Abstract
We report the identification of Schizosaccharomyces pombe mde10+ as a gene possessing a FLEX element, which forms a binding site for the meiosis-specific transcription factor Mei4. In fact, mde10+ is transcribed only in diploid cells that are induced to meiosis in a Mei4-dependent manner. Western blot analysis indicated that the epitope-tagged Mde10 protein accumulates transiently during meiosis and then rapidly decreases. Mde10 is a multidomain protein containing a metalloprotease catalytic domain, a disintegrin domain, a cysteine-rich domain, and membrane-spanning regions, all of which are shared by members of the mammalian ADAM family. A fusion protein of Mde10 and green fluorescent protein localized to the endoplasmic reticulum during meiosis and was located at the peripheral region of spores at the end of meiosis. An mde10Δ deletion mutant showed no apparent defects in meiosis, sporulation, or spore germination. However, the mutant spores exhibited an aberrant surface appearance, in which the ragged outer spore wall was lost to a large extent. Furthermore, mde10Δ spores were found to be less tolerant to ethanol and diethyl ether than were wild-type spores. The mutagenic replacement of the conserved glutamic acid in the putative protease active site with an alanine residue did not affect the surface morphology or the resistance of spores to environmental stress. Our observations indicate that Mde10 is important in the development of the spore envelope, although this function of Mde10 seems to be independent of its metalloprotease activity.
Gametes in multicellular organisms differentiate into morphologically and functionally specialized cells—most typically, into sperms and eggs. Sporulation in single-celled eukaryotes such as yeasts is a morphogenetic process that is equivalent to gametogenesis because an ascospore is a highly specialized cell and its formation is preceded by meiosis. In morphogenesis and cell differentiation in multicellular organisms, the control of gene expression is extremely important; in addition, DNA microarray analyses have demonstrated that transcriptional regulation occurs during gametogenesis in yeast (5, 40).
The forkhead DNA-binding protein Mei4 of the fission yeast Schizosaccharomyces pombe has been identified as a meiosis-specific transcription factor that plays an important role in the progression of meiosis and sporulation (17). To date, approximately 30 genes whose transcription is dependent on Mei4 have been found, including spo4+ and spo6+, which encode the Cdc7/Dbf4-related protein kinase complex (30-32, 46). An in vitro binding assay with recombinant Mei4 proteins and the 5′ upstream region of spo6+ has demonstrated that a short DNA stretch of 14 nucleotides, designated the FLEX element, constitutes the recognition site for Mei4 (17). The FLEX element contains a heptamer core sequence, GTAAACA, which is highly conserved among the recognition sequences of mammalian transcription factors with a forkhead DNA-binding domain, such as human FREAC proteins (37).
A search of the S. pombe genome sequence database (The Wellcome Trust Sanger Institute, Hinxton, United Kingdom) with a query of the FLEX nucleotides (1) revealed an additional set of genes. The DNA microarray data (28) indicated that at least 23 genes among this additional set are upregulated by the induction of meiosis. We noted that the mde10+ gene (SPAC17A5.04) encodes a metalloprotease of the ADAM (for “a disintegrin and metalloprotease”) family. The ADAM proteins are highly conserved zinc-binding proteases and are mostly membrane-anchored glycoproteins (3). ADAM proteins are commonly composed of a prodomain, a metalloprotease domain, a disintegrin domain, a cysteine-rich domain, and an epidermal growth factor-like domain. They also contain a signal peptide and thus are transported to the plasma membrane via the endoplasmic reticulum (ER) and the Golgi apparatus. In their carboxy-terminal region, they usually contain one transmembrane domain, from which the cytoplasmic tail extends towards the C terminus. ADAM proteins are important in diverse cellular and developmental processes, such as the binding and fusion of sperms and eggs, and the processing of some cytokines and their receptor proteins (47, 48). Roughly half of the reported metazoan ADAM proteins are widely expressed in somatic tissues, although the others are exclusively or predominantly expressed in the testis and epididymis (41). ADAM-related proteases have not been found in unicellular eukaryotes; in fact, the genome of Saccharomyces cerevisiae contains no genes encoding ADAM-related proteins.
In this report, we describe the characterization of the mde10+ gene product and the phenotype of mde10 null mutants. We show that Mde10 is expressed in meiotic cells and is not essential for growth. Mde10 is localized to the ER during meiosis and at the spore surface at the end of meiosis. Spores from mde10Δ mutants exhibit abnormal surface morphology and are less tolerant to organic solvents. These data suggest that the fission yeast ADAM-related protein Mde10 is involved in the morphogenesis of spores.
MATERIALS AND METHODS
Yeast strains and culture conditions.
The S. pombe strains used in this study are listed in Table 1. The cells were grown on yeast extract agar complete medium or SD minimal medium. The liquid medium, MML+N, was also used for growth. Mating and sporulation were induced either on a malt extract agar medium or on a synthetic sporulation agar (SSA) media. Sporulation was also induced in MML−N and synthetic sporulation liquid (SSL) media. In some experiments, synchronous meiosis was conducted with pat1 mutants (18, 19, 35). The temperature-sensitive pat1-114 mutant cells were transferred to PM−N and then shaken at 24°C for 15 h to arrest the cell cycle at the G1 phase. Cells were then shifted up at 32°C to start meiosis in a synchronous fashion. Culture media used in this study have been described previously (8, 14, 29). Yeast transformation was carried out by the lithium acetate method (36).
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| L968 | h90 | U. Leupold |
| TN8 | h90 leu1 | T. Nakamura |
| C996-11D | h90 leu1 | C. Shimoda |
| C525-1A | h90 mei4::ura4+ura4-D18 ade6-M210 leu1 | C. Shimoda |
| JY878 | h90 ade6-M216 ura4-D18 leu1 | M. Yamamoto |
| NT10-2 | h90 mde10::ura4+ura4-D18 | This study |
| NT10-3 | h+s/h−mde10::ura4+/mde10::ura4+ade6-M210/ade6-M216 leu1/leu1 ura4-D18/ura4-D18 | This study |
| NT10-9 | h90 mde10::ura4+ura4-D18 leu1 | This study |
| NT10-10 | h90 ade6-M216 ura4-D18 leu1-32 mde10-HA ≪LEU2 | This study |
| NT10-11 | h90 leu1-32 mde10-GFP ≪LEU2 | This study |
| NT3-CH | h+s/h−ade6-M210/ade6-M216 leu1/leu1 | This study |
| JZ670 | h−/h−pat1-114/pat1-114+ade6-M210/ade6-M216 leu1/leu1 | M. Yamamoto |
| AB4 | h−/h−mei4::ura4+/mei4::ura4+ade6-M210/ade6-M216 leu1/leu1 ura4-D18/ura4-D18 pat1-114/pat1-114 | H. Abe |
Gene disruption of mde10+.
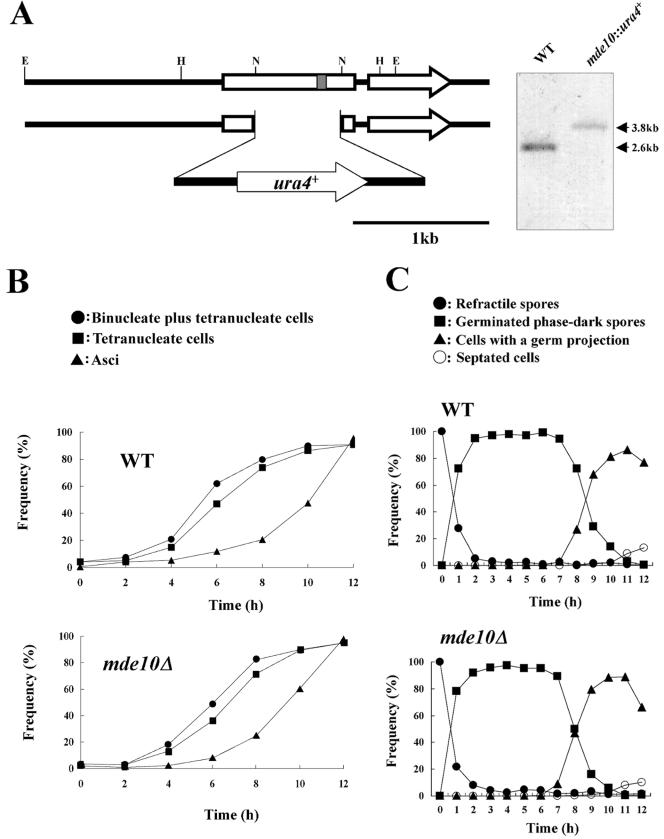
The mde10::ura4+ null allele was produced by one-step gene replacement (38) as follows. A 5,275-bp DNA fragment containing the mde10+ open reading frame (ORF) was amplified by PCR with the forward primer 5′-CCACCGTTTGGGCCG-3′ and the reverse primer 5′-CCAAGATGCGAGGCC-3′. The amplified fragment was cut by EcoRI and then cloned into pBluescript-II SK− (Stratagene, La Jolla, Calif.). To disrupt mde10+, a 0.6-kb NdeI fragment was replaced by the 1.8-kb ura4+ cassette (13). The resultant 3.8-kb EcoRI fragment containing the mde10::ura4+ allele was introduced into the recipient haploid strain JY878, and a few stable Ura+ transformants were isolated. Disruption was confirmed by Southern blot analysis of genomic DNA that had been completely digested with EcoRI, with an EcoRI fragment containing mde10+ as a probe (see Fig. 6B). Some of the stable Ura+ transformants were crossed with a wild-type homothallic strain, and the hybrid diploids were sporulated. In most asci, the ura4 marker was segregated regularly.
FIG.6.
Construction and phenotype of mde10 null mutants. (A) Construction of mde10::ura4+. A 0.6-kb NdeI fragment, including the zinc-binding catalytic site (hatched box), was replaced by a 1.6-kb ura4+ cassette to disrupt mde10+. Genomic Southern blot analysis verified that the mde10+ gene had been disrupted. Genomic DNA was digested with EcoRI and probed with an EcoRI fragment containing mde10+. Restriction sites are E (EcoRI), H (HindIII), and N (NdeI). (B) Kinetics of meiosis in diploid strains heterozygous at the mating type locus. Wild-type (WT) (NT3-CH) and mde10Δ homozygous (NT10-3) strains were cultured in the sporulation medium (MML−N). At intervals, a sample of the cells was fixed and stained with DAPI. The progression of meiotic nuclear divisions was monitored by counting the number of nuclei per cell. For each sample, at least 300 cells were counted. (C) Kinetics of spore germination in mde10Δ mutants. A wild-type and an mde10Δ mutant strain were sporulated in SSL. Isolated single spores prepared by Urografin density gradient centrifugation were suspended in MML+N growth medium and incubated at 30°C. Spores were observed under phase-contrast and fluorescence microscopes and categorized into four classes: ungerminated refractile spores, germinated phase-dark spores, spores with a projection, and septated cells. At least 300 cells were counted.
Plasmid construction.
The mde10+ gene was amplified from genomic DNA by PCR with the primers 5′-GGGGATTACGACTTTTGCTATTGCAGTGCT-3′ and 5′-CGTCGCCGACAAGGTTGAATGGGCCCGTGA-3′. The amplified fragment was first cloned into the pGEM-T easy vector (Promega), and then the SalI/BamHI fragment containing mde10+ was subcloned into pAL-KS, yielding the plasmid pAL(mde10+).
To construct the mde10-GFP fusion gene with transcription driven by the native mde10+ promoter, a genomic DNA stretch from nucleotide −1049 upstream of the initiation codon to the end of the ORF was amplified by PCR with a pair of oligonucleotides, 5′-GCCGTCGACCGATCATAGCTTTCACTAGAT-3′ (the SalI site is underlined) and 5′-GGGGCGGCCGCCCCAGAAAAACCATGCTAT-3′ (the NotI site is underlined). The PCR product was digested with SalI and NotI and then ligated into the same site of pTN143 (20), which contains the Aequorea gene for green fluorescent protein (GFPS65T). The resulting plasmid, referred to as pAL(Mde10-GFP), contained the fusion gene encoding GFP fused to the C terminus of Mde10. Similarly, we constructed a vector expressing a GFP-tagged variant of Mde10 in which the codons for the 19 C-terminal amino acid residues were deleted. The PCR primers used in this case were 5′-GCCGTCGACCGATCATAGCTTTCACTAGAT-3′ (the SalI site is underlined) and 5′-GGGGCGGCCGCCAAGCGATGGTTGTTTAGA-3′ (the NotI site is underlined). The PCR products were digested with SalI and NotI and ligated into the same sites of pTN143, yielding the plasmid pAL(Mde10ΔC-GFP).
A single copy of the mde10-GFP fusion gene was integrated into the mde10 locus on chromosome I as follows. Using S. pombe genome DNA as a template, the DNA fragments carrying the mde10 ORF were amplified with a pair of primers, 5′-GCCGTCGACCATGCGGCTCGTTTTACTGTT-3′ (the SalI site is underlined) and 5′-GGGGCGGCCGCCCCAGAAAAACCATGCTAT-3′ (the NotI site is underlined). The PCR products were digested with SalI and NotI and then cloned into the same sites of the integration plasmid pTN219 (31) to yield pIL(Δmde10-GFP). The resulting fusion gene lacks the promoter sequence. Plasmid pIL(Δmde10-GFP) was cleaved by HindIII and then introduced into the leucine auxotrophic (leu1-32) strain TN8. A homologous recombination event can generate a single copy of the intact med10-GFP gene at the chromosomal mde10 locus, and a few Leu+ integrants were obtained. Accurate integration was confirmed by PCR analysis.
Mde10 tagged with the hemagglutinin (HA) epitope was constructed as follows. First, using the S. pombe cDNA library (T. Nakamura, unpublished data) as a template, the mde10+ cDNA was amplified with two oligonucleotides. The PCR products were digested with BamHI and NotI and then ligated into the same sites of the expression vector pREP41. This plasmid was digested with SalI and NotI, and the DNA fragment was then ligated into the corresponding site of pTN218 (32). The resulting plasmid pIL2(mde10-3HA) carrying the mde10 ORF fused to three copies of the HA epitope at the C terminus was used to integrate mde10-3HA into the chromosomal mde10 locus.
Site-directed mutagenesis.
The glutamic acid residue at position 230 in the metalloprotease active site of Mde10 was replaced with an alanine residue by PCR with the following four primers: F3, 5′-ATGTTGTTATTAAGCTTCCTTTGAAAAAAT-3′; MAS, 5′-ATGTGTCCTATTGCGTGCGCGACA-3′; MS, 5′-TGTCGCGCACGCAATAGGACACAT-3′; B3, 5′-CGCGGATCCTCACCAGAAAAACCATGCTAT-3′. The first PCR was conducted with two primer sets, F3-MAS and MS-B3, with the pAL(mde10+) plasmid as a template. The amplified fragments were mixed and then subjected to a second PCR with the primer pair F3 and B3. The PCR products were ligated into the pGEM-T easy vector (Promega). A 1.4-kb HindIII fragment harboring the mutated mde10 allele was subcloned into pAL(mde10+) to replace the wild-type allele, yielding plasmid pAL(mde10EA).
Southern and Northern blot analyses.
Total genomic DNA was digested with restriction enzymes, fractionated on a 0.8% agarose gel, and then transferred onto nylon membranes (Biodyne A; Nihon Pall Co., Tokyo, Japan). Total RNA was prepared from S. pombe cultures (22) and fractionated on a 1% gel containing 3.7% formaldehyde (44). DNA probes were labeled with [α-32P]dATP by the random hexanucleotide labeling procedure (10). Ribosomal RNAs were stained with ethidium bromide and used as loading controls and size markers.
Western blot analysis.
The pIL2(mde10-3HA) plasmid carrying 5′-truncated (promoter deletion) mde10 with the C-terminal 3HA tag was linearized by digestion with HindIII and then transformed into strain JY878. One chromosomal integrant, referred to as NT10-10, was isolated and used for Western blot analysis. Cell extracts were prepared as described by Masai et al. (27), except that β-mercaptoethanol was not added to the lysis buffer. Crude proteins were fractionated on sodium dodecyl sulfate (SDS)-10% polyacrylamide gels and transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). The blotted membranes were probed with the mouse anti-HA monoclonal antibody 12CA5 (Boehringer Mannheim, Mannheim, Germany) at a 1:1,200 dilution. The membranes were also blotted with the anti-α-tubulin antibody TAT-1 (49) as a control for protein loading. Each band was visualized by staining with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (Promega) and by chemiluminescence (Perkin Elmer Life Sciences, Boston, Mass.).
Immunofluorescence microscopy.
For cell fixation, we followed the method of Hagan and Hyams (15). Spo14-HA was visualized by immunofluorescence microscopy with the rat anti-HA antibody 3F10 (Boehringer Mannheim) and Alexa 594-conjugated goat anti-rat immunoglobulin G (Molecular Probes, Eugene, Oreg.), as reported by Nakamura-Kubo et al. (33). To visualize the nuclear chromatin region, we stained the cells with 4′,6-diamidino-2-phenylindole (DAPI) at 1 μg/ml. Stained cells were observed under a fluorescence microscope (model BX50; Olympus, Tokyo, Japan) equipped with a charge-coupled device camera (Cool-SNAP; Roper Scientific, San Diego, Calif.).
Isolation of spores by Urografin gradient centrifugation.
The homothallic haploid strains L968 and NT10-2 were grown in SSL medium for 7 days at 28°C. Ascal walls were spontaneously dissolved, and single spores were liberated. Spores were isolated by linear 25 to 55% Urografin (Schering Co.) density gradient centrifugation (34). After centrifugation at 25,000 × g for 30 min in an ultracentrifuge (Hitachi CP70G), the cells formed two layers in the gradient. The lower band was exclusively composed of single spores, whereas the upper band contained vegetative cells and zygotes. Single spores were recovered from the lower band and were washed several times with deionized water.
Electron microscopy.
Cells were mounted in a thin layer on copper grids and plunged into liquid propane cooled with liquid N2. Frozen cells were transferred to 2% OsO4 in anhydrous acetone, kept at −80°C for 48 h in a solid CO2-acetone bath, and then transferred to −35°C for 2 h, 4°C for 2 h, and room temperature for 2 h. After three washes in anhydrous acetone, the samples were infiltrated with increasing concentrations of Spurr's resin in anhydrous acetone and finally with 100% Spurr's resin. These samples were then polymerized in capsules at 50°C for 5 h and 60°C for 50 h. Thin sections were cut on a Reichest Ultracut UCT and then stained with uranyl acetate and lead citrate. Sections were viewed on a Hitachi H-7600 transmission electron microscope (TEM) at 100 kV.
For scanning electron microscopy (SEM), the samples were dried in a critical point dryer (model HCP-2; Hitachi) and then coated with a 9-nm-thick layer of platinum-palladium with an ion sputter (model E-1030; Hitachi). The samples were viewed on a JSM-840 (JEOL) instrument.
EndoH treatment.
S. pombe cells expressing Mde10-3HA were cultured in MML−N at 30°C for 10 h. After heat treatment at 90°C for 5 min, the cells were disintegrated with glass beads by vigorous shaking on a vortex mixer in 100 μl of 50 mM Tris-HCl (pH 8.0) containing 1 mM phenylmethylsulfonyl fluoride. Cell extracts were treated with or without 0.05 U of endoglycosidase H (EndoH; Boehringer Mannheim)/ml at 37°C for 18 h in a solution of 0.3% SDS, 0.15 M sodium citrate (pH 5.5), and 5 mM NaN3. The Mde10-3HA protein was detected by SDS-polyacrylamide gel electrophoresis (PAGE) followed by Western blotting with the rat anti-HA antibody 12CA5.
Resistance of spores to diethyl ether and ethanol.
Resistance of the spores to diethyl ether was assessed basically according to the method of Law and Segall (26). Homothallic strains to be tested were sporulated in SSL medium at 28°C for 7 days. An aliquot of the spore suspension was mixed with an equal volume of diethyl ether and was vigorously vibrated on a vortex mixer for 10 min at room temperature. For ethanol treatment, spores were suspended in 40% ethanol and left to stand in an incubator at 24°C. At regular intervals, a small portion of spore suspension was mixed with >10-fold volume of distilled water. After the appropriate dilution was achieved, the cells were grown on SD minimal plates. After 3 days of incubation, the colonies were counted as a measure of viable cells.
RESULTS
Mde10 is an ADAM family protein.
The S. pombe protein Mei4 is a meiosis-specific transcriptional activator containing a forkhead DNA-binding domain (1, 17). The potential cis-acting element for Mei4 has been defined as a 14-bp double-stranded DNA stretch called the FLEX element (17). With this sequence serving as a query, the S. pombe genome sequence database was searched and at least nine target genes (mde1+ to mde9+) were found (1). On the basis of these data and Mei4-dependent genes found by other groups, the refined FLEX element sequence is now thought to be GTAAACAAACAAA(A/C) (Fig. 1A). Using these sequences and their complementary sequences as queries, the S. pombe genome sequence database was searched (H. Abe, unpublished data) (The Wellcome Trust Sanger Institute S. pombe Genome Project web site, http://www.sanger.ac.uk/Projects/S_pombe/). As a result, 32 genes were found to contain sequences fully matching the consensus sequences in their 5′ noncoding region; in other words, these sequences were rarely present in ORFs or in introns. Among these genes, we noted one potential target gene (SPAC17A5.04c) in which the FLEX consensus element was located from bases −104 to −117 upstream of the translational start point. This gene was found to be transcribed by Mei4 (see below), and we designated it mde10+. In a systematic search of protein localization in S. pombe with a GFP fusion library, clone A799, showing spore rim localization, contains a part of the mde10 ORF (6).
FIG. 1.
Promoter and coding sequences of the mde10+ gene. (A) FLEX element. FLEX consensus sequences comprising 14 nucleotides (no. 1 to 14) are compiled from 28 known genes, the transcription of which is dependent on Mei4. Completely identical nucleotides are denoted by capital letters, other nucleotides are denoted by lower-case letters. For comparison, the corresponding FLEX element of mde10+ is presented. (B) Nucleotide sequence of mde10+ and its deduced amino acid sequence. The FLEX consensus sequence in the 5′ promoter region is boxed, potential N-linked glycosylation sites, NX(S/T), are underlined twice, a putative intron is underlined once, the potential signal sequence is shaded, and two predicted membrane-spanning regions are indicated by dotted underlines. The asterisk indicates a termination codon.
The mde10+ ORF was found to be split by one short intron of 71 nucleotides, of which the 5′ and 3′ junctions and the branch point matched the respective consensus sequences (Fig. 1B). As an in-frame termination codon is present in the intron, we thought that splicing out the intron might be essential for the function of mde10+. The putative mde10+ gene product is a 56.4-kDa protein composed of 512 amino acids (Fig. 1B). The Mde10 protein shares significant sequence similarity with the mammalian metalloproteases referred to as ADAM family proteins (Fig. 2). For example, Mde10 has 24% amino acid identity (40% similarity) to mouse ADAM24 and 22% identity (39% similarity) to human ADAM28.
FIG. 2.
mde10+ encodes an ADAM-related protein. (A) Comparison of domain structures between Mde10 and mouse ADAM28. S, signal peptide; P, prodomain; M, metalloprotease domain; D, disintegrin domain; C, cysteine-rich domain; E, epidermal growth factor-like domain; T, transmembrane domain; CYT, cytoplasmic tail. The homology between Mde10 and mouse ADAM28 in the metalloprotease and disintegrin domains is given as the percent identity. (B) Amino acid sequence alignment of the metalloprotease catalytic site and the disintegrin and cysteine-rich domains. The arrowhead indicates the boundary between the disintegrin and cysteine-rich domains. The multiple-sequence alignment was performed with CLUSTALW. Amino acids identical to Mde10 are highlighted in white against a black background. The consensus sequences for the zinc-binding site and the integrin-binding site are shown.
The ADAM proteins are multidomain proteins comprising a metalloprotease catalytic domain, a disintegrin domain, and a cysteine-rich domain. Most of the ADAM proteins are integral membrane proteins. In addition to its overall sequence similarity to ADAM proteins, Mde10 contains a metalloprotease domain as well as disintegrin and cysteine-rich domains (Fig. 2). Moreover, Mde10 was predicted by the SOSUI program (16) to have a signal sequence and two potential membrane-spanning α helices (Fig. 2). The putative catalytic domain of Mde10 contains a well-conserved amino acid motif, HEXXHXXGXXHD, in which the three histidine residues are involved in zinc binding and the glutamic acid residue acts as an active site (23). These structural features imply that Mde10 is a novel member of the ADAM family. Notably, there is no ortholog of mde10+ in the budding yeast S. cerevisiae. The genome sequencing project of Neurospora crassa has revealed an ADAM-related gene (12). To date, this Neurospora ADAM protein is the only ortholog among other lower eukaryotes, although metalloproteases are present in a wide variety of both prokaryotes and eukaryotes (11, 24, 25, 39).
Expression of Mde10 and its cellular localization.
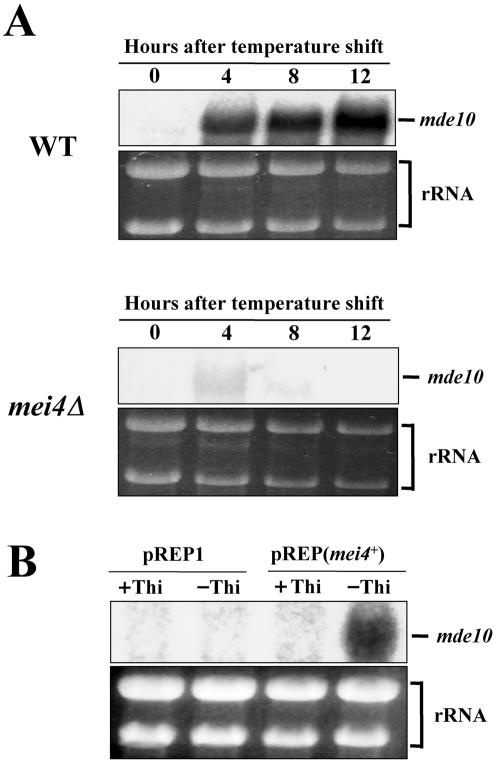
To study the transcriptional control of mde10+, we used Northern blotting to analyze mde10 mRNA in the mei4+ and mei4Δ backgrounds. Meiosis was induced in a synchronous fashion with pat1-114 mutants. As Fig. 3A (upper panel) shows, an mde10 mRNA of 2.0 kb was barely detectable in vegetative cells (at 0 h), but this mRNA rapidly accumulated after meiosis was induced (4 to 12 h). This transcription of mde10+ was completely abolished by the mei4 null mutation (Fig. 3, lower panel). We also tested whether or not mde10+ is ectopically transcribed when Mei4 is overexpressed by the nmt1 promoter during vegetative growth. Figure 3B shows that mde10 mRNA accumulated to high levels after 19 h of incubation in the absence of thiamine (−Thi), which is known to switch on the nmt1 promoter. Taken together, these experiments support the notion that the transcription of mde10+ depends upon Mei4.
FIG. 3.
Mei4-dependent transcription of mde10+. (A) Transcription of mde10+ during pat1-driven synchronous meiosis. Two diploid strains homozygous for pat1-114, JZ670 (mei4+) and AB4 (mei4Δ), were used. The approximate quantity of RNA loaded was monitored by staining gels with ethidium bromide to reveal total rRNA. WT, wild type. (B) Transcription of mde10+ is induced by ectopic overproduction of Mei4. C525-1A transformed with pREP(mei4+) was grown in PM medium without thiamine (−Thi) to induce the nmt1 promoter. Cultures in PM supplemented with thiamine (+Thi) were run in parallel as a control. After incubation for 19 h, total RNA was subjected to Northern blot analysis.
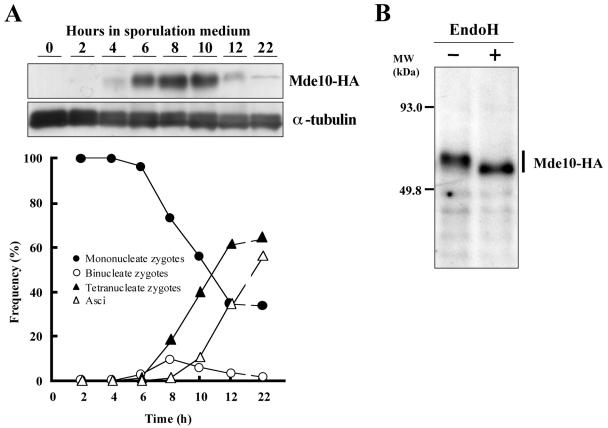
Next, changes in Mde10 protein levels during meiosis and sporulation were examined by Western blot analysis. The homothallic strain NT10-10, harboring an mde10-HA allele on the chromosome, was incubated in nitrogen-free sporulation medium (MML−N). As Fig. 4A shows, Mde10-HA accumulated transiently at 6 to 10 h and then rapidly decreased. Meiosis I began around 6 h, and mature asci appeared about 2 h later (Fig. 4), roughly indicating that Mde10 was expressed during meiosis but expression decreased after the appearance of spores. These results suggest that mde10+ is induced transcriptionally by Mei4 and that the protein accumulates transiently during meiosis.
FIG. 4.
Time course of the expression of Mde10 during meiosis. (A) Changes in Mde10-3HA during meiosis and sporulation. A homothallic strain (NT10-10) carrying the mde10-3HA allele was cultured in a nitrogen-free sporulation medium. Aliquots were removed every 2 h, and protein extracts were analyzed by immunoblotting with the mouse anti-HA antibody 12CA5 as well as with an anti-α-tubulin antibody as a loading control. The progression of meiotic nuclear divisions was monitored by counting the number of nuclei per cell. (B) Mobility shift of Mde10-3HA after treatment with EndoH. +, present; −, absent.
The apparent molecular mass of Mde10-HA determined by SDS-PAGE is about 62 kDa. This mass is a little higher than that predicted from the amino acid sequence. All known ADAM proteins are membrane-anchored glycoproteins and therefore have glycosylation modifications. In fact, the deduced amino acid sequence of Mde10 (Fig. 1B) contains three potential N-linked glycosylation sites, NX(S/T). To determine whether Mde10 is modified by N-linked glycosylation, Mde10-HA proteins from S. pombe extracts were treated with EndoH, which cleaves N-linked oligosaccharides. The digested products were run on an SDS-PAGE gel and then blotted with antibodies specific for HA. Figure 4B shows that EndoH treatment causes a mobility shift, suggesting that Mde10 is N glycosylated.
Mde10 is localized to the ER during meiosis and ends up at the spore surface.
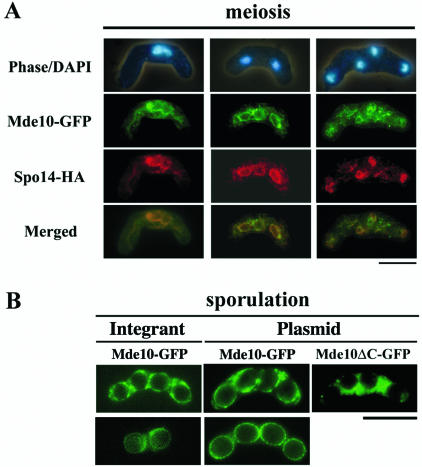
We thought that the intracellular localization of Mde10 may provide a clue to its biological function. To this end, an mde10+-GFP fusion gene borne on a multicopy plasmid, pAL(mde10-GFP), was introduced into the homothallic mde10 disruptant strain NT10-9. The defective phenotypes (see below) were only slightly suppressed, implying that the GFP fusion construct was not fully functional. The Mde10-GFP protein was detected only in cells committed to meiosis and not in vegetative cells (data not shown), consistent with the results of the Western blot analysis (Fig. 5A). The GFP fluorescent signal during meiosis appeared mainly as lines and dots in the cytoplasm that mostly surrounded the nucleus, suggesting that the protein was localized to the ER. This interpretation was confirmed by the immunostaining of cells expressing Spo14-HA, which is used as an ER marker protein (33). The Mde10-GFP and Spo14-HA cells overlapped to a large extent (Fig. 5A). At the postmeiotic stage, the GFP signal disappeared inside the nascent spores, and notably, the fluorescence was specifically localized at the periphery of the spores (Fig. 5B).
FIG. 5.
Localization of Mde10 during meiosis and sporulation. (A) The homothallic haploid strain NT10-9 carrying the pAL(mde10-GFP) plasmid was cultured in MML−N to induce meiosis. Cells fixed at different stages of meiosis were stained with DAPI. Spo14-3HA was visualized by an anti-HA antibody under a fluorescence microscope. Merged images indicate that Mde10 and Spo14 are colocalized. Bar, 10 μm. (B) Localization of Mde10 in mature spores. A homothallic mde10+ strain, JY878, transformed with either pAL(mde10-GFP) or pAL(mde10ΔC-GFP), and the mde10-GFP integrant strain NT10-11 were sporulated on malt extract agar. After incubation for 2 days, localization of the GFP fusion proteins was observed under a fluorescence microscope. Bar, 10 μm.
To exclude the possibility that the above observations were artifacts arising from the overproduction of Mde10-GFP, the localization experiments were repeated in a strain carrying a single copy of the mde10-GFP fusion gene on the chromosomal mde10 locus. Compared with the overproducing cells, the GFP fluorescence in the single-copy cells was not intensive. However, Mde10-GFP was clearly localized to the ER-like structures during meiosis (data not shown) and was concentrated at the surface of spores at the end of meiosis (Fig. 5B). Taking these results together, the Mde10 protein seems to be present primarily in the ER in meiotic stage cells, but as spores are formed it localizes to the surface of the spores.
In cells expressing an Mde10 mutant protein in which the C-terminal transmembrane domain was deleted, this spore surface localization was abolished and the fluorescence was dispersed throughout the cytoplasm of the mother cell outside the newly formed spores (Fig. 5B). As mentioned below, this deletion of the carboxy-terminal putative transmembrane domain also resulted in the loss of the mde10+ function. These observations suggest that the C-terminal transmembrane domain is essential for the proper localization of Mde10 to the spore surface and that this localization might be necessary for its function.
Mde10 is not required for the progression of meiosis, sporulation, or germination.
To elucidate the role of Mde10, a null mutant of mde10+ was generated by gene disruption with a 1.6-kb ura4+ cassette (Fig. 6A). The mde10Δ cells formed colonies on yeast extract agar complete medium, and their growth rate was comparable to that of wild-type cells (data not shown). We next monitored meiosis and sporulation in the mde10Δ disruptant. Meiotic nuclear divisions in the diploid strain NT10-3, which harbors a homozygous mde10::ura4+ allele, proceeded similar to that observed in the isogenic wild-type strain NT3-CH (Fig. 6B). The final yield of asci reached ∼90% in both strains. These observations indicate that mde10+ may not be required for either meiosis or sporulation.
Next, we examined whether spore germination would be affected by the mde10+ disruption. Single spores isolated by micromanipulation from homothallic mde10Δ cells were allowed to germinate on growth medium. The frequency of colony formation was ∼95% for mde10Δ spores and ∼92% for wild-type spores. Spore germination progresses through successive steps, namely, a loss of refractility (darkening) of spores, protrusion of a new cell body, nuclear division, and finally, septation. Spores purified by Urografin density gradient centrifugation were suspended in liquid growth medium, MML+N, and then incubated. The kinetics of these events in mde10Δ spores did not differ from that in wild-type spores (Fig. 6C). We therefore conclude that the mde10+ gene is not required for spore germination.
The spore wall structure of mde10Δ is aberrant.
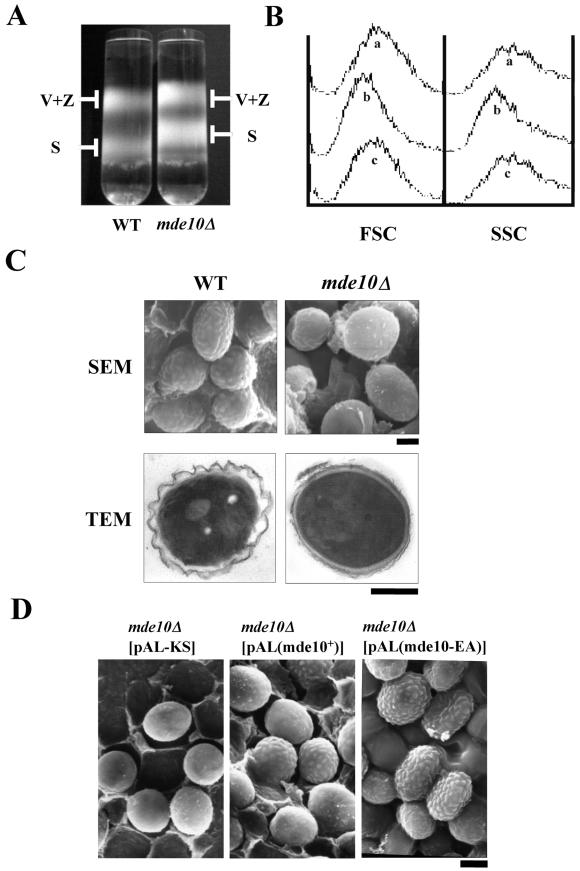
Centrifugal sedimentation of spores in the Urografin density gradient revealed that mde10Δ spores formed a band at a density lower than that of wild-type spores, although the density of vegetative cells did not differ between mde10Δ and wild-type strains (Fig. 7A). Next, the short and long axes of these spores were measured. It was found that mde10Δ spores were smaller than wild-type spores by approximately 10% of the cell volume. The smaller size of the mde10Δ spores was also observed by flow cytometry as parameters of FSC (forward scatter) and SSC (side scatter). These parameters are thought to relate to the dimension of cells. The shift in the FSC or SSC peaks caused by the mde10Δ mutation could be counteracted by introducing a multicopy plasmid pAL(mde10+) into the cells (Fig. 7B).
FIG.7.
Characterization of mde10Δ spores. (A) Density gradient centrifugation of spores. Wild-type (L968) and mde10Δ (NT10-2) cultures in SSL were subjected to 25 to 55% Urografin density gradient centrifugation. Note that the mde10Δ spores formed a band at a lower density than did the wild-type spores, whereas the upper band of vegetative cells and zygotes was the same in both strains. S, spores; V+Z, vegetative cells and zygotes. (B) Flow cytometric analysis of the spores. A homothallic mde10+ strain (C996-11D) harboring pAL-SK (a) and a homothallic mde10Δ strain (NT10-9) harboring pAL-SK (b) or pAL(mde10) (c) were cultured in SSL sporulation medium for 7 days. Cell populations were inspected by flow cytometry for their FSC and SSC parameters (FACSCalibur; Becton Dickinson). (C) Electron microscopy of mde10Δ spores. L968 (wild type [WT]) and NT10-2 (mde10Δ) were cultured in SSL for 7 days (TEM) or SSA for 3 days (SEM). Bars, 1 μm. (D) Complementation of the mde10Δ defect in spore surface structure by the introduction of plasmids expressing mde10+. Homothallic mde10Δ mutants harboring either pAL-KS, pAL(mde10+), or pAL(mde10-EA) were sporulated on SSA for 3 days. Spores were observed by SEM. Bar, 1 μm.
As these results implied that changes in the morphology of mde10Δ spores had taken place, we studied the fine structure of the spores by electron microscopy. Notably, the surface appearance of mde10Δ spores was markedly altered, as revealed by SEM (Fig. 7C). In contrast to wild-type spores, which have an irregular surface, these mde10Δ spores exhibited a rather smooth surface. TEMs of mde10Δ spores revealed that the outer layer of the spore walls was not wavy and was thinner than that of the wild-type spores. Thus, the mde10Δ spores seem to be defective in terms of wall construction.
S. pombe spores are relatively resistant to some organic solvents (7, 42). This resistance is thought to be dependent on the spore walls. Because the mde10Δ spores had an altered surface structure, we considered that they might have lost this resistance. We therefore tested the tolerance of the mde10Δ spores to ethanol and to diethyl ether. The spores were suspended in 40% ethanol and incubated at 24°C. At intervals, aliquots of the spore suspension were diluted with sterile water and were spread on SD plates to score viability. The resistance of mde10Δ spores to ethanol was ∼10 times lower than that of wild-type spores (data not shown). The reduced resistance of mde10Δ spores to ethanol could be recovered by introducing a multicopy plasmid carrying mde10+ into the cells (Fig. 8). We also tested the spores' tolerance to diethyl ether. The spores were incubated for 10 min in diethyl ether mixed with an equal volume of H2O under conditions of vigorous shaking. The viability of mde10Δ spores decreased to 1% of the wild-type level (data not shown). Taken together, these results indicate that the mde10+ gene is necessary for maintaining the tolerance of mature spores to ethanol and diethyl ether.
FIG. 8.
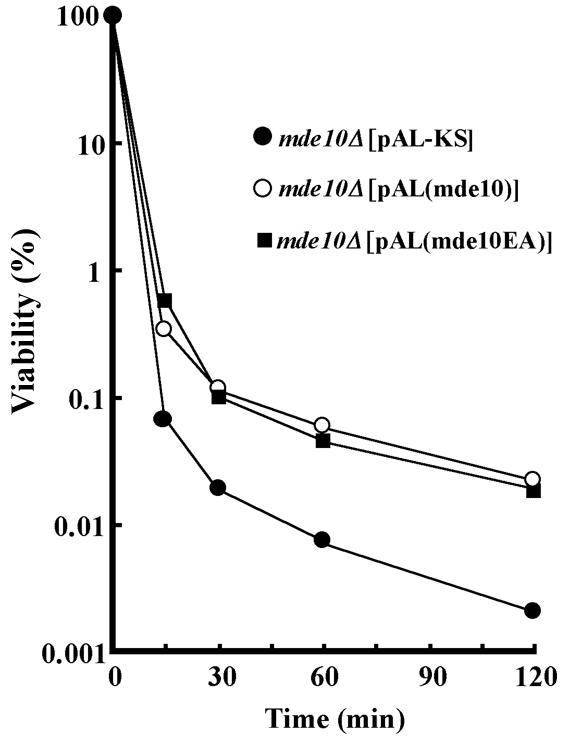
Loss of ethanol resistance in mde10Δ spores and complementation by mde10+-expressing plasmids. A homothallic mde10Δ strain (NT10-9) harboring pAL-SK, pAL(mde10+), or pAL(mde10-EA) was cultured in SSL for 7 days. The proportions of spores in the cell population were higher than 90%. Cells were treated with 40% ethanol at 24°C. At the indicated times, a portion of the cell suspension was rapidly diluted with sterile distilled water and then spread on SD minimal medium. After incubation for 7 days, the number of colonies was counted.
The metalloprotease catalytic domain is not necessary for the morphogenetic roles of Mde10.
As the defects in spore surface morphology and in the ethanol resistance of mde10Δ could be complemented by plasmids harboring the wild-type mde10+ gene, we used this assay to examine several deletion and point mutations. As mentioned above, the catalytic domain of Mde10 is highly conserved; therefore, we mutated the glutamic acid residue at position 230, which is presumed to be the active site, to alanine. Surprisingly, this E230A mutant (mde10-EA) was able to complement the impaired resistance to ethanol (Fig. 8) and also led to a recovery of the outer wavy layer of mde10Δ spore walls (Fig. 7D). Furthermore, a deletion mutant in which most of the catalytic domain was truncated still complemented these phenotypes of the null allele (data not shown). These facts strongly suggest that the metalloprotease catalytic activity is not required for the construction of spore walls by Mde10.
DISCUSSION
The mde10+ gene is a novel target of the Mei4 transcription factor.
In this article, we have demonstrated that mde10+ is transcribed under the control of the meiosis-specific transcription factor Mei4 (17). This forkhead family transcription factor recognizes a cis-acting element, referred to as the FLEX sequence (1). The FLEX element is usually located at a position 1 kb upstream from the initiation codon. In the case of mde10+, the distance between the initiation codon and the FLEX sequence is about 110 nucleotides. Ectopic overexpression of Mei4 in vegetative cells strongly induced the transcription of mde10+. This fact strongly supports the idea that Mei4 directly stimulates the transcription of mde10+. The analysis of S. pombe genes by DNA microarray also indicates that the mde10+ gene is extensively expressed under meiotic conditions (28). The identification and characterization of mde10+ has revealed the utility of this genome-wide screening for FLEX-containing genes, which has led to the discovery of new meiosis- and sporulation-specific genes. Characterization of other candidate genes possessing a FLEX element is now in progress.
The mde10+ gene encodes an ADAM family metalloprotease homolog.
Although ADAM family proteases have been found in a wide variety of higher animals, only a few homologous proteins have been reported from lower eukaryotes (12). ADAM family proteins are membrane-anchored glycoproteins composed of multiple conserved domains. Among these domains, Mde10 contains a metalloprotease catalytic site, as well as disintegrin and cysteine-rich domains (Fig. 2). In addition, Mde10 is equipped with an N-terminal signal sequence, a C-terminal membrane-spanning domain, and three potential N glycosylation sites. These structural features imply that Mde10 is a member of the ADAM family. As S. pombe is a genetically tractable microorganism, it will be possible to analyze the biological functions of Mde10 intensively, with implications for other ADAM family proteins.
The canonical metalloprotease active site is also conserved in the Mde10 amino acid sequence, although the protease activity of Mde10 has not yet been examined in biochemical experiments. Surprisingly, our mutational analyses suggested that the protease activity is not necessary for the observed function of Mde10, that is, proper construction of the spore surface and its tolerance to ethanol (Fig. 7 and 8). Many ADAM proteins that do not retain the metalloprotease catalytic site have been reported. For example, 9 of 24 ADAM proteins in mice lack a catalytic site (2). Therefore, there exists a group of ADAM family proteins whose functions do not depend on metalloprotease activity. As Mde10 accumulates long before the synthesis of spore walls, Mde10 probably has another nonessential function during meiosis. It is possible that such unknown roles of Mde10 depend on its metalloprotease activity.
Among the three conserved domains of Mde10, the disintegrin domain shows the highest homology to the corresponding domain of mammalian ADAM proteins (Fig. 2). A consensus motif, RXXXXXXDLPEF, in the disintegrin domain of the mammalian proteins has been identified as the integrin-interacting site (9). There is a similar, but not identical, motif in the corresponding domain of Mde10 (Fig. 3); however, the deletion of a small section of this motif (10 amino acid residues from R to P) resulted in no apparent phenotype (Nakamura, unpublished). The function of the disintegrin domain in the ADAM family has been associated with forms of cell adhesion such as sperm-egg fusion and myoblast fusion (4, 51). The roles played by the cysteine-rich region are not well understood. Although the C-terminal half of the cysteine-rich domain in Mde10 could not be aligned easily with that of the ADAM proteins unless a few gaps were inserted, there are 11 cysteine residues in this domain composed of 88 residues. As suggested for mammalian ADAM proteins, these domains of Mde10 might function in spore-to-spore adhesion or spore-to-substrate binding. Comprehensive mutagenesis of these domains in Mde10 may provide useful information with regard to their biological and biochemical roles.
Mde10 localizes to the spore surface.
Most ADAM proteins are known to localize to the plasma membrane; for example, Testase-1 (ADAM24) is a plasma membrane-anchored protease of sperms and is involved in egg-sperm fusion (50). From the sequence data, Mde10 was predicted to have two hydrophobic putative membrane-spanning domains in addition to its N-terminal signal sequence. The Mde10-GFP fusion protein was clearly shown to be localized to the ER during meiosis (Fig. 5A). Thus, it is likely that Mde10 is transported via the ER-Golgi system and is finally targeted to the plasma membrane. The C-terminal transmembrane domain was found to be essential for the proper localization of Mde10 to the spore periphery (Fig. 5B). Another interesting structural feature of Mde10 is the absence of the C-terminal cytoplasmic tail. Generally, this domain is thought to interact with cytoplasmic signaling components such as protein kinase C (21). As Mde10 lacks a cytoplasmic tail domain, the question of how Mde10 can respond to intracellular signals remains to be solved.
Mde10 is required for the construction of the spore envelope.
Our electron microscopic observations showed that the spore surface morphology is defective in mde10Δ spores. The mde10Δ spores exhibited a smooth surface, in contrast to the rough surface of wild-type spores. In S. cerevisiae, spore walls differ from vegetative cell walls and they give spores resistance to severe environmental stressors (43). In particular, the outermost proteinaceous layer is responsible for resistance to spore wall lytic enzymes, high temperature, and chemicals in S. cerevisiae (43). The gene meu10, which when mutated affects spore wall components, is known to be associated with tolerance to the following stressors: heat shock, cold temperature, and ethanol exposure (45). It is likely that mde10Δ spores are more sensitive to organic solvents than are wild-type spores.
On the basis of these observations, we can summarize the fate and role of Mde10 during meiosis and sporulation as follows. The mde10+ gene is transcribed under the control of Mei4, and its protein products are first targeted to the ER by the aid of its signal sequence. Membrane vesicles carrying Mde10 are then transported to the precursor forms of spores, in other words, to the prespores. Most probably, Mde10-carrying vesicles are derived from a source that is external to these prespores, because C-terminally truncated Mde10 is dispersed outside these structures (see Fig. 5B). If this is indeed the case, then the Mde10-carrying vesicles might fuse with the outer leaflet of the forespore membrane. Once on the spore surface, Mde10 is involved in the proper construction of the spore envelope. It should be noted, however, that most of these steps remain speculative and remain to be investigated in further studies.
Acknowledgments
We are grateful to Taro Nakamura of this laboratory for invaluable discussions. We thank K. Gull of the University of Manchester for the anti-α-tubulin antibody TAT-1, Y. Hiraoka of Kansai Advanced Research Center and M. Nakamura-Kubo for plasmids, and N. Uchida for assistance in electron microscopy.
This study was supported by a Grant-in-Aid for Scientific Research on Priority Areas (Genome Biology) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to C.S.
REFERENCES
- 1.Abe, H., and C. Shimoda. 2000. Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor mei4 and a genome-wide search for its target genes. Genetics 154:1497-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becherer, J. D., and C. P. Blobel. 2003. Biochemical properties and functions of membrane-anchored metalloprotease-disintegrin proteins (ADAMs). Curr. Top. Dev. Biol. 54:101-123. [DOI] [PubMed] [Google Scholar]
- 3.Blobel, C. P., and J. M. White. 1992. Structure, function and evolutionary relationship of proteins containing a disintegrin domain. Curr. Opin. Cell Biol. 4:760-765. [DOI] [PubMed] [Google Scholar]
- 4.Cao, Y., Z. Zhao, J. Gruszczynska-Biegala, and A. Zolkiewska. 2003. Role of metalloprotease disintegrin ADAM12 in determination of quiescent reserve cells during myogenic differentiation in vitro. Mol. Cell. Biol. 23:6725-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685-696. [DOI] [PubMed] [Google Scholar]
- 6.Ding, D. Q., Y. Tomita, A. Yamamoto, Y. Chikashige, T. Haraguchi, and Y. Hiraoka. 2000. Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells 5:169-190. [DOI] [PubMed] [Google Scholar]
- 7.Egel, R. 1977. Selective spore survival during replica-plating of fission yeast. Arch. Microbiol. 112:109-110. [DOI] [PubMed] [Google Scholar]
- 8.Egel, R., and M. Egel-Mitani. 1974. Premeiotic DNA synthesis in fission yeast. Exp. Cell Res. 88:127-134. [DOI] [PubMed] [Google Scholar]
- 9.Eto, K., C. Huet, T. Tarui, S. Kupriyanov, H. Z. Liu, W. Puzon-McLaughlin, X. P. Zhang, D. Sheppard, E. Engvall, and Y. Takada. 2002. Functional classification of ADAMs based on a conserved motif for binding to integrin alpha 9beta 1: implications for sperm-egg binding and other cell interactions. J. Biol. Chem. 277:17804-17810. [DOI] [PubMed] [Google Scholar]
- 10.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 11.Fujimura-Kamada, K., F. J. Nouvet, and S. Michaelis. 1997. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor. J. Cell Biol. 136:271-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L. J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Stange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. [DOI] [PubMed] [Google Scholar]
- 13.Grimm, C., J. Kohli, J. Murray, and K. Maundrell. 1988. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol. Gen. Genet. 215:81-86. [DOI] [PubMed] [Google Scholar]
- 14.Gutz, H., H. Heslot, U. Leupold, and N. Loprieno. 1974. Schizosaccaromyces pombe, p. 395-446. In R. C. King (ed.), Handbook of genetics, vol. 1. Plenum Press, New York, N.Y.
- 15.Hagan, I. M., and J. S. Hyams. 1988. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 89:343-357. [DOI] [PubMed] [Google Scholar]
- 16.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 17.Horie, S., Y. Watanabe, K. Tanaka, S. Nishiwaki, H. Fujioka, H. Abe, M. Yamamoto, and C. Shimoda. 1998. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell. Biol. 18:2118-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iino, Y., Y. Hiramine, and M. Yamamoto. 1995. The role of cdc2 and other genes in meiosis in Schizosaccharomyces pombe. Genetics 140:1235-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iino, Y., and M. Yamamoto. 1985. Mutants of Schizosaccaromyces pombe which sporulate in the haploid state. Mol. Gen. Genet. 198:416-421. [DOI] [PubMed] [Google Scholar]
- 20.Ikemoto, S., T. Nakamura, M. Kubo, and C. Shimoda. 2000. S. pombe sporulation-specific coiled-coil protein Spo15p is localized to the spindle pole body and essential for its modification. J. Cell Sci. 113:545-554. [DOI] [PubMed] [Google Scholar]
- 21.Izumi, Y., M. Hirata, H. Hasuwa, R. Iwamoto, T. Umata, K. Miyado, Y. Tamai, T. Kurisaki, A. Sehara-Fujisawa, S. Ohno, and E. Mekada. 1998. A metalloprotease-disintegrin, MDC9/meltrin-gamma/ADAM9 and PKCdelta are involved in TPA-induced ectodomain shedding of membrane-anchored heparin-binding EGF-like growth factor. EMBO J. 17:7260-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, R., G. F. Sprague, Jr., and I. Herskowitz. 1983. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc. Natl. Acad. Sci. USA 80:3035-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, W., and J. S. Bond. 1992. Families of metalloendopeptidases and their relationships. FEBS Lett. 312:110-114. [DOI] [PubMed] [Google Scholar]
- 24.Kornitzer, D., D. Teff, S. Altuvia, and A. B. Oppenheim. 1991. Isolation, characterization, and sequence of an Escherichia coli heat shock gene, htpX. J. Bacteriol. 173:2944-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumagai, H., Y. Kawamura, K. Yanagisawa, and H. Komano. 1999. Identification of a human cDNA encoding a novel protein structurally related to the yeast membrane-associated metalloprotease, Ste24p. Biochim. Biophys. Acta 1426:468-474. [DOI] [PubMed] [Google Scholar]
- 26.Law, D. T., and J. Segall. 1988. The SPS100 gene of Saccharomyces cerevisiae is activated late in the sporulation process and contributes to spore wall maturation. Mol. Cell. Biol. 8:912-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masai, H., T. Miyake, and K. Arai. 1995. hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 14:3094-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mata, J., R. Lyne, G. Burns, and J. Bahler. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32:143-147. [DOI] [PubMed] [Google Scholar]
- 29.Moreno, S., A. Klar, and P. Nurse. 1990. Molecular genetic analysis of fission yeast Schizosaccaromyces pombe. Methods Enzymol. 194:793-823. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura, T., M. Kishida, and C. Shimoda. 2000. The Schizosaccharomyces pombe spo6+ gene encoding a nuclear protein with sequence similarity to budding yeast Dbf4 is required for meiotic second division and sporulation. Genes Cells 5:463-479. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura, T., M. Nakamura-Kubo, A. Hirata, and C. Shimoda. 2001. The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1+-encoding syntaxin-like protein. Mol. Biol. Cell 12:3955-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura, T., M. Nakamura-Kubo, and C. Shimoda. 2002. Novel fission yeast Cdc7-Dbf4-like kinase complex required for the initiation and progression of meiotic second division. Mol. Cell. Biol. 22:309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura-Kubo, M., T. Nakamura, A. Hirata, and C. Shimoda. 2003. The fission yeast spo14(+) gene encoding a functional homologue of budding yeast Sec12 is required for the development of forespore membranes. Mol. Biol. Cell 14:1109-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishi, K., C. Shimoda, and M. Hayashibe. 1978. Germination and outgrowth of Schizosaccharomyces pombe ascospores isolated by Urografin density gradient centrifugation. Can. J. Microbiol. 24:893-897. [DOI] [PubMed] [Google Scholar]
- 35.Nurse, P. 1985. Mutants of the fission yeast Shizosaccaromyces pombe which alter the shift between cell proliferation and sporulation. Mol. Gen. Genet. 198:497-502. [Google Scholar]
- 36.Okazaki, K., N. Okazaki, K. Kume, S. Jinno, K. Tanaka, and H. Okayama. 1990. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 18:6485-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierrou, S., M. Hellqvist, L. Samuelsson, S. Enerback, and P. Carlsson. 1994. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 13:5002-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 39.Rudner, D. Z., P. Fawcett, and R. Losick. 1999. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. USA 96:14765-14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlecht, U., and M. Primig. 2003. Mining meiosis and gametogenesis with DNA microarrays. Reproduction 125:447-456. [DOI] [PubMed] [Google Scholar]
- 41.Seals, D. F., and S. A. Courtneidge. 2003. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 17:7-30. [DOI] [PubMed] [Google Scholar]
- 42.Shimoda, C. 1980. Differential effect of glucose and fructose on spore germination in the fission yeast, Schizosaccharomyces pombe. Can. J. Microbiol. 26:741-745. [Google Scholar]
- 43.Smits, G. J., H. van den Ende, and F. M. Klis. 2001. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147:781-794. [DOI] [PubMed] [Google Scholar]
- 44.Thomas, P. S. 1980. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc. Natl. Acad. Sci. USA 77:5201-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tougan, T., Y. Chiba, Y. Kakihara, A. Hirata, and H. Nojima. 2002. Meu10 is required for spore wall maturation in Schizosaccharomyces pombe. Genes Cells 7:217-231. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe, T., K. Miyashita, T. T. Saito, T. Yoneki, Y. Kakihara, K. Nabeshima, Y. A. Kishi, C. Shimoda, and H. Nojima. 2001. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 29:2327-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfsberg, T. G., P. D. Straight, R. L. Gerena, A. P. Huovila, P. Primakoff, D. G. Myles, and J. M. White. 1995. ADAM, a widely distributed and developmentally regulated gene family encoding membrane proteins with a disintegrin and metalloprotease domain. Dev. Biol. 169:378-383. [DOI] [PubMed] [Google Scholar]
- 48.Wolfsberg, T. G., and J. M. White. 1996. ADAMs in fertilization and development. Dev. Biol. 180:389-401. [DOI] [PubMed] [Google Scholar]
- 49.Woods, A., T. Sherwin, R. Sasse, T. MacRae, A. J. Baines, and K. Gull. 1989. Definition of individual components within the cytosleleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491-500. [DOI] [PubMed] [Google Scholar]
- 50.Zhu, G. Z., D. G. Myles, and P. Primakoff. 2001. Testase 1 (ADAM 24) a plasma membrane-anchored sperm protease implicated in sperm function during epididymal maturation or fertilization. J. Cell Sci. 114:1787-1794. [DOI] [PubMed] [Google Scholar]
- 51.Zhu, X., N. P. Bansal, and J. P. Evans. 2000. Identification of key functional amino acids of the mouse fertilin beta (ADAM2) disintegrin loop for cell-cell adhesion during fertilization. J. Biol. Chem. 275:7677-7683. [DOI] [PubMed] [Google Scholar]