Abstract
The Sm-binding site of the kinetoplastid spliced leader RNA has been implicated in accurate spliced leader RNA maturation and trans-splicing competence. In Trypanosoma brucei, RNA interference-mediated knockdown of SmD1 caused defects in spliced leader RNA maturation, displaying aberrant 3′-end formation, partial formation of cap 4, and overaccumulation in the cytoplasm; U28 pseudouridylation was unaffected.
Spliced leader RNA plays a central role in kinetoplastid gene expression, as it is trans-spliced onto every nucleus-derived mRNA. Spliced leader RNA maturation is a multistep process involving precursor 3′ extension removal (16, 18), 5′-end methylation (2), pseudouridylation of U28 (ψ28) (8), and trafficking to the cytoplasm mediated by exportin 1 (21). The details of spliced leader RNA nuclear import have not yet been described, and the order and intracellular locations of the various modifications have not been defined unambiguously.
A key structural element of spliced leader RNA is the Sm-binding site (11, 16), RAU4-6GR, that is also present on many of the small nuclear RNAs (snRNAs) involved in splicing. The Sm-binding site interacts with a heptameric complex of Sm proteins (20). The mammalian U1 snRNP is assembled via three RNA-free Sm subcomplexes, D1-D2, F-E-G, and D3-B, that form a ring around the single-stranded Sm-binding site (15). None of the subcomplexes can associate with the Sm-binding site in isolation (13). The importance of the spliced leader RNA Sm-binding site has been demonstrated by mutagenesis in Leishmania tarentolae (16) and Leptomonas collosoma (11), which affected spliced leader RNA maturation and abolished trans-splicing. Conversely, Sm-binding site mutants of Leptomonas seymouri were competent for trans-splicing, but only when delivered on a low-copy-number episome (9). Transcription studies in Trypanosoma brucei demonstrated that the Sm-binding site forms a boundary for cap 4 formation (10).
We challenged the role of SmD1 protein (12) in kinetoplastid spliced leader RNA biogenesis by performing a knockdown experiment with RNA interference (RNAi). Our working model of the spliced leader RNA maturation pathway (21) predicted that 5′ and 3′ processing of spliced leader RNA would be impaired by the absence of SmD1 and that spliced leader RNA would accumulate in the cytosol. Our results support posttranscriptional cap 4 modification of spliced leader RNA.
Loss of the SmD1 mRNA is lethal to T. brucei.
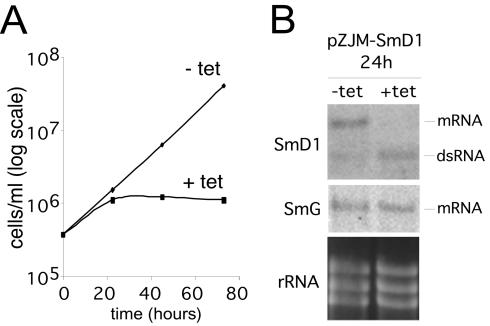
An inducible-RNAi plasmid containing SmD1 gene sequence was transfected into T. brucei. Upon induction with tetracycline, clonal lines containing the SmD1 RNAi construction ceased division after 24 h, whereas the growth of control 29-13 and noninduced cells was unaffected (Fig. 1A). RNA blotting (Fig. 1B) revealed increasing levels of induced double-stranded RNA and demonstrated that the SmD1 mRNA disppeared at 24 h. Reprobing the blot for a different Sm protein transcript (12) showed no loss of SmG mRNA at 24 h, confirming the specificity of the RNAi effect. Ethidium bromide staining of rRNA bands controlled for the quality and quantity of each RNA sample loaded.
FIG. 1.
SmD1 mRNA is required for growth of T. brucei. (A) Comparative growth curves of induced and uninduced T. brucei (clonal line E) containing a tetracycline (tet)-regulated RNAi construction directed against SmD1 mRNA. (B) SmD1 mRNA is abolished by RNAi at 24 h postinduction. Total-cell RNA from induced and noninduced cultures in separate experiments was size separated on a 1% agarose-formaldehyde gel, blotted, and probed for the presence of SmD1 mRNA. As a control, the gel was stained with ethidium bromide to visualize rRNA bands and the blot was reprobed for SmG mRNA.
SmD1 is necessary for accurate 3′-end formation.
In L. tarentolae, spliced leader RNA Sm-binding site mutants lose 3′ processing, accumulating in the form of primary transcription products with staggered 3′ ends (18). The Sm-protein complex has been implicated in nuclear import of trafficked snRNAs. Thus, the 3′-end phenotype and subcellular localization of spliced leader RNA in the SmD1 protein in the induced RNAi cell line were examined.
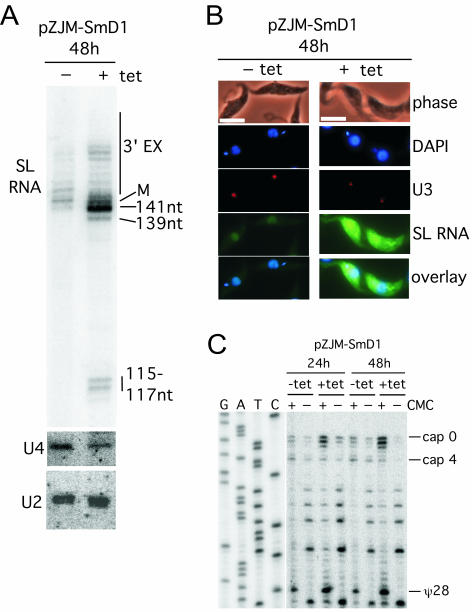
High-resolution RNA blots were hybridized initially with a stem-loop I probe (5′-CTACTGGGAGCTTCTCATCA; Fig. 2A), which yielded a complex pattern of bands ranging from 3′-extended products to transcripts below mature spliced leader RNA size. In the uninduced RNA population, mature spliced leader RNA was identified as the band at 142 nucleotides. Based on 5′-end determination by primer extension (data not shown and Fig. 2C), higher-molecular-weight bands represent intermediates at various stages of 3′ extension (21). In RNA from tetracycline-induced cells, 3′-extended forms of spliced leader RNA were detected, as were additional longer forms. A major spliced leader RNA band was present at 141 nucleotides with greater relative abundance than the uninduced RNA sample, and minor products were present at single-nucleotide intervals above and below the major band. Two lower-molecular-weight bands migrating at approximately 115 to 117 nucleotides were prominent in the induced RNA.
FIG. 2.
Knockdown of SmD1 mRNA affects spliced leader RNA abundance, size, and subcellular distribution. (A) High-resolution 6% acrylamide-8 M urea gels were used to separate RNA transcripts to the single-nucleotide level. Blots were probed with spliced leader (SL) RNA, U2, and U4 snRNA probes. M, mature size (142 nucleotides). (B) Fluorescence in situ hybridization analysis for intracellular localization of spliced leader RNA with a fluorescein isothiocyanate-labeled probe was performed on uninduced (without tetracycline, −tet) and induced (+tet) cells at 48 h. Bar, 10 μm. U3 snoRNA was localized with a Tamara-labeled oligonucleotide probe; nuclear and kinetoplast DNAs were stained with DAPI. (C) ψ28 occurs independent of SmD1. ψ28 was assayed on RNA from induced (+tet) and uninduced (−tet) populations with carboxymethyl cellulose (CMC) modification (+ or −) and primer extension. Products extended with the 5′-end-labeled TbSL stem-loop I oligonucleotide were resolved alongside a corresponding DNA sequencing ladder through a 6% acrylamide-8 M urea gel. Additional primer extension stops at three AU and one AC dinucleotide were seen inconsistently in different experiments.
Hybridization of the blot for the spliceosomal U4 snRNA (21), which contains an Sm-binding site, showed a twofold reduction in induced cells, consistent with a stability defect in the absence of SmD1 protein; no 3′-extended forms of U4 snRNA were detected. Spliceosomal U2 snRNA, which also contains an Sm-binding site, was detected by the oligonucleotide 5′-TATCAGGAGTTACTCTGATAAGAACA, revealing equivalent levels in both uninduced and induced cell lines and no longer forms. The unaltered U2 snRNA level is consistent with normal core U2 snRNP assembly in U2 Sm-binding site mutants (6). The relative abundances of U2 snRNA and small ribosomal RNAs visualized by ethidium bromide staining (data not shown) indicated that spliced leader RNA transcript was hyperaccumulating in the induced lines.
The accumulation of spliced leader RNA relative to U2 snRNA may be due to the hundredfold difference in gene copy number (4, 19) coupled with different rates of transcription (14), variable half-lives, and loss of consumption of spliced leader RNA in the trans-splicing reaction. As a substrate in trans-splicing, spliced leader RNA has a shorter half-life than the other spliceosomal components, estimated to be between 3 min (7) and 1.4 h (5) in T. brucei.
SmD1 depletion results in cytoplasmic accumulation of spliced leader RNA.
The distribution of spliced leader RNA was analyzed by fluorescence in situ hybridization in the induced SmD1 RNAi cell line. In the absence of the SmD1 mRNA, spliced leader RNA signal became more intense throughout individual cells in an increasing percentage of the population over time (14% of cells at 24 h and 25% at 48 h), consistent with the increased abundance of spliced leader RNA observed in RNA blots. Higher magnification showed that spliced leader RNA was overaccumulating in the cytoplasm (Fig. 2B). The relative abundance and distribution of U3 snoRNA, which does not contain an Sm-binding site, were unchanged in the induced cells. All directly compared images were captured at the same shutter speed and processed in parallel. We have demonstrated previously that nuclear spliced leader RNA signal is not due to DNA or cytosolic signal to mRNA or autofluorescence (21).
These results demonstrate overaccumulation of spliced leader RNA in SmD1 T. brucei knockdown lines, consistent with the molecular phenotype of the Sm-binding site mutations in the L. tarentolae spliced leader RNA (16). Substrate spliced leader RNA overaccumulation in the cytoplasm indicated that the SmD1 protein is necessary for spliced leader RNA maturation prior to nuclear import; additional Sm-dependent spliced leader RNA modifications may be required as import signals.
Exon pseudouridinylation occurs independent of Sm-binding site interaction.
Although ψ28 of the spliced leader does not appear to be critical for trans-splicing (17) or association with polysomes (21), it may modulate or enhance mRNA interactions with the splicing or translation machinery. To determine whether pseudouridylation of spliced leader RNA is SmD1 dependent, total-cell RNA samples from induced and uninduced SmD1 RNAi lines were subjected to carboxymethyl cellulose treatment (1, 8). Irreversible carboxymethyl cellulose modification of ψ28 was visualized by primer extension with the oligonucleotide TbSL stem-loop I (Fig. 2C). As expected, RNA from uninduced cells yielded a termination at position 29 of the exon, indicative of ψ28. RNA from 48-h-induced cells also showed reverse transcriptase termination at this position, indicating that pseudouridylation was unimpaired in the absence of SmD1. An internal control for the integrity of RNA and efficacy of RNAi induction was apparent in the form of extension products terminating at the 5′ end of carboxymethyl cellulose-treated spliced leader RNA. In uninduced RNA, a fully modified cap 4 termination product was visible, whereas in induced RNA cap 0/cap 1 termination products were evident.
The localization of SLA1 RNA (8) combined with temporal and Sm-independent acquisition of the pseudouridylation of spliced leader RNA supports modification of ψ28 in the nucleus, most likely in the nucleolus, prior to cytosolic trafficking. However, the presence of SLA1 RNA in the nucleolus may be necessitated by its own processing requirements and does not preclude nucleoplasmic or cytosolic spliced leader RNA pseudouridylation. Furthermore, inhibition of cap 4 methylation in induced cell lines mirrored results obtained in Sm-binding site mutagenesis studies of spliced leader RNA in L. tarentolae and Leptomonas collosoma, in contrast to the cotranscriptional cap 4 acquisition model (10).
SmD1 defects define steps in the spliced leader RNA processing model.
Our working model for biogenesis of kinetoplastid spliced leader RNA is as follows. Spliced leader RNA is transcribed as a 3′-extended precursor (18) and receives an m7G cap cotranscriptionally (10). The acquisition of ψ28 on spliced leader RNA is directed by SLA1 RNA (8) in the nucleolus. The m7G cap of nascent spliced leader RNA is recognized by homologues of cap-binding complex and exported to the cytoplasm via a nuclear export complex containing exportin 1 (21). Sm-independent methylations to spliced leader RNA occur posttranscriptionally in the nucleus or cytoplasm. Spliced leader RNA is bound by Sm proteins in the cytosol, an interaction required for accurate 3′-end trimming and further Sm-dependent 5′-end methylation. Localization of these SmD1-dependent events has not been determined experimentally, but they could occur in the cytoplasmic and/or nuclear compartments. Spliced leader RNA returns to the nucleus to participate in trans-splicing in an Sm complex-dependent manner.
Several components of the nuclear import pathway have been described in eukaryotes. For snRNAs, binding of the Sm complex followed by acquisition of the m2,2,7G cap, recognized by snurportin 1, creates the nuclear import signal (3). A similar pathway may function in the nuclear import of kinetoplastid U1, U2, and U4 snRNAs, which possess m2,2,7G caps and Sm-binding sites. Spliced leader RNA may require an alternative nuclear import mechanism due to the absence of an m2,2,7G cap. The location(s) of 5′ cap methylation acquisition has not been determined, and thus an intermediate cap structure may distinguish substrate spliced leader RNA that is destined to be imported into the nucleus from the fully mature cap 4 on trans-spliced mRNA in the cytoplasm. For translation, precursor spliced leader might not be recognized by cytosolic cap-binding proteins such as eIF4E, consistent with the absence of polyribosome formation on mRNAs lacking mature cap 4 structures (22). The identification and experimental analysis of nuclear import factors, 5′ cap structure of nuclear import-blocked spliced leader RNA, and the cap 4 methyltransferases will clarify the roles of Sm proteins and subsequent modifications in spliced leader RNA nuclear import.
Acknowledgments
This work was supported by NIH grants AI34536 to D.A.C. and AI054496 to N.R.S. and by grant MSM123100002 from the Ministry of Education of the Czech Republic to J.L. G.M.Z. is a predoctoral trainee of Microbial Pathogenesis Training Grant 2-T32-AI-07323.
We thank George Cross for providing T. brucei 29-13 cells, Paul T. Englund for the pZJM vector, and Kent Hill for use of the Zeiss Axiocam fluorescence microscope.
REFERENCES
- 1.Bakin, A., and J. Ofengand. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyl transferase center: analysis by the application of a new sequencing technique. Biochemistry 32:9754-9762. [DOI] [PubMed] [Google Scholar]
- 2.Bangs, J. D., P. F. Crain, T. Hashizume, J. A. McCloskey, and J. C. Boothroyd. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267:9805-9815. [PubMed] [Google Scholar]
- 3.Bordonné, R. 2000. Functional characterization of nuclear localization signals in yeast Sm proteins. Mol. Cell. Biol. 20:7943-7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Lange, T., A. Y. Liu, L. H. Van der Ploeg, P. Borst, M. C. Tromp, and J. H. Van Boom. 1983. Tandem repetition of the 5′ mini-exon of variant surface glycoprotein genes: a multiple promoter for VSG gene transcription? Cell 34:891-900. [DOI] [PubMed] [Google Scholar]
- 5.Ehlers, B., J. Czichos, and P. Overath. 1987. RNA turnover in Trypanosoma brucei. Mol. Cell. Biol. 7:1242-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Günzl, A., M. Cross, Z. Palfi, and A. Bindereif. 1993. Assembly of the U2 small nuclear ribonucleoprotein from Trypanosoma brucei. A mutational analysis. J. Biol. Chem. 268:13336-13343. [PubMed] [Google Scholar]
- 7.Laird, P. W., J. C. Zomerdijk, D. de Korte, and P. Borst. 1987. In vivo labelling of intermediates in the discontinuous synthesis of mRNAs in Trypanosoma brucei. EMBO J. 6:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang, X.-H., Y. Xu, and S. Michaeli. 2002. The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the spliced leader RNA. RNA 8:237-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lücke, S., G. L. Xu, Z. Palfi, M. Cross, V. Bellofatto, and A. Bindereif. 1996. Spliced leader RNA of trypanosomes: in vivo mutational analysis reveals extensive and distinct requirements for trans splicing and cap 4 formation. EMBO J. 15:4380-4391. [PMC free article] [PubMed] [Google Scholar]
- 10.Mair, G., E. Ullu, and C. Tschudi. 2000. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem. 275:28994-28999. [DOI] [PubMed] [Google Scholar]
- 11.Mandelboim, M., C. L. Estraño, C. Tschudi, E. Ullu, and S. Michaeli. 2002. On the role of exon and intron sequences in trans-splicing utilization and cap 4 modification of the trypanosomatid Leptomonas collosoma spliced leader RNA. J. Biol. Chem. 277:35210-35218. [DOI] [PubMed] [Google Scholar]
- 12.Palfi, Z., S. Lücke, H.-W. Lahm, W. S. Lane, V. Kruft, E. Bragado-Nilsson, B. Séraphin, and A. Bindereif. 2000. The spliceosomal snRNP core complex of Trypanosoma brucei: cloning and functional analysis reveals seven Sm protein constituents. Proc. Natl. Acad. Sci. USA 97:8967-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raker, V. A., G. Plessel, and R. Lührmann. 1996. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 15:2256-2269. [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts, T. G., J. M. Dungan, K. P. Watkins, and N. Agabian. 1996. The SLA RNA gene of Trypanosoma brucei is organized in a tandem array which encodes several small RNAs. Mol. Biochem. Parasitol. 83:163-174. [DOI] [PubMed] [Google Scholar]
- 15.Stark, H., P. Dube, R. Lührmann, and B. Kastner. 2001. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409:539-542. [DOI] [PubMed] [Google Scholar]
- 16.Sturm, N. R., and D. A. Campbell. 1999. The role of intron structures in trans-splicing and cap 4 formation for the Leishmania spliced leader RNA. J. Biol. Chem. 274:19361-19367. [DOI] [PubMed] [Google Scholar]
- 17.Sturm, N. R., J. Fleischmann, and D. A. Campbell. 1998. Efficient trans-splicing of mutated spliced leader exons in Leishmania tarentolae. J. Biol. Chem. 273:18689-18692. [DOI] [PubMed] [Google Scholar]
- 18.Sturm, N. R., M. C. Yu, and D. A. Campbell. 1999. Transcription termination and 3′-end processing of the spliced leader RNA in kinetoplastids. Mol. Cell. Biol. 19:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tschudi, C., F. F. Richards, and E. Ullu. 1986. The U2 RNA analogue of Trypanosoma brucei gambiense: implications for a splicing mechanism in trypanosomes. Nucleic Acids Res. 14:8893-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walke, S., E. Bragado-Nilsson, B. Séraphin, and K. Nagai. 2001. Stoichiometry of the Sm proteins in yeast spliceosomal snRNPs supports the heptamer ring model of the core domain. J. Mol. Biol. 308:49-58. [DOI] [PubMed] [Google Scholar]
- 21.Zeiner, G. M., N. R. Sturm, and D. A. Campbell. 2003. Exportin 1 mediates nuclear export of the kinetoplastid spliced leader RNA. Eukaryot. Cell 2:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeiner, G. M., N. R. Sturm, and D. A. Campbell. 2003. The Leishmania tarentolae spliced leader contains determinants for association with polysomes. J. Biol. Chem. 278:38269-38275. [DOI] [PubMed] [Google Scholar]