [38].Saaresranta T, Aittokallio T, Utriainen K, Polo O. Medroxyprogesterone improves nocturnal breathing in postmenopausal women with chronic obstructive pulmonary disease. Respir Res. 2005;6:28. doi: 10.1186/1465-9921-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Saaresranta T, Polo O. Long-term progestin therapy for female chronic respiratory insufficiency? Respir Med. 2004;98:194–5. doi: 10.1016/j.rmed.2003.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A representative MS/MS spectrum (Supplemental Figure A) and the data interpretation used to identify the nitrotyrosine - containing 28 amino acid peptide from progestin and adipoQ receptor family member III is described here in detail. A singly charged ion ([M + H]+, m/z = 3340.6) in the MS spectrum was selected to accumulate MS/MS spectra (n = 100) with a normalized collision energy = 35 arbitrary units, followed by a Bioworks TuboSequest analysis in Swiss-Prot Database. The singly charged b - and y - product ions (y9, y11, y12, y14, y15, y16, y18, y19, y21, y22, y26, y26, y26, b9, b12, b18, b21, b23, b25, b26, and b27) are labeled in the MS/MS spectrum with the corresponding amino acid sequence LYTY*EQIPGSLKDNPYITDGYRAYLPSR shown, where Y* = nitrotyrosine. Furthermore, the singly charged a - productions (a7, a9, a11, a12, a13, a14, a16, a19, and a24) were detected in the MS/MS spectrum. Three ion - series of singly charged b -, y -, and a -ions determined, with a high level of confidence, this nitropeptide amino acid sequence and the assignment of the nitrotyrosine site. This peptide sequence matched exactly to residues 30 – 57 of the protein, and tyrosine nitration was assigned to Y 33 (Table 1). Similar approaches were used to interpret the eight other MS/MS spectra from human ex - smoker BALF shown in Supplemental Figures B–H.
Progestin and adipoQ receptor family member III
Zinc finger protein 432
Proteasome alpha 2
NADH-ubiquinone oxidoreductase B14
Slit homolog 1 protein
Lysozyme
Aldose 1- epimerase
PTS system lactose - specific EIICB component