Abstract
Fruiting body development in fungi is a complex cellular differentiation process that is controlled by more than 100 developmental genes. Mutants of the filamentous fungus Sordaria macrospora showing defects in fruiting body formation are pertinent sources for the identification of components of this multicellular differentiation process. Here we show that the sterile mutant pro11 carries a defect in the pro11 gene encoding a multimodular WD40 repeat protein. Complementation analysis indicates that the wild-type gene or C-terminally truncated versions of the wild-type protein are able to restore the fertile phenotype in mutant pro11. PRO11 shows significant homology to several vertebrate WD40 proteins, such as striatin and zinedin, which seem to be involved in Ca2+-dependent signaling in cells of the central nervous system and are supposed to function as scaffolding proteins linking signaling and eukaryotic endocytosis. Cloning of a mouse cDNA encoding striatin allowed functional substitution of the wild-type protein with restoration of fertility in mutant pro11. Our data strongly suggest that an evolutionarily conserved cellular process controlling eukaryotic cell differentiation may regulate fruiting body formation.
Fruiting bodies are highly complex multicellular structures that are generated during the sexual life cycle of filamentous fungi. In ascomycetous fungi, such as Neurospora crassa, Sordaria macrospora, and Aspergillus nidulans, fruiting bodies are termed ascomata and enclose sexual meiosporangia (asci) and meiospores (ascospores). While ascospores arise from hyphae during the meiotic cycle, all tissues forming the fruiting body arise from haploid, nondikaryotic hyphae. This feature distinguishes fruiting body formation in ascomycetes clearly from that in basidiomycetous fungi, in which dikaryotic hyphae are involved in both, the meiotic cycle and fruiting body formation (for a review, see reference 26).
The sexual life cycle of ascomycetes can be either heterothallic (self-sterile) or homothallic (self-fertile). Heterothallic fungi, such as N. crassa, exist in two mating types, A and a, and mating only occurs between sexual structures of opposite mating type. However, in S. macrospora, a homothallic fungus, every strain is able to complete the sexual cycle without a mating partner. Fruiting body development starts with the formation of female gametangia, which are enveloped by sterile hyphae to form closed spherical prefruiting bodies (protoperithecia), which can be considered primordial stages of mature fruiting bodies. Subsequent cell differentiation gives rise to an outer-pigmented peridial tissue and inner ascus initials embedded in sterile paraphyses, which leads to the formation of mature flask-like fruiting bodies, called perithecia.
It has been demonstrated for a number of ascomycetes that several genes control not only sexual development but also asexual sporulation (1). In contrast to other fungal model organisms, S. macrospora produces only meiotically derived ascospores, while asexual spores, such as conidia, are absent. Since there is no interference between sexual and asexual developmental programs, S. macrospora is an ideal model to identify genes involved in the sexual differentiation process. Molecular genetic procedures have been applied to characterize the genes that regulate this developmental process in S. macrospora (24, 31, 46). To isolate additional regulatory genes from S. macrospora, we used ethyl methanesulfonate mutagenesis to generate mutants with defects in fruiting body formation. Within our collection of mutants, we identified a class of strains which are arrested in the transition from the protoperithecium to the perithecium. Sterile mutants of this type were all designated with the prefix pro- to indicate that they form protoperithecia only. Using the pro1 mutant, we previously identified a Zn2C6 transcription factor which is essential for the morphological transition from protoperithecia to perithecia (24).
In this study, the molecular and functional analysis of mutant pro11 is illustrated, which serves the identification of the pro11 gene controlling essential steps in fruiting body development. The predicted gene product is highly homologous to mammalian brain proteins belonging to the striatin family. Members of this family, including striatin, zinedin, and SG2NA, are found in the cytosol and are also associated with membranes. In addition, they are endowed with diverse protein-protein association modules, for instance, a caveolin-binding motif, a coiled-coil structure, a Ca2+-calmodulin-binding domain, and multiple WD40 repeats homologous to WD40 repeats in the beta subunit of trimeric G-proteins. Due to these domains, proteins of the striatin family are engaged in multiple protein associations and are thought to act as scaffolds as well as signaling proteins (5, 10, 11, 16).
Here, we demonstrate that a mouse striatin gene is able to rescue the sterile phenotype of the S. macrospora pro11 mutant, strongly suggesting functional conservation of the fungal pro11 gene and the mammalian striatin gene. Complementation studies with C-terminally truncated versions of the PRO11 protein revealed different functional roles of this multidomain protein, and experimental evidence is provided for a membrane association of the PRO11 polypeptide, which shares this feature with animal homologues.
MATERIALS AND METHODS
Sordaria strains, media, growth conditions, and transformation.
Sordaria macrospora L3346 held in our laboratory collection displays a wild-type phenotype. The mutant pro11 (strain S24117) was isolated from strain L3346 after ethyl methanesulfonate mutagenesis. For ethyl methanesulfonate mutagenesis, protoplast suspensions from the wild-type strain were incubated for 45 to 60 min in a 5% (wt/vol) ethyl methanesulfonate solution (Sigma) and plated on CM medium (31) supplemented with 10.8% sucrose for regeneration (0.1% survival rate). After 24 h, individual clones were transferred to BMM fructification medium (14) to monitor clones with phenotypic variations in the development of fruiting bodies. The mutant pro11 was isolated from one of these clones, and a single-spore isolate was generated for further molecular analysis. Unless otherwise stated, standard growth conditions, genetic techniques, DNA-mediated transformation, and complementation analysis for S. macrospora were carried out as described previously (24, 31, 34, 47). Cotransformation was performed together with vector pANsCos1 (32).
Plasmids.
Cloning of S. macrospora DNA fragments with vectors pBCKS+, pQE31 (Qiagen), pDrive (Qiagen), pANsCos1 (32), and pEHN2, a derivative of plasmid pIG1783 without the egfp gene (33), was performed by standard techniques (39). Vector pEHN2 is a fungal expression vector with the strong gpd promoter of Aspergillus nidulans upstream of a single NotI cloning site. The cosmids and plasmids used in this investigation are listed in Table 1.
TABLE 1.
Cosmids and plasmids used for transformation experiments
| Plasmid | Vector | Insert |
|---|---|---|
| pANsC7 | pANsCos1 | Cosmid, S. macrospora 34-kb genomic DNA |
| pSB16 | pBCKS+ | 3.6-kb ApaI/EcoRI fragment of pANsC7, pro11 ORF |
| pSB10 | pBCKS+ | 1.8-kb ApaI/XbaI fragment of pANsC7, pro11 ORF (5′ end) |
| pSB10H(+4) | pBCKS+ | Derivative of pSB10, frameshift mutation (+4 bp) at position 961 of the pro11 ORF |
| pSB22 | pBCKS+ | Derivative of pSB16, frameshift mutation (−82 bp), positions 1469-1550 of pro11 ORF |
| pAK2 | pQE31 | 857-bp cDNA, pro11 ORF (5′ end) |
| pIG1807-24 | pEHN2 | NotI fragment of pro11 ORF |
| pIG1808-23 | pEHN2 | NotI fragment of mouse striatin ORF |
Preparation of nucleic acids and PCR.
Genomic DNA for PCR and Southern blots was isolated from S. macrospora as described previously (35). Isolation of total RNA was done from batch cultures showing prefruiting body and fruiting body formation after 3 and 6 days in Fernbach flasks, respectively. This total RNA was used for polyadenylated RNA isolation. Reverse transcriptase-coupled reverse transcription-PCR and primer extension were performed as described previously (35). The mouse striatin cDNA was amplified from mouse brain cDNA, kindly provided by M. Kilimann (Ruhr-University Bochum, Germany) with PCR primers stria3 (5′-GGCGGCCGCGACGAGCAGGCGGGGCCCGGCGT; NotI cloning site italic) and stria4 (5′-GGCGGCCGCTCATACAAAGACCTTAGCCAGGG; NotI cloning site italic), synthesized according to the mouse striatin cDNA sequence (NM_011500). Sequencing was done by the MWG-Biotech Customer Service (Ebersberg, Germany).
Antibody production.
His tag fusion derivatives of a pro11 gene fragment encoding the N terminus of the PRO11 polypeptide were synthesized in Escherichia coli strain M15. For this purpose, an 857-bp SalI/PstI fragment was amplified with primers p11-31 (5′-GTCGACATGGGCACCAACGGCGTTCATGTC; SalI cloning site italic) and p11-32 (5′-CTGCAGTCTAGACGAACGCAACGCTCCATGGG; PstI cloning site italic), with S. macrospora cDNA as a template.
For sequence verification, the amplified fragment was subcloned into the vector pDrive (Qiagen). After sequence verification, the amplified fragment was subsequently cloned into vector pQE31 (Qiagen, Germany) to generate plasmid pAK2 (Table 1). This plasmid was used to express the 5′ end of the cDNA of the pro11 gene in E. coli. Proteins containing an N-terminal His tag were purified on Ni-nitrilotriacetic acid-agarose columns (Qiagen) according to the supplier's instructions. For antiserum production, purified eluates of E. coli strains expressing the 5′ end of the pro11 open reading frame were dialyzed against 50 mM ammonium carbonate and evaporated. The production of polyclonal antiserum against PRO11 in rabbits was performed by Eurogentec (Belgium).
Protein fractionation and Western analysis.
Cell fractionation procedures for S. macrospora were performed by a series of centrifugation steps as described by Bowman et al. (8). The 10,000 × g, 100,000 × g (100K), and soluble protein fractions were separated in sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gels. The proteins were transferred to polyvinylidene difluoride Western blotting membranes (Roche, Germany) with a semidry blotting system (Biometra, Germany). Detection was carried out with a polyclonal anti-PRO11 antibody, a polyclonal antibody against actin of Aspergillus niger (Acris Antibodies, Bad Nauheim, Germany), and an antibody against mitochondrial porin from Saccharomyces cerevisiae (REF; generous gift of W. Kunau, Bochum University) with the chemiluminescence Western blotting kit (Roche, Germany) following the manufacturer's instructions.
Light microscopy.
Light microscopic observations were conducted with a Zeiss Axiophot microscope with appropriate filter sets. For light microscopy, nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole, 0.5 μg/μl).
DNA and protein sequence analysis.
Sordaria macrospora protein sequences were used as query sequences in Blast searches (2, 17). Preliminary A. fumigatus genome sequences were obtained from the the Institute for Genomic Research (TIGR) (http://www.tigr.org). Sequences of Magnaporthe grisea were obtained from the Magnaporthe Sequencing Project (Release I, Ralph Dean, Fungal Genomics Laboratory at North Carolina State University, http://www.fungalgenomics.ncsu.edu) and the Whitehead Institute/MIT Center for Genome Research (www-genome.wi.mit.edu). Sequences of the basidiomycete Phanerochaete chrysosporium were obtained from the DOE Joint Genome Institute (http://www.jgi.doe.gov/programs/whiterot.htm).
Amino acid sequences and sequence alignments were done with the Clustal W program (44). Prediction of coiled-coil domains of the proteins was performed with the programs Coils (23), Paircoil (7), and Multicoil (48) at the ExPASY server (http://ca.expasy.org/); prediction of WD40 repeats was carried out at the Biomolecular Engineering Research Center (BMERC) (http://bmerc-www.bu.edu/bioinformatics/WD40repeat.html). and prediction of potential calmodulin binding sites was done at the Calmodulin Target Database (http//calcium.oci.utoronto.ca).
Nucleotide sequence accession number.
Sequence data for the S. macrospora pro11 gene have been submitted to the EMBL database under accession number AJ564211.
RESULTS
Characterization of the sterile mutant pro11.
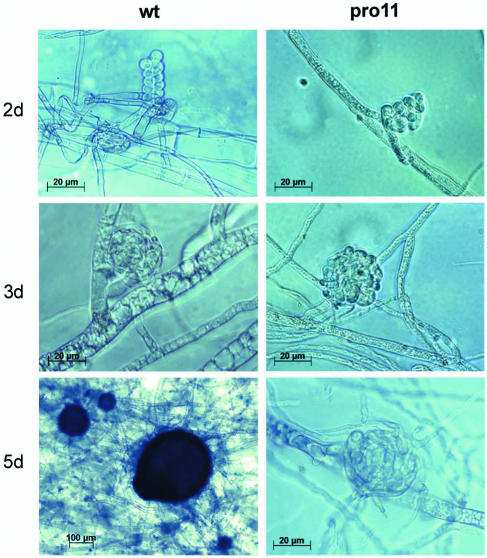
The sterile mutant pro11 was isolated from the S. macrospora wild-type strain after ethyl methanesulfonate mutagenesis and displays defects in early sexual development. Although the mutant strain showed the formation of ascogonia and protoperithecia, it was unable to perform the transition to mature fruiting bodies (Fig. 1). As can also be seen, this process occurred in the wild-type strain after 5 days on fructification medium. Consequently, any meiotic or postmeiotic division, which is a prerequisite for the formation of sexual spores in the wild type, was lacking in the pro11 mutant.
FIG. 1.
Sexual developmental stages of the wild-type (wt) and mutant (pro11) strains. Differential interference microscopy identified ascogonia (wild type, pro11), protoperithecia (wild type, pro11), and young perithecia (only wild-type strain). Strains were grown on fructification medium and examined after growth at 25°C for the number of days indicated.
The mycelium of pro11 showed an increased density of aerial hyphae, giving the mycelium a cotton-like appearance. Furthermore, pro11 displayed vegetative growth similar to the wild-type strain, with a growth rate of 2.9 cm/day on fructification medium, and there was no general impairment in essential vegetative functions. When inoculated on fructification medium, pro11 produced only 140 protoperithecia/cm2 and thus displays a significant reduction in protoperithecium formation compared to the wild-type strain (231 protoperithecia/cm2). Fertile fruiting bodies were absent in the mutant strain, which showed an arrest in the transition from protoperithecia to perithecia. Conventional genetic analysis of more than 10 ordered tetrads from the cross pro11 × wild type revealed a Mendelian 1:1 segregation of the wild-type and the mutant phenotypes, indicating that a single gene is responsible for the mutant phenotype.
Complementation analysis identifies a gene for a WD40 repeat protein.
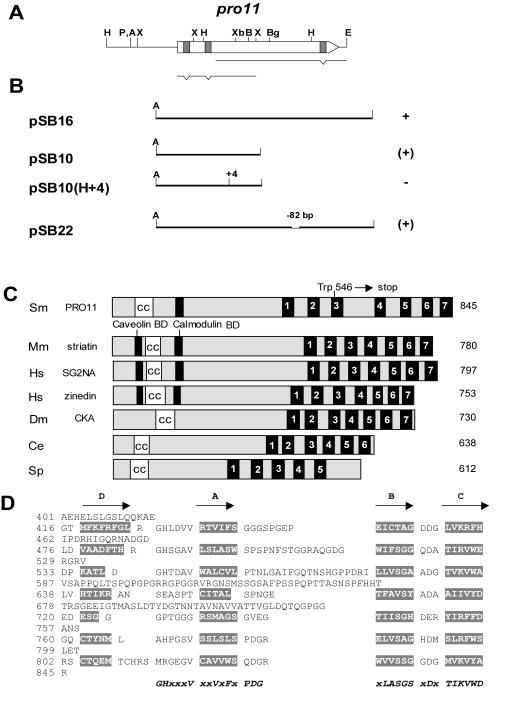
The pro11 gene was cloned by a recently described genomic complementation strategy (24) with an indexed cosmid library of S. macrospora (34). A single clone with an insert size of 34 kb, designated pANsC7, yielded fertile transformants that showed perithecium and ascospore formation. In subsequent complementation experiments, we succeeded in isolating a 3.6-kb ApaI/EcoRI subfragment of cosmid pANsC7 (Fig. 2A) that was able to complement pro11.
FIG. 2.
Complementation analysis to restore fertility in pro11 and structural comparison of eukaryotic WD40 repeat proteins with PRO11 (AJ564211). (A) Molecular organization of the pro11 gene. Restriction map of the sequenced 4.0-kb fragment carrying the pro11 gene. The location of the open reading frame is indicated by an arrow, and the three introns are marked by grey boxes. Fragments generated by reverse transcription-PCR are shown below. (B) Recombinant plasmids carrying inserts of the pro11 gene were used for complementation experiments. The ability of transformants to restore fertility (wild-type phenotype) is indicated by a +, while no complementation is shown by −; (+) denotes transformants which produced few fertile perithecia but showed a hyphal morphology similar to that of the mutant strain. Abbreviations: A, ApaI; B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; P, PstI; X, XhoI; Xb, XbaI. (C) Schematic representation of PRO11 and some closely related proteins. White boxes represent coiled-coil domains, and numbered black boxes show the position of WD40 repeats. Black boxes indicate the position of calmodulin binding sites and caveolin binding domains in striatin, SG2NA, and zinedin. Accession numbers: Mm, Mus musculus striatin (055106); Hs, Homo sapiens SG2NA (Q13033) and H. sapiens zinedin (NP_037535); Dm, Drosophila melanogaster CKA (Q9VLT9); Ce, Caenorhabditis elegans protein encoded by the K07C5.8 gene (Q17406); Sp, Schizosaccharomyces pombe protein encoded by the SPBC1773.01 gene (O94560). (D) Alignment of the seven predicted WD40 repeats in PRO11. Putative β strands are shaded and their positions are indicated with arrows (A to D). The WD40 repeat core consensus is given at the bottom. The coordinates of the amino acid positions are marked on the left.
The pro11 gene was mapped to the only major open reading frame present on the complementing fragment. The pro11 open reading frame comprised 2,716 bp interrupted by three introns of 69 bp, 54 bp, and 55 bp, whose presence was confirmed by sequencing of corresponding cDNAs. The pro11 gene encodes a putative protein of 845 amino acids with a predicted molecular mass of 90.6 kDa and an average pI of 5.9. A polyadenylation site was found 170 bp downstream of the stop codon, and primer extension analysis revealed two transcription start sites 145 bp and 387 bp upstream of the translation start codon. These should result in transcript sizes of 2.5 kb and 2.7 kb, respectively. The pro11 gene was only very weakly expressed, and we were not able to detect signals in Northern hybridization with enriched polyadenylated mRNA (data not shown). However, in reverse transcription-PCR experiments, we were able to amplify the cDNA lacking any intron sequences. This clearly demonstrated transcriptional expression of the pro11 gene. These data are further supported by experiments with specific antibodies against the PRO11 polypeptide.
To verify that we did not identify a suppressor gene of the pro11 mutation, the mutant allele was sequenced. It differs from the wild-type pro11 gene by a single point mutation within the coding region, resulting in an in-frame stop codon (TGG Trp to TGA stop) at amino acid position 546 of PRO11 (Fig. 2C). In addition, transformation experiments were carried out with clones containing truncated versions of the pro11 gene (Fig. 2B). Only the full-length version of the pro11 gene was able to fully complement the sterile phenotype of strain pro11. Truncated versions containing the coding region for the N-terminal part of PRO11 or a deletion in the C-terminal coding part of the pro11 open reading frame (pSB10 and pSB22, respectively) resulted in only partial complementation (Fig. 2B). Partially complemented transformants were able to form fertile fruiting bodies with asci and ascospores. However, the number of fruiting bodies and ascospores was greatly reduced, and the transformants still showed an increased density of aerial hyphae. However, no complementation was observed when the N-terminal part of the pro11 open reading frame contained a frameshift mutation (pSB10H [+4]).
Characterization of the WD40 repeat protein PRO11.
Database searches revealed that the PRO11 protein showed significant sequence similarity to three mammalian brain proteins, all WD40 proteins of the striatin family, which includes striatin (055106), SG2NA (Q13033), and zinedin (NP_037535). These multimodular WD40 repeat and calmodulin binding proteins are thought to act as scaffolds, as signaling proteins, or as catalytic subunits of protein phosphatase 2A (5, 10, 11, 29). In addition, the PRO11 protein shows distinct homology with the recently identified connector of kinase to AP-1 (CKA) protein of Drosophila melanogaster (Q9VLT9), a multidomain WD40 protein, which regulates the Jun N-terminal kinase signal transduction pathway and, thereby, epithelial sheet movement that occurs during dorsal closure (12). In addition to proteins of the striatin family, PRO11 was found to be homologous to WD40 proteins of unknown function (Fig. 2C).
Within filamentous ascomycetes, the pro11 gene seems to be very conserved. Blast searches through the genomic sequences of N. crassa, Magnaporthe grisea, Aspergillus fumigatus, and the basidiomycete Phanerochaete chrysosporium indicated that they contain a single gene encoding a protein which displays 94.6%, 62.8%, 48.4%, and 28.4% amino acid identity, respectively, to PRO11. In contrast, no pro11 homologue could be identified in the genome of the yeast Saccharomyces cerevisiae.
We analyzed the predicted PRO11 protein and found that a 41-amino-acid region near the N terminus of the protein (positions 56 to 96) has the potential to form a coiled-coil structure (Fig. 2C). This domain is one of the common structural motifs mediating protein-protein interactions (22). MultiCoil and Paircoil scores indicate that this region has a >80% probability of forming a dimer (7, 48). A putative Ca2+-calmodulin binding site was predicted in the N terminus between amino acid 80 and amino acid 91. In addition, computer analysis revealed that an expanded C-terminal domain of PRO11 (419 amino acids, positions 426 to 844) consists of seven copies of the degenerate WD40 repeat (Fig. 2D). WD40 repeats are domains of approximately 40 to 60 amino acids. Each repeat contains a variable domain and a core region, which includes preferentially conserved amino acids typically bracketed by the dipeptides Gly-His and Trp-Asp, and correspond to a permutated structural repeat of four antiparallel β strands (41). All of these features are present in the predicted WD40 repeats of PRO11 (Fig. 2D).
Subcellular localization of the WD40 repeat protein in fungal cells.
It is important to consider the subcellular localization of the PRO11 WD40 protein because mammalian homologues, such as striatin, SG2NA, and zinedin, are neuronal proteins that are strictly expressed in the somatodendritic compartments. These are found in the cytosol and are also associated with membranes (11, 16, 38).
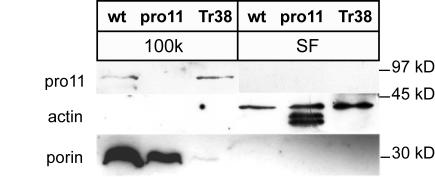
To obtain further information about the subcellular location of the fungal homologue PRO11, we raised an antibody against the N-terminal part of PRO11. In a Western blot analysis, the PRO11 antibody detected a band of about 90 kDa in the wild-type strain as well as in transformants expressing the pro11 open reading frame, but no bands in pro11 (Fig. 3). The absence of the PRO11 protein in the mutant can be explained by the missense mutation within the mutated gene affecting the stability of the truncated protein. Further analysis of the 100K and soluble fractions showed that only the 100K membrane fraction contained detectable amounts of the PRO11 protein in the wild-type strain (Fig. 3). This, however, does not exclude that trace amounts of PRO11 are present in other protein fractions as well. As a control, the soluble protein as well as the 100K membrane fraction were characterized with antibodies against actin and the mitochondrial membrane protein porin (15), respectively (Fig. 3).
FIG. 3.
Western blot analysis of polypeptides from different strains. Aliquots (30 μg) of protein fractions, as indicated, were fractionated on denaturing SDS-8% PAGE. Western blot detection of PRO11 protein was performed with a polyclonal PRO11 antibody. As a control, actin and porin were detected with specific antibodies in different protein fractions. Size markers are indicated. Abbreviations: wt, wild-type strain; pro11, pro11 mutant; Tr38, pro11 transformant carrying plasmid pIG1807-24; SF, soluble fraction; 100k, protein fractions as described in the text.
Cloning of the mouse striatin gene and functional substitution of the pro11 gene.
WD40 proteins of known crystal structure form a β-propeller fold, which is thought to create a stable platform formation of protein complexes. Despite possessing a common sequence motif, WD40 repeat proteins exhibit a high degree of functional diversity (41, 42). The best way to find functionally related WD40 proteins is by sequence comparisons of the WD40 repeat protein, ignoring the nonsurface residues (41) (those within strands A, B, and C and in the B to C turn were replaced with X's; Fig. 2D). Based on this type of analysis, the C terminus of PRO11 (445 amino acids) is highly similar to the mammalian proteins striatin, SG2NA, and zinedin.
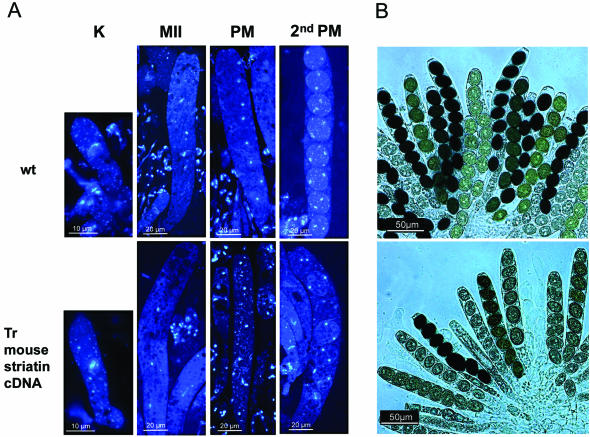
Proteins with very similar surfaces are expected to have related functions, and therefore we raised the question of whether the mammalian striatin gene was able to complement developmental defects of the S. macrospora mutant pro11. For this purpose, we amplified the coding region of the mouse striatin gene from a brain cDNA preparation and cloned the resulting PCR fragment into the fungal expression vector pEHN2. The corresponding construct, pIG1808-23 (Table 1), which contains the mouse striatin gene under the control of the gpd promoter of Aspergillus nidulans, was transformed into pro11. Transformants carrying the construct developed fertile perithecia containing rosettes of asci (Fig. 4). DAPI staining of nuclei proved that several asci contained eight ascospores with two nuclei, showing that karyogamy, meiosis I and II, and two postmeiotic mitoses as essential processes of the sexual cycle were fully restored in the transgenic strains (Fig. 4A). The frequency of unusual asci with less than eight ascospores was, however, increased (Fig. 4B). In some instances, the separation of nuclei after meiosis II or postmeiotic mitosis seemed to be impaired and led to multinuclear ascospores. This complementation analysis strongly supports the assumption that the mouse striatin gene is a functional homologue of the fungal pro11 gene.
FIG. 4.
Ascus development in the wild type and a representative pro11 transformant (Tr) carrying plasmid pIG1808-23 and expressing the mouse striatin gene. (A) DAPI staining identified nuclei during karyogamy (K), meiosis II (MII), and postmeiotic mitosis (PM). The second postmeiotic mitosis (2nd PM) occurs after spore wall formation. (B) Rosettes from a mature perithecium are shown. Note that some of the asci of the transformant carry less than eight ascospores, which are regularly seen in a wild-type ascus.
DISCUSSION
PRO11 shares several functional features with animal WD40 repeat proteins.
The pro11 gene, which was isolated by complementing the sterile S. macrospora mutant pro11, encodes a protein with three recognizable structural domains: starting from the N terminus, these are the coiled-coil region, the putative calmodulin binding site, and a C-terminal domain with seven WD40 repeats. These are characterized by highly variable consensus sequences (given at http://bmerc-www.bu.edu/bioinformatics/WD40repeat.html), of which the most conservative is shown in Fig. 2C. Interestingly, PRO11 revealed significant amino acid similarity to the mammalian WD40 repeat proteins striatin, SG2NA, and zinedin. Since the pro11 gene occurs as a single copy in the S. macrospora genome (S. Pöggeler, unpublished data), gene multiplication might have occurred during evolution, possibly with concomitant functional specialization of the three genes in mammals (11).
Proteins orthologous to striatin, SG2NA, and zinedin are present in metazoa throughout their evolution; they have been detected in nonmammalian vertebrates (Danio rerio and Xenopus laevis) and in invertebrates (Drosophila melanogaster and Caenorhabditis elegans) (27). In D. melanogaster, it was demonstrated that the striatin homologue CKA regulates AP-1 activity by organizing a molecular complex of kinases and transcription factors, thus coordinating the spatial and temporal expression of AP-1-regulated genes (12). A recent search for calmodulin binding proteins in the genome of Arabidopsis thaliana revealed that 29 animal calmodulin binding proteins, including striatin, SG2NA, and zinedin, have no homologues in A. thaliana. However, with the exception of S. cerevisiae, striatin gene homologues have been detected in various fungal genomes.
N-terminal part of PRO11 is sufficient to restore fertility.
The mutant pro11 gene contains a single G to A substitution, changing a conserved Trp (TGG) codon of the third WD40 repeat into a stop (TGA) codon. This removes all but the first two of the WD40 domains. Mutant pro11 does not seem to produce any detectable amount of a truncated protein, as was shown by Western blot analysis. Somehow, the mutation either prevents translation of the mRNA or destabilizes the translated protein. Interestingly, a truncated version of the wild-type pro11 gene, encoding only the N terminus of the PRO11 protein, was capable of partially complementing the developmental defects of pro11. Thus, the N-terminal domains of PRO11 retain significant functions on their own, and it appears to be more beneficial for PRO11 to be without a WD40 repeat domain than to have a defective one. A comparable observation was described before for the WD40 repeat protein COP1, a photomorphogenesis repressor of A. thaliana. Similar to PRO11, the COP1 protein contains a C-terminal WD40 repeat domain. In A. thaliana, all characterized lethal alleles of the cop1 gene contain mutations within the WD40 repeat domain, whereas weak mutant alleles lack the entire WD40 domain (18, 25). Similarly, in the human lisencephaly-1 gene encoding a WD40 repeat protein whose haploinsufficiency causes a devastating brain malformation known as lissencephaly, mutations cluster in exons which code for WD40 repeats 4, 5, and 6. Several of these are missense mutations resulting in protein truncations (9, 21, 45).
As discussed for the COP1 protein, there may be two possible explanations for the observation that the N-terminal portion of PRO11 itself retains some function, whereas a defective WD40 domain disrupts the function of the N-terminal domain. One possibility is that the truncation of the WD40 repeat in the protein encoded by the mutant pro11 gene disrupts the function of the N-terminal portion, possibly due to an unfavorable conformation. Alternatively, some other WD40 repeat proteins in the fungal cell may be able to take the place of the C-terminal domain. This substitution cannot take place in mutants with a defective WD40 repeat motif as a result of steric hindrance.
Animal homologues of PRO11 are functionally conserved in eukaryotic cell development.
Functional conservation of mammalian and fungal genes at the cellular level has previously been demonstrated for yeasts and filamentous fungi. For example, the human homologue of the Schizosaccharomyces pombe cdc2 gene, encoding a serine-threonine protein kinase, was cloned by expressing a human cDNA in fission yeast (20) and the rat c15 gene is capable of rescuing the nuclear movement defect of a nudC3 mutant from Aspergillus nidulans (30).
Similarly, the mouse striatin cDNA was able to restore the wild-type developmental program in pro11. Beside structural similarities, these data clearly indicate direct functional homology between the cognate proteins, implying that the cellular function of the PRO11 protein may be evolutionarily conserved from fungi to humans.
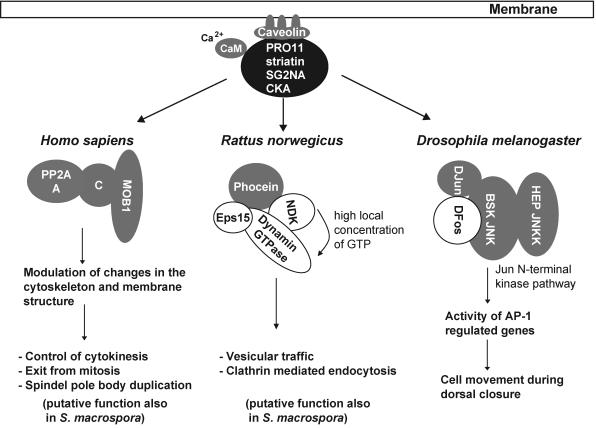
Animal homologues of PRO11 have been identified as components in three different protein complexes (Fig. 5). (i) In mammals, striatin, SG2NA, and zinedin are neuronal proteins strictly expressed in somatodendritic compartments and primarily localized to the cytosol and membranes (10, 11). PRO11 is physically present at several sites of the fungal cell, suggesting that the protein functions in multiple cellular processes. Members of the striatin family are thought to be involved in visual and neuron-specific Ca2+/calmodulin signal transduction cascades and have been shown to form a complex with protein phosphatase 2A. As depicted in Fig. 5, it was hypothesized that striatin and SG2NA may target the catalytic subunit C of protein phosphatase 2A to components of Ca2+-dependent signaling pathways (11, 29). As a result, members of this protein family may be involved in Ca2+-dependent neural functions, such as neurotransmission, maintenance, or dismantling of cytoskeletal elements (38). Moreover, it was demonstrated that the proteins of the striatin family directly bind caveolin-1, a membrane associated protein (16). Caveolins are scaffolding proteins able to collect a large number of signaling proteins bearing a caveolin-binding motif and confer the shape and structural organization of caveolae during caveola-mediated endocytosis (13, 40). Although endocytosis has not conclusively been demonstrated for filamentous fungi, the recently sequenced genome of Neurospora crassa led to the identification of genes encoding putative components of a complex endocytic protein machinery (36).
FIG. 5.
Scheme showing the signal transduction pathways of animal striatin-like proteins. All binding partners of the striatin-like proteins from mammals (Homo sapiens and Rattus norwegicus) and Drosophila melanogaster are shown in grey. With the exception of caveolin, genes encoding homologues for all depicted polypeptides have been identified in the fully sequenced genome of Neurospora crassa.
Using affinity-purified antisera against SG2NA, Moreno and coworkers (28) identified the mammalian homologue of the yeast protein MOB1 as a member of the striatin- and SG2NA-protein phosphatase 2A complexes. It was suggested that mammalian MOB1 might be a substrate of protein phosphatase 2A (28).
(ii) As shown in Fig. 5, an additional function was assumed from the data of a two-hybrid screen with striatin as a bait. Baillart et al. (4) identified phocein, an intracellular protein which bears a few homologies with the σ-subunits of clathrin adaptor proteins. With phocein as a bait, nucleoside diphosphate kinase and Eps15a multidomain protein, involved in clathrin-mediated endocytosis, were identified as interacting partners. Since nucleoside diphosphate kinase and Eps15 have been shown to be functional partners of dynamin, a GTPase that plays a critical role in endocytosis, the hypothesis that phocein and striatin might play a role in membrane and vesicular traffic is strengthened (3, 19, 37, 43).
(iii) D. melanogaster mutants lacking the striatin homologue CKA fail in dorsal closure during embryogenesis, causing the embryos to develop a hole in the dorsal epidermis. The Drosophila CKA protein is an essential component of the Jun N-terminal kinase pathway, which is required for this developmental process (12). Interestingly. CKA homologues from C. elegans to mammals have been reported to replace the D. melanogaster CKA in activating the Jun N-terminal kinase signal transduction pathway (12), indicating that striatin binding partners are not limited to calmodulin, caveolin, protein phosphatase 2A, MOB1, and phocein.
In consideration of our data as a whole, it seems reasonable to assume that the S. macrospora multidomain protein PRO11 regulates signaling by organizing protein complexes of kinases, phosphatases, and transcription factors. In this context, it is noteworthy that we have identified the S. macrospora homologues of phocein and MOB1, showing a high degree of similarity (33% and 49%, respectively) to the corresponding human proteins (S. Pöggeler unpublished data). Furthermore, a search of the genome sequence of N. crassa revealed that, with the exception of caveolin, all putative binding partners of PRO11 are encoded by this closely related species (Fig. 5). Therefore, the pathways in which the PRO11 protein might be involved are control of cytokinesis, vesicular traffic, and endocytosis.
Mutant alleles of the striatin, SG2NA, and zinedin genes are not known. However, downregulation of striatin in vivo with antisense oligonucleotides results in decreased locomotor activity in adult rats. In contrast, in vitro downregulation in embryonic motoneurons leads to a blockade of dendritic but not axonal growth, indicating that striatin might play a role in the establishment of polarity within developing neurons (6).
The S. macrospora sexual pathway provides an optimal experimental system for studying the mechanism and regulation of developmental processes that are usually obscured by complex cell-cell interactions involved in animal and plant development.
Acknowledgments
We thank Silke Nimtz, Ingeborg Godehardt, Susanne Schlewinski, and Swenja Ellssel for excellent technical assistance, Gabriele Frenβen-Schenkel for help in the artwork; and Sylvain Bukow and Stefanie Bunse for help with some experiments. We are indebted to Manfred Kilimann for the gift of mouse cDNA and W. Kunau for providing antibodies against mitochondrial porin.
This work was funded by a grant from the Deutsche Forschungsgemeinschaft (SFB 480) to S.P and U.K.
Dedicated to K. Esser on the occasion of his 80th birthday.
REFERENCES
- 1.Adams, T. H., J. K. Wieser, and J.-H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Baillat, G., S. Gaillard, F. Castets, and A. Monneron. 2002. Interactions of phocein with nucleoside-diphosphate kinase, Eps15, and Dynamin I. J. Biol. Chem. 277:18961-18966. [DOI] [PubMed] [Google Scholar]
- 4.Baillat, G., A. Moqrich, F. Castets, A. Baude, Y. Bailly, A. Benmerah, and A. Monneron. 2001. Molecular cloning and characterization of phocein, a protein found from the Golgi complex to dendritic spines. Mol. Biol. Cell 12:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartoli, M., A. Monneron, and D. Ladant. 1998. Interaction of calmodulin with striatin, a WD-repeat protein present in neuronal dendritic spines. J. Biol. Chem. 273:22248-22253. [DOI] [PubMed] [Google Scholar]
- 6.Bartoli, M., J. P. Ternaux, C. Forni, P. Portalier, P. Salin, M. Amalric, and A. Monneron. 1999. Down-regulation of striatin, a neuronal calmodulin-binding protein, impairs rat locomotor activity. J. Neurobiol. 40:234-243. [PubMed] [Google Scholar]
- 7.Berger, B., D. B. Wilson, E. Wolf, T. Tonchev, M. Milla, and P. S. Kim. 1995. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA 92:8259-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman, E. J., B. J. Bowman, and C. W. Slayman. 1981. Isolation and characterization of plasma membranes from wild type Neurospora crassa. J. Biol. Chem. 256:12336-12342. [PubMed] [Google Scholar]
- 9.Cardoso, C., R. J. Leventer, J. J. Dowling, H. L. Ward, J. Chung, K. S. Petras, J. A. Roseberry, A. M. Weiss, S. Das, C. L. Martin, D. T. Pilz, W. B. Dobyns, and D. H. Ledbetter. 2002. Clinical and molecular basis of classical lissencephaly: Mutations in the LIS1 gene (PAFAH1B1). Hum. Mutat. 19:4-15. [DOI] [PubMed] [Google Scholar]
- 10.Castets, F., M. Bartoli, J. V. Barnier, G. Baillat, P. Salin, A. Moqrich, J. P. Bourgeois, F. Denizot, G. Rougon, G. Calothy, and A. Monneron. 1996. A novel calmodulin-binding protein, belonging to the WD-repeat family, is localized in dendrites of a subset of CNS neurons. J. Cell Biol. 134:1051-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castets, F., T. Rakitina, S. Gaillard, A. Moqrich, M.-G. Mattei, and A. Monneron. 2000. Zinedin, SG2NA, and striatin are calmodulin-binding, WD repeat proteins principally expressed in the brain. J. Biol. Chem. 275:19970-19977. [DOI] [PubMed] [Google Scholar]
- 12.Chen, H. W., M. J. Marinissen, S. W. Oh, X. Chen, M. Melnick, N. Perrimon, J. S. Gutkind, and S. X. Hou. 2002. CKA, a novel multidomain protein, regulates the Jun N-terminal kinase signal transduction pathway in Drosophila. Mol. Cell. Biol. 22:1792-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conner, S. D., and S. L. Schmid. 2003. Regulated portals of entry into the cell. Nature 422:37-44. [DOI] [PubMed] [Google Scholar]
- 14.Esser, K. 1982. Cryptogams — cyanobacteria, algae, fungi, lichens. Cambridge University Press, London, England.
- 15.Freitag, H., M. Janes, and W. Neupert. 1982. Biosynthesis of mitochondrial porin and insertion into the outer mitochondrial membrane of Neurospora crassa. Eur. J. Biochem. 126:197-202. [DOI] [PubMed] [Google Scholar]
- 16.Gaillard, S., M. Bartoli, F. Castets, and A. Monneron. 2001. Striatin, a calmodulin-dependent scaffolding protein, directly binds caveolin-1. FEBS Lett. 508:49-52. [DOI] [PubMed] [Google Scholar]
- 17.Gish, W., and D. J. States. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266-272. [DOI] [PubMed] [Google Scholar]
- 18.Holm, M., C. Hardtke, R. Gaudet, and X. W. Deng. 2001. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20:118-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishnan, K. S., R. Rikhy, S. Rao, M. Shivalkar, M. Mosko, R. Narayanan, P. Etter, P. S. Estes, and M. Ramaswami. 2001. Nucleoside diphosphate kinase, a source of GTP, is required for dynamin-dependent synaptic vesicle recycling. Neuron 30:197-210. [DOI] [PubMed] [Google Scholar]
- 20.Lee, M. G., and P. Nurse. 1987. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature 327:31-35. [DOI] [PubMed] [Google Scholar]
- 21.Leventer, R. J., C. Cardoso, D. H. Ledbetter, and W. B. Dobyns. 2001. LIS1 missense mutations cause milder lissencephaly phenotypes including a child with normal IQ. Neurology 57:416-422. [DOI] [PubMed] [Google Scholar]
- 22.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375-382. [PubMed] [Google Scholar]
- 23.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 24.Masloff, S., S. Pöggeler, and U. Kück. 1999. The pro1+ gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 152:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNellis, T., A. G. von Armin, T. Araki, Y. Komeda, S. Misera, and X. W. Deng. 1994. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6:487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, D. 1998. Fungal morphogenesis. Cambridge University Press, Cambridge, England.
- 27.Moqrich, A., M. G. Mattei, M. Bartoli, T. Rakitina, G. Baillat, A. Monneron, and F. Castets. 1998. Cloning of human striatin cDNA (STRN), gene mapping to 2p22-p21, and preferential expression in brain. Genomics 51:136-139. [DOI] [PubMed] [Google Scholar]
- 28.Moreno, C. S., W. S. Lane, and D. C. Pallas. 2001. A mammalian homolog of yeast MOB1 is both a member and a putative substrate of striatin family-protein phosphatase 2A complexes. J. Biol. Chem. 276:24253-24260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreno, C. S., S. Park, K. Nelson, D. Ashby, F. Hubalek, W. S. Lane, and D. C. Pallas. 2000. WD40 repeat proteins Striatin and S/G2 nuclear autoantigen are members of a novel family of calmodulin-binding proteins that associate with protein phosphatase 2A. J. Biol. Chem. 275:5257-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris, S. M., P. Anaya, X. Xiang, N. R. Morris, G. S. May, and L. Yu-Lee. 1997. A prolactin-inducible T cell gene product is structurally similar to the Aspergillus nidulans nuclear movement protein NUDC. Mol. Endocrinol. 11:229-236. [DOI] [PubMed] [Google Scholar]
- 31.Nowrousian, M., S. Masloff, S. Pöggeler, and U. Kück. 1999. Cell differentiation during sexual development of the fungus Sordaria macrospora requires ATP citrate lyase activity. Mol. Cell. Biol. 19:450-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osiewacz, H. D. 1994. A versatile shuttle cosmid vector for the efficient construction of genomic libraries and for the cloning of fungal genes. Curr. Genet. 26:87-90. [DOI] [PubMed] [Google Scholar]
- 33.Pöggeler, S., S. Masloff, B. Hoff, S. Mayrhofer, and U. Kück. 2003. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr. Genet. 43:54-61. [DOI] [PubMed] [Google Scholar]
- 34.Pöggeler, S., M. Nowrousian, S. Jacobsen, and U. Kück. 1997. An efficient procedure to isolate fungal genes from an indexed cosmid library. J. Microbiol. Methods 29:49-61. [Google Scholar]
- 35.Pöggeler, S., S. Risch, U. Kück, and H. D. Osiewacz. 1997. Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic ascomycete. Genetics 147:567-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Read, N., and D. Kalkman. 2003. Does endocytosis occur in fungal hyphae? Fungal Genet. Biol. 39:199-203. [DOI] [PubMed] [Google Scholar]
- 37.Salcini, A. E., M. A. Hilliard, A. Croce, S. Arbucci, P. Luzzi, C. Tacchetti, L. Daniell, P. De Camilli, P. G. Pelicci, P. P. Di Fiore, and P. Bazzicalupo. 2001. The Eps15 C. elegans homologue EHS-1 is implicated in synaptic vesicle recycling. Nat. Cell Biol. 3:755-760. [DOI] [PubMed] [Google Scholar]
- 38.Salin, P., P. Kachidian, M. Bartoli, and F. Castets. 1998. Distribution of striatin, a newly identified calmodulin-binding protein in the rat brain: an in situ hybridization and immunocytochemical study. J. Comp. Neurol. 397:41-59. [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Smart, E. J., G. A. Graf, M. A. McNiven, W. C. Sessa, J. A. Engelman, P. E. Scherer, T. Okamoto, and M. P. Lisanti. 1999. Caveolins, liquid-ordered domains, and signal transduction. Mol. Cell. Biol. 19:7289-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat a common architecture for diverse functions. Trends Biochem. Sci. 25:325-330. [DOI] [PubMed] [Google Scholar]
- 42.Sprague, E. R., M. J. Redd, A. D. Johnson, and C. Wolberger. 2000. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 19:3016-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tebar, F., T. Sorkina, A. Sorkin, M. Ericsson, and T. Kirchhausen. 1996. Eps15 is a component of clathrin-coated pits and vesicles and is located at the rim of coated pits. J. Biol. Chem. 271:28727-28730. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vallee, R. B., C. Tai, and N. E. Faulkner. 2001. LIS1: cellular function of a disease-causing gene. Trends Cell Biol. 11:155-160. [DOI] [PubMed] [Google Scholar]
- 46.van Heemst, D., F. James, S. Pöggeler, V. Berteaux-Lecellier, and D. Zickler. 1999. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell 98:261-271. [DOI] [PubMed] [Google Scholar]
- 47.Walz, M., and U. Kück. 1995. Transformation of Sordaria macrospora to hygromycin B resistance: characterization of transformants by electrophoretic karyotyping and tetrad analysis. Curr. Genet. 29:88-95. [DOI] [PubMed] [Google Scholar]
- 48.Wolf, E., P. S. Kim, and B. Berger. 1997. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6:1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]