Abstract
To investigate the characteristics of main Streptococcus pneumoniae clones of serotype 6D (ST282 and ST3171) in South Korea, antimicrobial susceptibility testing was performed, and 11 genes around the cps locus were sequenced on ST2826D, ST31716D, and ST816A isolates. The antimicrobial susceptibility patterns were very similar between clones belonging to the same clonal complex, ST816A and ST2826D; nonsusceptibilities to penicillin and cefuroxime, high MICs of ceftriaxone, and high resistance rates to trimethoprim-sulfamethoxazole. However, ST31716D isolates showed resistance to only macrolides and clindamycin. The sequences of 11 genes around the cps locus indicated the same genetic backgrounds between the ST816A and ST2826D isolates. On the other hand, ST31716D isolates showed nucleotide and amino acid differences from ST816A and ST2826D isolates in most genes, indicating a different genetic background. The mosaic structure of dexB gene in ST2826D isolates indicated that recombination might occur in the dexB gene. Our results suggest that the multidrug-resistant ST2826D pneumococcal clone has emerged by serial genetic recombination, including capsular switch.
INTRODUCTION
Streptococcus pneumoniae is a common and important pathogen that causes invasive and noninvasive bacterial diseases in infants, children, and adults. S. pneumoniae comprises more than 90 serotypes. Although serogroup 6 was classically considered to consist of serotypes 6A and 6B, another two serotypes, 6C and 6D, have recently been described (14, 22). It has been postulated that serotypes 6C and 6D emerged by the replacement of wciNβ in the capsular loci of serotypes 6A and 6B, respectively (3, 22). However, two recent papers reported that the serotypes 6C and 6D emerged just by the replacement of the wciNβ and wciP mutation (5, 23). Bratcher et al. (5) proposed that serotype 6C has been produced by introduction of a DNA fragment spanning the capsular polysaccharide synthesis (cps) locus irrespective of serotypes 6A and serotype 6D has resulted from a recombination between the cps loci of serotypes 6B and 6C. In addition, we also showed the emergence of cps loci of serotypes 6C and 6D by complicated recombination (23).
To date, S. pneumoniae serotype 6D isolates have been reported in several regions including Fiji, South Korea, China, Japan, Hong Kong, Denmark, Poland, Australia, and Peru (3, 4, 7, 12, 16, 18, 20, 21, 25). Especially, serotype 6D is relatively prevalent in South Korea, comprising >10% of serogroup 6 isolates (1, 8). Choi et al. (8) reported that serotype 6D isolates from South Korea consist of three sequence types (STs) in multilocus sequence typing (MLST) analysis: ST189, ST282, and ST3171. Interestingly, ST189 and ST282 are single-locus variants (SLVs) of ST81, differing in the aroE locus. Although ST81 belongs to the Spain23F-1 clone, a globally disseminated multidrug-resistant (MDR) pneumococcal clone (15), it was also found in serotype 6A from South Korea (2). However, relationships between ST282 showing serotype 6D (ST2826D) and ST816A isolates have not been investigated.
In the present study, we sequenced the region around the cps locus to identify the evolutionary scenario resulting in the newly found serotype 6D. In addition, we compared the antimicrobial resistances among serotypes and genotypes.
MATERIALS AND METHODS
S. pneumoniae isolates.
Forty-five S. pneumoniae isolates from South Korea were investigated: 26 ST816A isolates, 16 ST2826D isolates (including ST3595 and ST4672, which are SLVs of ST282), and 3 ST31716D isolates (Table 1). All isolates were collected as part of several surveillance studies of ANSORP (Asian Network for Surveillance of Resistant Pathogens) or KONSID (Korean Network for Studies of Infectious Diseases). They were isolated from seven tertiary-care hospitals from 1997 to 2009. Most of them (31 isolates [68.9%]) were isolated from sputum, and three and one isolates were obtained from cerebrospinal fluid and blood, respectively (Table 1). Isolates obtained between 2008 and 2009 were identified to be invasive, but the clinical significance for the others is not known. Serotyping was done using the capsular Quellung reaction with commercial antisera (Statens Serum Institute, Copenhagen, Denmark), as recommended by the manufacturer. In addition, a previously described serotype-specific PCR method (13) was used to identify serotype 6D. MLST was performed as described previously (11).
Table 1.
S. pneumoniae serotype 6D isolates used in this study, their MLST data, and antimicrobial susceptibilities
| Serotype | STa | Isolate | Yr | Specimenb | Penicillin resistancec | Resistance profiled |
|---|---|---|---|---|---|---|
| 6A | 81 (4-4-2-4-4-1-1) | Kor 146 | 1998 | Transtracheal aspirate | I | FUR-ERY-AZI-CLA-CD |
| 420 | 1999 | ND | I | FUR-ERY-SXT | ||
| K10-25 | 2000 | Eye discharge | I | FUR-ERY-AZI-CLA | ||
| Kor 14 | 2000 | Sputum | I | FUR-ERY | ||
| K08-52 | 2008 | Sputum | R | FUR-ERY-AZI-CLA-CD | ||
| K01-39 | 2008 | CSF | R | FUR-ERY | ||
| K07-13 | 2008 | Sputum | I | FUR-SXT | ||
| K08-9 | 2008 | CSF | I | FUR | ||
| K08-12 | 2008 | Sputum | I | FUR-ERY-AZI-CLA-CD | ||
| K13-1 | 2008 | Sputum | I | FUR-ERY-AZI-CLA-CD | ||
| K13-4 | 2008 | Sputum | I | FUR-ERY-AZI-CLA-CD | ||
| K13-17 | 2008 | Sputum | I | FUR-ERY-AZI-CLA-CD | ||
| K13-50 | 2008 | Sputum | I | FUR-ERY-AZI-CLA-CD | ||
| K16-94 | 2008 | CSF | I | FUR-ERY-AZI-CLA-CD-SXT | ||
| K13-75 | 2009 | Sputum | R | FUR-ERY-AZI-CLA-CD | ||
| K13-59 | 2009 | Sputum | R | FUR-ERY-AZI-CLA-CD | ||
| K13-70 | 2009 | Sputum | I | FUR-ERY-AZI-CLA-CD | ||
| K13-72 | 2009 | Sputum | I | FUR-ERY-AZI-CLA-CD | ||
| K13-78 | 2009 | Sputum | I | FUR-ERY-AZI-CLA-CD | ||
| K13-85 | 2009 | Sputum | I | FUR-ERY-AZI-CLA-CD-SXT | ||
| K13-87 | 2009 | Sputum | I | FUR-ERY-AZI-CLA-CD | ||
| K13-98 | 2009 | Sputum | I | FUR-ERY-AZI-CLA-CD-SXT | ||
| K13-102 | 2009 | Sputum | S | ERY | ||
| K13-120 | 2009 | Sputum | I | ERY-AZI-CLA-CD | ||
| K13-130 | 2009 | Sputum | I | FUR-ERY-AZI-CLA-CD | ||
| K15-64 | 2009 | Sputum | I | FUR-ERY | ||
| 6D | 282 (30-4-2-4-4-1-1) | 04-8 | 2004 | Sputum | R | FUR-ERY-AZI-CLA |
| 05-246 | 2005 | Sputum | R | FUR-ERY-AZI-CLA | ||
| 05-387 | 2005 | Pus | R | FUR-ERY -CLA | ||
| 06-265 | 2006 | Sputum | I | FUR-ERY -CLA | ||
| 07-056 | 2007 | Sputum | R | FUR-ERY-AZI | ||
| 07-077 | 2007 | Sputum | R | FUR-ERY-AZI-CLA-CIP | ||
| 07-107 | 2007 | Transtracheal aspirate | I | FUR-ERY-AZI-LEV-GAT-CIP | ||
| K15-99 | 2008 | ND | I | FUR-ERY-AZI-CLA-CD-SXT | ||
| K15-129 | 2008 | ND | I | FUR-ERY-AZI-CLA | ||
| K15-115 | 2009 | ND | I | FUR-ERY-AZI-CLA-SXT | ||
| K15-17 | 2009 | Sputum | I | FUR-ERY | ||
| K13-108 | 2009 | Sputum | I | FUR-LEV-GAT-CIP | ||
| K13-109 | 2009 | Sputum | I | LEV-GAT-CIP | ||
| K13-110 | 2009 | Sputum | I | FUR-LEV-GAT-CIP | ||
| 4762 (30-4-2-4-30-1-1) | K15-60 | 2008 | Sputum | I | FUR | |
| 3595 (30-4-2-1-4-4-1) | B0704-047 | 2007 | Blood | I | FUR-ERY-AZI | |
| 3171 (8-13-9-6-78-119-14) | Kor 74 | 1997 | Throat swab | S | ERY-AZI-CLA-CD | |
| 05-447 | 2005 | Transtracheal aspirate | S | ERY-AZI-CLA-CD | ||
| K13-22 | 2009 | Sputum | S | ERY-AZI-CLA-CD |
aroE-gdh-gki-recP-spi-xpt-ddl.
ND, not described; CSF, cerebrospinal fluid.
MIC breakpoints of penicillin susceptibility: susceptible (S), ≤0.06 mg/liter; intermediate (I), 0.12 to 1 mg/liter; and resistant (R), ≥2 mg/liter.
FUR, cefuroxime; AMX, amoxicillin; A/C, amoxicillin-clavulanate; AXO, ceftriaxone; ERY, erythromycin; AZI, azithromycin; CLA, clarithromycin; CD, clindamycin; LEV, levofloxacin; MOX, moxifloxacin; GAT, gatifloxacin; CIP, ciprofloxacin; SXT, trimethoprim-sulfamethoxazole.
Antimicrobial susceptibility testing.
MIC was determined for all S. pneumoniae isolates by the broth microdilution method, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (9). In vitro susceptibility was tested for 14 antimicrobial agents: penicillin, amoxicillin, amoxicillin-clavulanate, ceftriaxone, cefuroxime, erythromycin, azithromycin, clarithromycin, levofloxacin, moxifloxacin, gati-floxacin, ciprofloxacin, clindamycin, and trimethoprim-sulfameth-oxazole. Penicillin resistance was evaluated by the current CLSI breakpoints for oral therapy (i.e., intermediate, 0.12 to 1 mg/liter; resistance, ≥2 mg/liter) (9). S. pneumoniae ATCC 49619, Staphylococcus aureus ATCC 29213, and Escherichia coli ATCC 25922 were used as control strains.
Sequencing.
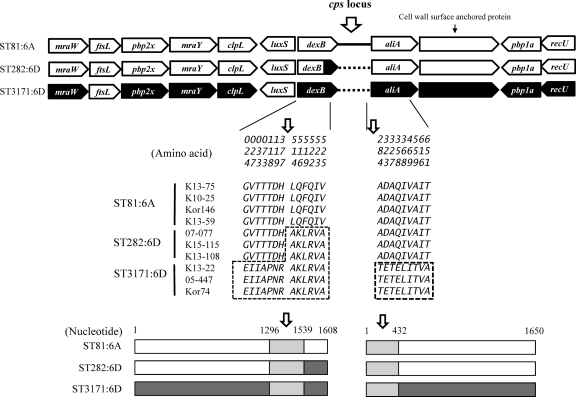
Eleven genes around the cps locus (mraW, ftsL, pbp2x, mraY, clpL, luxS, dexB, aliA, cell wall surface-anchored protein gene [SPN23F_03400 in ATCC 700669], pbp1a, and recU) of four ST816A, three ST2826D, and three ST31716D isolates (Fig. 1) were sequenced using the primers listed in Table 2. The isolates were selected according to locality and isolation year.
Fig 1.
Structure of regions around cps locus in ST816A, ST2826D, and ST31716D isolates. The amino acid and nucleotide variations in dexB and aliA genes are indicated. Filled arrows indicate the tentative recombination site in ST2826D isolates.
Table 2.
Primers used in this study
| Gene(s) | Primer | Sequence (5′–3′) |
|---|---|---|
| mraW and ftsL | mraW-F | GACAAGTGCAAGCTGGTCG |
| ftsL-R | GAGTTGCTCTCTTACATAGGA | |
| mraY | mraY-F | GTTGTTCAGAAGCAAGATGTTC |
| mraY-R | GAGCAGCCTAAAGTTAGCTTTC | |
| mraY-R1 | CCC ATC AAT TGG TTA AAT AAA TC | |
| clpL | clpL-F1 | TAGGGTAGCGGTTTAACTAGTC |
| clpL-R1 | GCGACGAACACGTTCTGTTAA | |
| clpL-F2 | CTTGCTAAGCAATTGGCACTC | |
| clpL-R2 | AGGACCACAGCCTCTTCTCT | |
| luxS | luxS-F | GCCTATATGTGTAAATCACGAGA |
| luxS-R | CCAATACGACCGCTTATATCG | |
| Cell wall surface- | SPN23-F1 | CTTGGAACTTCAAGACAAGGC |
| anchored protein genea | SPN23-R1 | ATGATCATCTCCGCTGACATG |
| SPN23-F2 | GAGACAACAGCAGAGTACTTG | |
| SPN23-R2 | ACGTCCGTTGAGCGTTACTG | |
| SPN23-F3 | GCTAGGTGGCTACAGCATGA | |
| SPN23-R3 | ATTTCCACCAGAATCACCACG | |
| SPN23-F4 | CTCAAGCAAGCAACTTGGAAA | |
| SPN23-R4 | TGTATTCAAAAATGGAGCTCGC | |
| SPN23-R4-1 | CAA AAT CAG AAT CCT CAA CC | |
| pbp1A | P1A-F2 | GAGCTCCAAGTTGGGCGAT |
| recU | recU-R | CCAAAACCAAAGCTGTAGCCA |
SPN23F_03400 in ATCC 700669.
GenBank accession numbers.
The sequences have been deposited in the GenBank database under accession numbers JN645687 to JN645796.
RESULTS
Antimicrobial susceptibility.
The results of antimicrobial susceptibility testing for the 45 S. pneumoniae isolates belonging to three groups are shown in Table 1. Notably, nearly all ST816A and ST2826D isolates were susceptible to penicillin, but three isolates of ST31716D were susceptible (Table 1), which was also demonstrated in the penicillin MIC distribution (see the supplemental material). MIC distribution for amoxicillin, amoxicillin/clavulanate, and ceftriaxone also differed among groups (see the supplemental material). Unlike penicillins and cephalosporins, all ST31716D isolates showed resistance to erythromycin, azithromycin, and clarithromycin (Table 1). Fluoroquinolone-resistant isolates were found only in ST2826D. Four or five ST2826D isolates from two different hospitals were resistant to fluoroquinolones. Although 24 (92.3%) and 9 (56.3%) of the ST816A and ST2826D isolates, respectively, were nonsusceptible to trimethoprim-sulfamethoxazole, all isolates of ST31716D were susceptible to it (Table 1; see also the supplemental material).
Structure of regions around cps locus.
The structure of the 11 genes around the cps locus in the three groups (ST816A, ST2826D, and ST31716D) is shown in Fig. 1. Most genes, except dexB, were nearly identical between ST816A and ST2826D isolates at both the amino acid and nucleotide levels. On the other hand, ST31716D isolates showed amino acid differences from ST816A and ST2826D isolates in most genes except ftsL and luxS. Amino acid variations were found: 2 in mraW, 62 in pbp2x, 15 in mraY, 13 in clpL, 9 in aliA, 28 in the cell wall surface-anchored protein gene, 57 in pbp1a, and 5 in recU. In addition, inserted sequences of 9 bp in the cell wall surface-anchored protein gene were found in only three ST31716D isolates.
Unlike the rest of the genes, dexB of ST2826D isolates showed a mosaic structure (Fig. 1). By amino acid 377 (nucleotide 1296) of dexB, ST2826D showed the same sequences with ST816A. However, from amino acid 514 (nucleotide 1539) onward, it shared the same sequences with ST31716D, which were clearly different from ST816A. For the aliA gene, all isolates of three groups showed the same sequences, but ST31716D showed sequence variations from amino acid 284 (nucleotide 432).
DISCUSSION
S. pneumoniae serotype 6D isolates have been found in several countries and are relatively abundant in South Korea (8, 23). In our study, two main clones (ST282 and ST3171) have been identified among S. pneumoniae serotype 6D isolates from Korea. In addition to these two clones, ST189, a SLV of ST81, was also identified previously (8). Besides these STs from Korea, very diverse STs have been identified worldwide, including Australia (ST4241), Finland (ST5163), Poland (ST948, ST2181, ST1612, and ST4734), Peru (ST6148), Japan (ST2924), Fiji (ST639, ST473, and ST4240), China (ST982 and ST4190), and Hong Kong (ST5085 and ST5086) (7, 12, 16, 18, 20, 24, 25). Of these, only four STs from Australia, China, and Hong Kong, ST4241 (7-13-9-6-10-6-14), ST982 (8-13-9-60-78-119-6), ST5085 (8-13-241-60-78-119-6), and ST5086 (8-13-9-60-78-1-6), might belong to the same clonal complex with ST3171 (8-13-9-6-78-119-14), differing at two or three alleles. Thus, it could be inferred that serotype 6D isolates might occur independently worldwide.
In a previous study, another 6D clone, ST189, was reported in South Korea (8). ST282 and ST189 belong to the same clonal complex, CC81, as does the representative resistant pneumococcal clone, Spain23F-1. Both ST282 and ST189 differ from ST81 at aroE. Since the ST1896D isolate was not found in our collections, we could not include it in our analysis. Two different clones of 6D pneumococci (ST282 and ST3171) from South Korea showed different antimicrobial susceptibility profiles, which was partially suggested in a previous study (8). Presently, we identified different resistance profiles in most antimicrobial agents tested between two main 6D pneumococcal isolates from South Korea, which may indicate different genetic and evolutionary backgrounds. In addition, the antimicrobial susceptibility profiles of ST2826D isolates were very similar to those of the ST816A isolates. With respect to antimicrobial resistance, ST2826D may be closer to ST816A than ST31716D.
The structure of the 11 genes around cps locus evidenced the close relatedness between ST2826A and ST816A. The complete sequence identity of four genes of the cps locus (wchA, wciN, wciO, and wciP) between ST2826D and ST31716D isolates was described in our previous study (23). ST2826D is thought to have originated as a result of capsular switch at ST69519A in the United States (6). Sequence comparison suggests that the genetic recombinant site is located between nucleotides 1296 and 1539 of dexB at the left side. Although the genetic recombinant site at the right side could not be identified, it is thought to be any site between rmlA and rmlC, which are the right and low divergent genes of the cps locus of serogroup 6 (17).
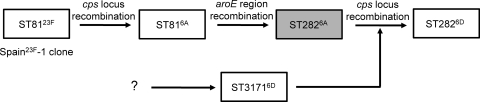
Figure 2 illustrates the evolutionary scenario on the emergence of ST2826D isolates in South Korea. ST81 is known as a genotype of the Spain23F pneumococcal clone, the most globally prevalent clone (15, 19). However, ST81 isolates showing serotype 6A (ST816A) are prevalent in South Korea (2). According to a recent study of genome sequencing of large ST81 pneumococcal collections worldwide, ST816A is a result of genetic recombination of cps locus, that is, a capsular switch (10). Since ST81 and ST282 were different only at the aroE locus, a genetic recombination of regions including aroE might explain the emergence of ST2826A from ST816A. Although the ST2826A isolate was not identified in South Korea, an ST2826A isolate has been found in Japan (23). Finally, ST2826D might have emerged by a genetic recombination of the cps locus of ST31716D, which has existed in South Korea since at least 1996 (Table 1) (8). A clonal complex including ST3171 might have represented serotype 6D long before that, because it shares four or five alleles with ST4241, ST982, ST5085, and ST5086, which also represent serotype 6D. Thus, serial recombination may have led to the emergence of high antimicrobial-resistant serotype 6D pneumococcal clone in South Korea.
Fig 2.
Hypothesis on the emergence of ST2826D clone.
In the present study, we demonstrate by the sequencing of genes around the cps locus and antimicrobial susceptibility testing that the ST2826D clone has emerged by genetic recombination of the cps locus from the ST31716D clone.
Supplementary Material
ACKNOWLEDGMENTS
The S. pneumoniae isolates used in this study were obtained from the Asian Bacterial Bank of the Asia Pacific Foundation for Infectious Diseases (Seoul, Korea).
This study was supported by a Samsung Biomedical Research Institute grant.
Footnotes
Published ahead of print 14 December 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Baek JY, Ko KS, Song J-H. 2011. Streptococcus pneumoniae serotype 6D cross-reacting with serotype 6A, 6B, and 6C factor sera. J. Clin. Microbiol. 49:765–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baek JY, et al. 2011. Comparison of genotypes of Streptococcus pneumoniae serotypes 6A and 6B before and after the introduction of PCV7 vaccination in Korea. Diagn. Microbiol. Infect. Dis. 69:370–375 [DOI] [PubMed] [Google Scholar]
- 3. Bratcher PE, Park IH, Hollingshead SK, Nahm MH. 2009. Production of a unique pneumococcal capsule serotype belonging to serogroup 6. Microbiology 155:576–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bratcher PE, Kim KH, King JH, Hong JY, Moon MH. 2010. Identification of natural pneumococcal isolates expressing serotype 6D by genetic, biochemical and serological characterization. Microbiol. 156:555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bratcher PE, et al. 2011. Evolution of the capsular gene locus of Streptococcus pneumoniae serogroup 6. Microbiology 157:189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brueggemann AB, Pai R, Crook DW, Beall B. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 3:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang B, et al. 2010. Isolation of Streptococcus pneumoniae serotypes 6C and 6D from the nasopharyngeal mucosa of healthy Japanese children. Jpn. J. Infect. Dis. 63:381–383 [PubMed] [Google Scholar]
- 8. Choi EH, et al. 2010. Prevalence and genetic structures of Streptococcus pneumoniae serotype 6D, South Korea. Emerg. Infect. Dis. 16:1751–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clinical Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing, 21st informational supplement, M100–S21. CLSI, Wayne, PA [Google Scholar]
- 10. Croucher NJ, et al. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Enright MC, Spratt BG. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049–3069 [DOI] [PubMed] [Google Scholar]
- 12. Ho P-L, Ang I, Chow K-H, Lai EL, Chiu SS. 2010. The prevalence and characteristics of Streptococcus pneumoniae isolates expressing serotypes 6C and 6D in Hong Kong prior to the introduction of the 7-valent pneumococcal conjugate vaccine. Diagn. Microbiol. Infect. Dis. 68:439–444 [DOI] [PubMed] [Google Scholar]
- 13. Jacobs MR, et al. 2009. Occurrence, distribution, and origins of Streptococcus pneumoniae serotype 6C, a recently recognized serotype. J. Clin. Microbiol. 47:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jin P, et al. 2009. First report of putative Streptococcus pneumoniae serotype 6D among nasopharyngeal isolates from Fijian children. J. Infect. Dis. 200:1375–1480 [DOI] [PubMed] [Google Scholar]
- 15. Klugman KP. 2002. The successful clone: the vector of dissemination of resistance in Streptococcus pneumoniae. J. Antimicrob. Chemother. 50(S2):1–5 [DOI] [PubMed] [Google Scholar]
- 16. Kuch A, Sadowy E, Skoczyńska A, Hryniewicz W. 2010. First report of Streptococcus pneumoniae serotype 6D isolates from invasive infections. Vaccine 28:6406–6407 [DOI] [PubMed] [Google Scholar]
- 17. Mavroidi A, et al. 2004. Evolutionary genetics of the capsular locus of serogroup 6 pneumococci. J. Bacteriol. 186:8181–8192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mercado E, et al. 2011. First report of Streptococcus pneumoniae serotype 6D in South America. J. Clin. Microbiol. 49:2080–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McGee L, et al. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nahm MH, et al. 2011. A report of Streptococcus pneumoniae serotype 6D in Europe. J. Med. Microbiol. 60:46–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oftadeh S, Satzke C, Gilbert GL. 2010. Identification of newly described Streptococcus pneumoniae serotype 6D by use of the Quellung reaction and PCR. J. Clin. Microbiol. 48:3378–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park IH, et al. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 45:1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song JH, Baek JY, Ko KS. 2011. Comparison of capsular genes of Streptococcus pneumoniae serotype 6A, 6B, 6C, and 6D isolates. J. Clin. Microbiol. 49:1758–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Satzke C, et al. 2010. Molecular epidemiology of Streptococcus pneumoniae serogroup 6 isolates from Fijian children, including newly identified serotypes 6C and 6D. J. Clin. Microbiol. 48:4298–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yao K, et al. 2011. Type distribution of serogroup 6 Streptococcus pneumoniae and molecular epidemiology of newly identified serotypes 6C and 6D in China. Diagn. Microbiol. Infect. Dis. 70:291–298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.