Abstract
Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae are important causes of meningitis and other infections, and rapid, sensitive, and specific laboratory assays are critical for effective public health interventions. Singleplex real-time PCR assays have been developed to detect N. meningitidis ctrA, H. influenzae hpd, and S. pneumoniae lytA and serogroup-specific genes in the cap locus for N. meningitidis serogroups A, B, C, W135, X, and Y. However, the assay sensitivity for serogroups B, W135, and Y is low. We aimed to improve assay sensitivity and develop multiplex assays to reduce time and cost. New singleplex real-time PCR assays for serogroup B synD, W135 synG, and Y synF showed 100% specificity for detecting N. meningitidis species, with high sensitivity (serogroup B synD, 99% [75/76]; W135 synG, 97% [38/39]; and Y synF, 100% [66/66]). The lower limits of detection (LLD) were 9, 43, and 10 copies/reaction for serogroup B synD, W135 synG, and Y synF assays, respectively, a significant improvement compared to results for the previous singleplex assays. We developed three multiplex real-time PCR assays for detection of (i) N. meningitidis ctrA, H. influenzae hpd, and S. pneumoniae lytA (NHS assay); (ii) N. meningitidis serogroups A, W135, and X (AWX assay); and (iii) N. meningitidis serogroups B, C, and Y (BCY assay). Each multiplex assay was 100% specific for detecting its target organisms or serogroups, and the LLD was similar to that for the singleplex assay. Pairwise comparison of real-time PCR between multiplex and singleplex assays showed that cycle threshold values of the multiplex assay were similar to those for the singleplex assay. There were no substantial differences in sensitivity and specificity between these multiplex and singleplex real-time PCR assays.
INTRODUCTION
Approaches for detection of bacterial meningitis pathogens are continuously being improved. Detection of the specific serogroup or serotype causing disease can lead to better understanding of disease epidemiology, which is essential for planning appropriate vaccination programs and monitoring their impact. Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae are human commensal bacteria that also cause a spectrum of invasive diseases that include not only meningitis but also pneumonia and sepsis (8, 14, 24, 25, 29). Each of the three organisms is categorized by the structure of the polysaccharide capsule into different serogroups or serotypes. Six N. meningitidis serogroups (A, B, C, W135, X, and Y), six H. influenzae serotypes (a, b, c, d, e, and f), and 23 S. pneumoniae serotypes are commonly associated with invasive disease (14, 15). N. meningitidis and S. pneumoniae are considered the leading causes of bacterial meningitis worldwide after the implementation of the H. influenzae type b (Hib) conjugate vaccine (16, 18–20, 26). The serotype or serogroup distributions of the three pathogens vary by geographic region (8, 16, 27).
Singleplex real-time PCR assays have been developed to rapidly detect N. meningitidis ctrA (the capsule transport gene), H. influenzae hpd (the protein D gene), and S. pneumoniae lytA (the autolysin gene) (4, 6, 22, 28). Singleplex real-time PCR assays are available to determine N. meningitidis serogroups by detecting serogroup-specific genes in the cap locus (serogroup A sacB, W135 synG, X xcbB, B synD, C synE, and Y synF) (22). However, the assay sensitivity for serogroups B, W135, and Y is low. We aimed to improve the sensitivity of the singleplex real-time PCR assays for serogroup B synD, W135 synG, and Y synF by redesigning the primers and probes. Bacterial detection using singleplex real-time PCR requires considerable time and costly reagents, especially when handling large quantities of specimens. To reduce the cost and time for detecting the causative bacteria in settings such as meningitis outbreaks and epidemics, we developed and validated three multiplex real-time PCR assays for the detection of (i) N. meningitidis ctrA, H. influenzae hpd, and S. pneumoniae lytA (NHS assay), (ii) N. meningitidis serogroups A, W135, and X (AWX assay), and (iii) N. meningitidis serogroups B, C, and Y (BCY assay), which are the major causes of meningococcal diseases in the United States.
MATERIALS AND METHODS
Bacterial strains.
The clinical isolates of N. meningitidis, H. influenzae, and S. pneumoniae used in this study were collected from 1997 to 2008 as part of the Active Bacterial Core surveillance (ABCs) of the Centers for Disease Control and Prevention's Emerging Infections Program (http://www.cdc.gov/abcs/index.htm). These isolates were collected from culture-confirmed cases of meningitis, pneumonia, or bacteremia. The N. meningitidis serogroup or H. influenzae serotype of each isolate was determined by slide agglutination as part of the routine testing. The S. pneumoniae serotypes were determined by PCR (10). Reference strains were purchased from the American Type Culture Collection (ATCC).
Growth conditions.
All H. influenzae isolates were grown on chocolate II agar supplemented with hemoglobin and IsoVitalex (BD, Sparks, MD) and incubated at 37°C for 18 to 24 h with 5% CO2. All N. meningitidis and S. pneumoniae isolates were grown on TSA II agar plates supplemented with 5% sheep blood (BD, Sparks, MD) and incubated at 37°C for 18 to 24 h with 5% CO2.
DNA extraction.
For isolates and clinical specimens from Brazil, DNA was extracted as described previously (4). For clinical specimens and transport medium samples that did not yield viable cultures (nonviable transport medium [NVTM] samples) from South Africa and specimens from nasal washes or throat swabs, DNA was extracted using a MagNA pure compact nucleic acid isolation kit I (Roche Diagnostics, Mannheim, Germany) with a MagNA pure compact instrument (Roche Applied Science, Mannheim, Germany). A known specimen (positive control) and a water sample (negative control) were also extracted each time the DNA extraction procedure was performed on unknown specimens, which ensures that the negative real-time PCR results were not due to DNA extraction failure and that the positive PCR results were not due to cross-contamination introduced during the DNA extraction process. To prepare crude cell lysates, bacterial cultures grown overnight on appropriate agar plates were collected and suspended in 1 ml of 10 mM Tris buffer (pH 8.0). The cell suspensions were boiled for 10 min and stored at −20°C.
Real-time PCR and lower limit of detection (LLD).
Real-time PCR was performed as described previously (22) using Invitrogen Platinum quantitative PCR Supermix-UDG master mix and run on either a Stratagene Mx3005P instrument (Agilent Technology, Santa Clara, CA) or an Applied Biosystems 7500 instrument (Applied Biosystems, Santa Clara, CA). PCR assembly and cycling conditions are described in Table S1 in the supplemental material.
To determine the LLD of each real-time PCR assay run on the Stratagene Mx3005P instrument (Agilent Technology, Santa Clara, CA), genomic DNA was extracted from clinical isolates, and the DNA concentration was determined using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and adjusted to 20 ng/μl; DNA was then serially diluted in 10-fold increments. Each DNA concentration was converted to a genome equivalent per microliter using a standard of 2.2 Mb per N. meningitidis genome, 1.83 Mb per H. influenzae genome, or 2.1 Mb per S. pneumoniae genome. The LLD for a real-time PCR assay was defined as the DNA concentration that yielded a cycle threshold (CT) value of 35.
Development of new singleplex real-time PCR assays for N. meningitis serogroup B synD, W135 synG, and Y synF.
Primer Express 3.0 (Applied Biosystems) was used to design appropriate primers and probes for the singleplex real-time PCR assays based on published sequences of synD in N. meningitidis B, synG in N. meningitidis W135, and synF in N. meningitidis Y (22). All primers and probes were optimized by testing concentrations in the range of 100 nM to 900 nM and 100 nM to 400 nM, respectively, except for the lytA real-time PCR assay (see Table S2 in the supplemental material). The lytA assay was performed as described previously (4), with modifications (http://www.cdc.gov/ncidod/biotech/strep/protocols.htm) (6). Fifty-eight isolates of non-N. meningitidis species collected through the ABCs or from the ATCC were tested to determine whether the new real-time PCR assays were specific for detecting the species N. meningitidis (Table 1; also see Table S3 in the supplemental material). The capability of the three new singleplex real-time PCR assays for detecting the specific N. meningitidis serogroup was evaluated by testing a convenience sample of 227 ABCs clinical isolates, including 22 serogroup A, 76 serogroup B, 24 serogroup C, 39 serogroup W135, and 66 serogroup Y isolates. The CT value of each new assay was compared with that of the corresponding previously developed assay by testing 61 ABCs N. meningitidis isolates, including serogroups B, W135, and Y. All real-time PCRs were run on a Stratagene Mx3005P instrument (Agilent Technology, Santa Clara, CA).
Table 1.
Detection of nontarget bacterial species by the real-time PCR assays in multiplex and singleplexa
| Assay | No. negative/total no. (%) | 95% CIb |
|---|---|---|
| N. meningitidis ctrA | 58/58 (100) | 93–100 |
| H. influenzae hpd | 60/60 (100) | 94–100 |
| S. pneumoniae lytA | 66/66 (100) | 95–100 |
| N. meningitidis serogroup | 58/58 (100) | 93–100 |
The negative rates for each assay were identical in multiplex and singleplex. The isolates tested are listed in Table S3 in the supplemental material.
95% CI, 95% confidence interval.
Validation of the three multiplex real-time PCR assays using ABCs isolates.
Sixty-seven isolates representing different bacterial species collected through the ABCs or from the ATCC were tested to confirm that the multiplex and singleplex real-time PCR assays were specific for detecting the target bacterial species (see Table S3 in the supplemental material). The three multiplex real-time PCR assays (NHS, AWX, and BCY assays) were then evaluated using a convenience sample of 109 ABCs clinical isolates. The NHS assay was tested using 80 ABCs isolates, including 31 H. influenzae, 23 N. meningitidis, and 26 S. pneumoniae isolates. The serogroup-specific AWX and BCY assays were tested using 52 ABCs isolates, including 8 serogroup B, 9 serogroup C, 10 serogroup Y, 9 serogroup A, 7 serogroup X, and 9 serogroup W135 isolates. The 23 N. meningitidis isolates used for the NHS assay validation were included in the 52 isolates used for validating the N. meningitidis serogroup-specific AWX and BCY assays. To test a wide range of template DNA concentrations, crude DNA prepared from these isolates was diluted (10−4, 10−5, 10−6, and 10−7); each DNA dilution was tested using the real-time PCR assays in both multiplex and singleplex platforms. All real-time PCRs were run on a Stratagene Mx3005P instrument (Agilent Technology, Santa Clara, CA). The CT values of each real-time PCR assay between multiplex and singleplex platforms were compared by pairwise comparison analysis. Dilutions that did not give any CT value were excluded from the analysis. In addition, pairwise comparison between duplicate singleplex assays for N. meningitidis ctrA, H. influenzae hpd, S. pneumoniae lytA, and serogroup B synD was also conducted to evaluate intra-assay variation (see Tables S4 and S5 in the supplemental material).
Clinical validation of multiplex real-time PCR NHS assay using specimens from routine surveillance for invasive disease caused by N. meningitidis, H. influenzae, and S. pneumoniae in South Africa.
A total of 121 specimens from South Africa (67 cerebrospinal fluid [CSF] specimens, 26 blood specimens, and 28 NVTM samples) were used to validate the NHS assay in multiplex and singleplex platforms on an Applied Biosystems 7500 instrument (Applied Biosystems, Santa Clara, CA). The clinical specimens were collected from patients of all ages from January 2008 through April 2009 as part of routine national laboratory-based surveillance for invasive disease due to N. meningitidis, H. influenzae, and S. pneumoniae. Clinical specimens and NVTM samples, with clinical and demographic data, were sent to the National Institute for Communicable Diseases (NICD) in Johannesburg, South Africa, from approximately 120 laboratories throughout South Africa. NVTM samples refer to transport medium samples that yielded no culture in the reference laboratory at the NICD. Brain heart infusion (BHI) broth was added to the nonviable transport medium samples, which were incubated overnight at 37°C with 5% CO2 for DNA extraction. A composite reference standard (positive defined as culture or latex agglutination positive; negative otherwise) was used to compare the sensitivities and specificities among the multiplex and singleplex real-time PCR assays (2).
Clinical validation of real-time PCR H. influenzae hpd assay in multiplex and singleplex using nasal washes and/or throat swabs from patients with respiratory infections at Lackland Air Force Base.
A total of 487 nasal washes and/or throat swabs were collected from 255 patients with upper respiratory infections as part of infectious disease surveillance at Lackland Air Force Base in San Antonio, TX. Nasal washes were collected in approximately 2 ml saline; throat swabs were stored in 5 ml BD viral transport medium. Genomic DNA was extracted from 400 μl of nasal wash or throat swab specimens and tested with various PCR assays. Each specimen was screened for a variety of viral and bacterial pathogens, including H. influenzae, at Lackland Air Force Base (21). A high rate of H. influenzae in patients with viral infections was identified by use of a conventional PCR assay targeting the fucK gene (23). These specimens were used to validate the H. influenzae hpd real-time PCR assay in multiplex (NHS assay) and singleplex platforms using an Applied Biosystems 7500 instrument (Applied Biosystems, Santa Clara, CA). The fucK PCR assay was used as the reference standard for comparison of the sensitivities and specificities of the H. influenzae hpd assay between multiplex and singleplex platforms.
Clinical validation of real-time PCR assays for N. meningitidis ctrA and serogroup C synE in multiplex and singleplex using clinical specimens from meningitis patients in Brazil.
CSF specimens from patients of all ages were collected from routine bacterial meningitis surveillance from 2007 to 2008 in Sao Paul, Brazil. A convenience sample of 100 CSF specimens was sent to the CDC in Atlanta, GA, as part of a quality control program agreed upon between the CDC and the Adolfo Lutz Institute in Sao Paul, Brazil, and was used to validate the N. meningitidis ctrA and serogroup C synE real-time PCR in multiplex (NHS and BCY) and singleplex assays using a Stratagene Mx3005P instrument (Agilent Technology, Santa Clara, CA). Since culture or latex agglutination results were not available for these CSF specimens, counterimmunoelectrophoresis (CIE) (13) conducted at the Adolfo Lutz Institute was used as the reference standard to compare the sensitivities and specificities of the N. meningitidis ctrA and serogroup C synE assays between multiplex and singleplex assays.
Statistical analysis.
For each pairwise comparison (between multiplex and singleplex or between singleplex duplicates), the Bland-Altman plot was used to depict the magnitude of disagreement between the two assays (ΔCTmultiplex-singleplex [ΔCTm-s] and ΔCTsingleplex1-singleplex2 [ΔCTs1-s2]) as a function of the average CT value of the two assays [(CTmultiplex or singleplex + CTsingleplex)/2]. The reference lines on the plot indicated the ideal zero difference and the observed mean CT difference (mean ΔCTm-s or mean ΔCTs1-s2), as well as the upper and lower limits of agreement defined by the mean CT difference ± 1.96 standard deviations (SD). If the CT differences (ΔCTm-s or ΔCTs1-s2) are normally distributed, then 95% of the CT differences (ΔCT) would fall between the lower and upper limits of the agreement (1–3). Summary statistics were calculated for each assay, including sample size (n), mean CT, SD, minimum CT, median CT, and maximum CT.
The sensitivity and specificity of multiplex and singleplex platforms for 4 real-time PCR assays (N. meningitidis ctrA, H. influenzae hpd, S. pneumoniae lytA, and serogroup C synE) were assessed using the specimens collected from Lackland Air Force Base, Brazil, and South Africa. Other real-time PCR assays were not assessed due to the insufficient sample size (see Table S6 in the supplemental material). For all PCR assays, a specimen was considered positive if its CT value was ≤35 and negative if its CT value was >40. If a CT value was >35 and ≤40, the specimen was diluted 10-fold and retested to determine if PCR inhibitors were present. The specimen was considered positive if the CT value of the diluted specimen was ≤35 and negative if the CT value was >35. The specimens that were negative by real-time PCR were tested for the presence of an RNase P-encoding gene to exclude PCR inhibition and DNA extraction failure. The sensitivity of a real-time PCR assay was defined as the proportion of real-time PCR-positive cases (multiplex or singleplex) out of positive cases determined by a reference standard [number of true positives/(number of true positives + number of false negatives)]. The specificity of a real-time PCR assay was defined as the proportion of real-time PCR-negative cases out of the negative cases determined by a reference standard [number of true negatives/(number of true negatives + number of false positives)]. Analyses were performed using SAS 9.2 (SAS Institute, Inc., Cary, NC).
RESULTS
Improved singleplex real-time PCR assays for detection of N. meningitidis serogroups B, W135, and Y.
To improve the sensitivity of N. meningitidis serogroup determination, we redesigned or modified the primers and probes for real-time PCR detection of N. meningitidis serogroups B, W135, and Y, based on published DNA sequences of the synD, synG, and synF genes, respectively (see Table S2 in the supplemental material) (22). The three new singleplex real-time PCR assays did not amplify DNA from any non-N. meningitidis species (Table 1; also see Table S3 in the supplemental material). Each assay was 100% specific for detection of its target serogroup (Table 2). The positive rates for detection of the target serogroup were 99%, 97%, and 100% for assays detecting serogroup B synD, W135 synG, and Y synF, respectively. The lower limit of detection (LLD) of each assay was significantly improved compared to that of the corresponding previously developed assay (Table 3). CT values of the new assays were also compared to those of the previously developed assays by testing an additional 21 serogroup B, 11 serogroup W135, and 29 serogroup Y isolates. Each new assay showed lower CT values than the previously developed assay, with a difference in CT value of 4 to 28 cycles for all isolates tested (data not shown).
Table 2.
Detection of N. meningitidis serogroups by the new singleplex real-time PCR assaysa
| Assay | Result for target N. meningitidis serogroup |
No. negative/no. tested for other N. meningitidis serogroups | |
|---|---|---|---|
| No. positive/total no. (%) | 95% CI | ||
| Serogroup B synD | 75/76 (99) | 93–100 | 0/151 |
| Serogroup W135 synG | 38/39 (97) | 87–100 | 0/188 |
| Serogroup Y synF | 66/66 (100) | 95–100 | 0/161 |
A total of 227 N. meningitidis isolates were tested, including 22 serogroup A, 76 serogroup B, 24 serogroup C, 39 serogroup W135, and 66 serogroup Y isolates.
Table 3.
Lower limits of detection of the real-time PCR assays
| Assay | Organism | Genome DNA equivalents/PCR |
||
|---|---|---|---|---|
| Multiplexa | New singleplex | Previous singleplex | ||
| N. meningitidis ctrA | N. meningitidis B | 18 | 23 | NAb |
| N. meningitidis Y | 14 | 12 | NA | |
| H. influenzae hpd | Hib | 42 | 46 | NA |
| HiNTc | 23 | 23 | NA | |
| S. pneumoniae lytA | S. pneumoniae type 4 | 1 | 1 | NA |
| S. pneumoniae type 6B | 1 | 1 | NA | |
| Serogroup B synD | N. meningitidis B | 9 | 9 | 66,171 |
| Serogroup C synE | N. meningitidis C | 29 | 17 | NA |
| Serogroup Y synF | N. meningitidis Y | 11 | 10 | 8,955 |
| Serogroup A sacB | N. meningitidis A | 210 | 221 | NA |
| Serogroup W135 synG | N. meningitidis W135 | 48 | 43 | 1,088,161 |
| Serogroup X xcbB | N. meningitidis X | 11 | 20 | NA |
The multiplex assays were the NHS assay, detecting N. meningitidis, H. influenzae, and S. pneumoniae; the BCY assay, detecting N. meningitidis serogroups B, C, and Y; and the AWX assay, detecting N. meningitidis serogroups A, W135, and X.
NA, not applicable.
HiNT, nontypeable H. influenzae.
Development of multiplex real-time PCR assays (NHS, AWX, and BCY).
Fluorescence-labeled probes were tested for each multiplex real-time PCR assay. Table S2 in the supplemental material shows the optimal combinations of probe-reporter dye for each multiplex assay. Each of the nine assays was 100% specific for detection of its target organism in both multiplex and singleplex (Table 1). The LLD of each assay in multiplex was similar to that in singleplex (Table 3). Seven of the assays were able to detect low numbers of DNA copies per PCR (less than 50 genome equivalents). The serogroup A sacB assay detected higher genome equivalents than the other assays tested in this study.
Pairwise comparison of real-time PCR assays between multiplex and singleplex platforms and between singleplex assay duplicates.
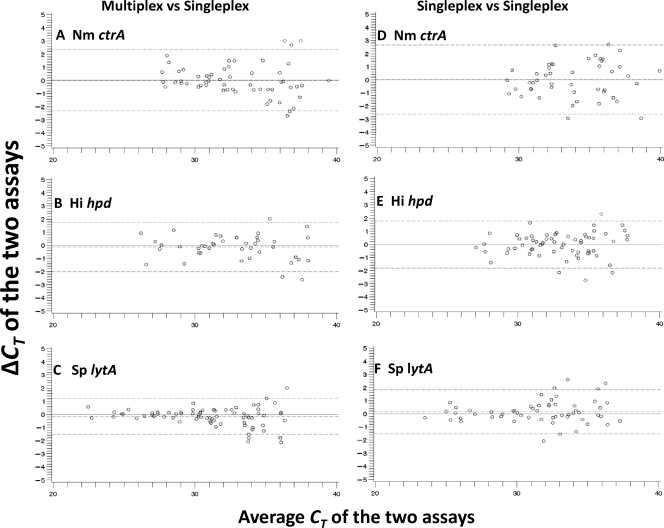
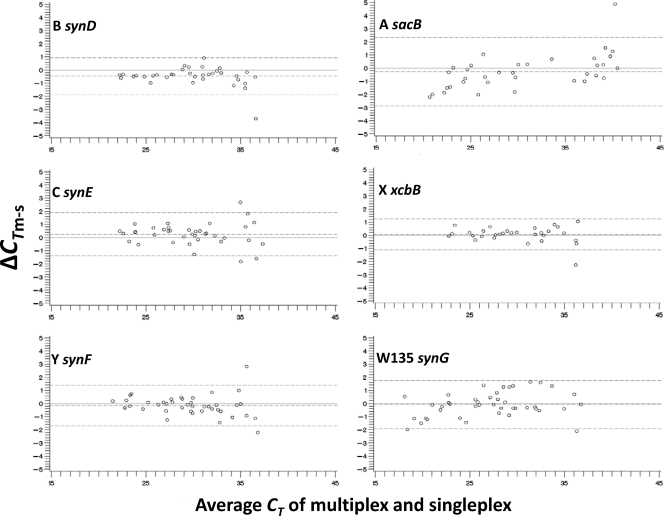
Pairwise comparison of the NHS, AWX, and BCY assays between multiplex and singleplex was first conducted using clinical isolates. ΔCTm-s of these assays was within a range of −3.7 to 4.9 cycles (Fig. 1 and 2; also see Table S4 in the supplemental material). Pairwise comparison between duplicate singleplex assays for N. meningitidis ctrA, H. influenzae hpd, and S. pneumoniae lytA (Fig. 1) and serogroup B synD (data not shown) was also conducted to evaluate intra-assay variation. ΔCTs1-s2 was within a similar range, from −2.9 to 3.4 (see Table S5 in the supplemental material). For each assay, both the CT differences and the standard deviations (SD) of the CT differences were similar between the multiplex-singleplex and singleplex-singleplex comparisons, indicating that the measurement errors were similar between the multiplex and singleplex platforms of these assays. In general, for each assay, the level of agreement between multiplex and singleplex was similar to that between singleplex duplicates, with most CT differences in each comparison falling within the limits of agreement (Fig. 1). The ΔCT of PCR for most DNA specimens tested was within the lower and upper limits of agreements of −2.9 and 2.3 cycles, respectively, between multiplex and singleplex and −2.6 and 2.7, respectively, between singleplex duplicates (see Tables S4 and S5). A few outlier results with greater ΔCTm-s or ΔCTs1-s2 values were observed when the DNA concentration was low (CT value of >35). The mean ΔCTm-s value of each assay was very close to zero; there are not substantial differences in real-time PCR between the multiplex and singleplex platforms.
Fig 1.
Pairwise comparison analyses for species-specific PCR assays for N. meningitidis (Nm) ctrA, H. influenzae (Hi) hpd, and S. pneumoniae (Sp) lytA carried out using ABCs isolates. Each assay was compared between multiplex and singleplex (A, B, and C) and between singleplex duplicates (D, E, and F). Shown is the Bland-Altman plot that depicts the cycle threshold difference (ΔCT) between the two assays (ΔCTmultiplex-singleplex [ΔCTm-s] or ΔCTsingleplex1-singleplex2 [ΔCTs1-s2]) as a function of the average CT value of the two assays [(CTmultiplex or singleplex + CTsingleplex)/2]. The reference lines on the plot indicate the ideal zero difference (solid line) and the observed mean CT difference (mean ΔCTm-s or mean ΔCTs1-s2) (middle dashed line), as well as the upper and lower limits of agreement defined by the mean ΔCT ± 1.96 standard deviations (top and bottom dashed lines) (1–3).
Fig 2.
Pairwise comparison analyses for N. meningitidis serogroup-specific PCR assays carried out using ABCs isolates. Each of the six PCR assays was compared between multiplex and singleplex platforms. The Bland-Altman plot for each assay is shown.
ΔCTm-s was also determined by testing clinical specimens from South Africa, Brazil, and Lackland Air Force Base (see Fig. S1 and Table S6 in the supplemental material). Extracted DNA from these specimens was tested without dilution. Only specimens that were positive by the reference standard test were included in the pairwise comparison (see Fig. S1 and Table S6). The greatest ΔCTm-s values were observed for the H. influenzae hpd assay using nasal washes and/or throat swabs collected at Lackland Air Force Base, but only 13 values (4.9%) were outside the limits of agreement (see Fig. S1). Although few specimens from South Africa were tested with the H. influenzae hpd and S. pneumoniae lytA assays, the level of agreement between multiplex and singleplex was the same as that observed when the U.S. clinical isolates were tested. However, a significantly lower average CT value (mean difference of −1.7, paired t test, P < 0.01) was observed in the multiplex assay than in the singleplex assay for the specimens from South Africa when tested with N. meningitidis ctrA (see Fig. S1). Similarly, when the clinical specimens from Brazil were tested with the serogroup C synE assay, a lower average CT value (mean difference of −1.3, paired t test, P < 0.01) was observed using the multiplex assay than using the singleplex (see Fig. S1).
Sensitivity and specificity of multiplex and singleplex real-time PCR assays for N. meningitidis ctrA, H. influenzae hpd, S. pneumoniae lytA, and serogroup C synE.
Different reference standards (fucK PCR, CIE, and culture/latex composite reference standards) were used to assess the sensitivity and the specificity of real-time PCR assays for N. meningitidis ctrA, H. influenzae hpd, S. pneumoniae lytA, and serogroup C synE in multiplex and singleplex platforms. As shown in Table 4, there was no significant difference in sensitivity and specificity between the multiplex and singleplex real-time PCR assays.
Table 4.
Sensitivities and specificities of the multiplex and singleplex real-time PCR assays for detection of meningitis pathogens from clinical specimens
| Specimen group (reference standard) | Assay (type)a | No. positive/no. tested | Sensitivity (95% CI) | No. negative/no. tested | Specificity (95% CI) |
|---|---|---|---|---|---|
| Lackland (fucK PCR) | H. influenzae hpd (M) | 206/244 | 84.4 (79.3–88.7) | 217/243 | 89.3 (84.7–92.9) |
| H. influenzae hpd (S) | 206/244 | 84.4 (79.3–88.7) | 218/243 | 89.7 (85.2–93.2) | |
| Brazil (CIEb) | N. meningitidis ctrA (M) | 72/75 | 96.0 (88.8–99.2) | 8/24 | 33.3 (15.6–55.3) |
| N. meningitidis ctrA (S) | 72/75 | 96.0 (88.8–99.2) | 9/24 | 37.5 (18.8–59.4) | |
| Serogroup C synE (M) | 71/75 | 94.7 (86.9–98.5) | 17/24 | 70.8 (48.9–87.4) | |
| Serogroup C synE (S) | 72/75 | 96.0 (88.8–99.2) | 17/24 | 70.8 (48.9–87.4) | |
| South Africa (compositec) | N. meningitidis ctrA (M) | 56/76 | 73.7 (62.3–83.1) | 43/44 | 97.7 (88.0–99.9) |
| N. meningitidis ctrA (S) | 58/76 | 76.3 (65.2–85.3) | 37/44 | 84.1 (69.9–93.4) | |
| H. influenzae hpd (M) | 16/21 | 76.2 (52.8–91.8) | 98/99 | 99.0 (94.5–100.0) | |
| H. influenzae hpd (S) | 18/21 | 85.7 (63.7–97.0) | 91/99 | 91.9 (84.7–96.4) | |
| S. pneumoniae lytA (M) | 16/17 | 94.1 (71.3–99.8) | 102/103 | 99.0 (94.7–100.0) | |
| S. pneumoniae lytA (S) | 16/17 | 94.1 (71.3–99.8) | 96/103 | 93.2 (86.5–97.2) |
M, multiplex; S, singleplex.
One specimen was not tested by CIE and was therefore excluded from this analysis.
Combined laboratory tests (culture and/or latex agglutination) were used as the composite reference standard, where a positive result was defined as culture or latex agglutination positive (negative otherwise). One specimen was not tested by culture or latex agglutination and was therefore excluded from this analysis.
DISCUSSION
Real-time PCR is a powerful tool for detection of human pathogens because of the high sensitivity and throughput capability. Introduction of fluorescent probes improves the assay specificity and enables development of a multiplex platform for amplification and detection of multiple target genes. Multiplex real-time PCR allows simultaneous detection of multiple organisms in a single reaction, which conserves limited quantities of clinical specimens and significantly reduces costs, such as reagents and person time, which is particularly important in resource-limited settings. However, concerns that the sensitivity of a single assay may be compromised when it is run with other PCR assays in a single reaction remain. In this study, we found that the sensitivity of three multiplex real-time PCR assays was indistinguishable from that of the singleplex real-time PCR assays for detection of bacterial meningitis pathogens as evaluated using ABCs clinical isolates as well as clinical specimens collected from different geographic locations. The sensitivities and specificities of each assay varied among the different laboratories due to the use of different reference standards. Culture is still considered the “gold standard.” Since none of the reference standards used in this study is considered the “gold standard,” the sensitivity and specificity in this analysis may not be accurately assessed.
A multiplex real-time PCR assay using N. meningitidis ctrA, H. influenzae bexA, and S. pneumoniae ply as target genes has previously been developed (9). However, the bexA gene is present only in encapsulated H. influenzae, and the bexA-based real-time PCR assay is not able to detect nontypeable H. influenzae. Additionally, the lytA PCR assay has been shown to be more specific than the ply assay for detection of S. pneumoniae (4). The multiplex assay described in this study improves upon these previously developed assays by using the protein D gene as a target for detection of both typeable and nontypeable H. influenzae and the lytA gene as a target for detection of S. pneumoniae (4, 6, 28). Both multiplex real-time PCR assays use the ctrA gene as a target for detection of N. meningitidis. While the ctrA-based real-time PCR assay is a suitable tool for capturing most of the encapsulated N. meningitidis strains, it fails to detect strains that lack ctrA or that have significant variation in this gene (5, 7, 11). A recently developed real-time PCR assay targeting the N. meningitidis sodC gene can capture groupable and nongroupable N. meningitidis and is very useful for analyzing isolates or specimens collected from nasopharyngeal carriage surveys (12). The performance of the sodC assay in the multiplex platform remains to be investigated.
A CSF specimen from a meningitis patient typically has bacterial counts in excess of 103 to 105 CFU per milliliter (17). The LLDs of the multiplex real-time PCR assays described in this study range from 1 to 210 genome equivalents per real-time PCR (250 to 52,500 CFU equivalents per milliliter), which is within the range of bacterial counts in a meningitis CSF specimen. The real-time PCR assays described in this study should be able to detect the target pathogen from meningitis CSF specimens with typical bacterial load. This study used a CT cutoff value of 35 for all real-time PCR assays. We recommend that specimens with high CT values (>35 and ≤40) should be analyzed individually. We have observed that nonspecific amplification results in high CT values and that specimens with high CT values often have poor replicate reproducibility, which leads to higher rates of false positives (data not shown). High CT values could also result from the presence of PCR inhibitors or target DNA degradation or low DNA quantity due to antibiotic treatment prior to CSF specimen collection or suboptimal specimen transport and storage conditions. Some of these possibilities can be evaluated by diluting specimens which have high CT values or by performing antibiotic detection. In any case, interpretation of specimens with high CT value should include consideration of clinical data and other laboratory results.
Several factors may influence the multiplex real-time PCR performance compared to the singleplex performance, including PCR reagents, DNA quality, and fluorescence reporter dyes. We observed lower sensitivity of some assays in multiplex than in singleplex when different commercial PCR reagents were used. In addition, the serogroup A sacB assay failed to detect N. meningitidis A when the probe was labeled with Cy5 in multiplex (data not shown) but performed well when the probe was labeled with Hex. This underscores the importance of assay optimization when developing multiplex real-time PCR assays from the singleplex real-time PCR assays. The optimal conditions for singleplex real-time PCR may not be optimal for the multiplex PCR. The performance of the multiplex real-time PCR assay does not seem to be affected by different PCR instruments or methods used for DNA extraction.
The three multiplex real-time PCR assays described here have demonstrated sensitivities similar to that of the singleplex real-time PCR for detecting bacterial meningitis pathogens in clinical isolates, CSF specimens, blood, nasal washes, throat swabs, and inoculated transport medium specimens. Multiplex real-time PCR also conserves valuable resources, which makes it particularly useful in resource-limited settings. These advantages make the multiplex real-time PCR assays developed in this study a widely useful tool for disease surveillance and research into the causative pathogens of bacterial meningitis.
Supplementary Material
ACKNOWLEDGMENTS
We greatly thank the Emerging Infection Program's Active Bacterial Core surveillance team for providing clinical isolates. We also thank the Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa for collecting specimens for surveillance, the Streptococcus Laboratory in the Respiratory Disease Branch at the CDC for providing DNA, Brian Plikaytis in the Division of Bacterial Disease at the CDC for consultation, and Mary McCauley at the CDC for her critical review.
Footnotes
Published ahead of print 14 December 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Altman DG, Bland JM. 1983. Measurement in medicine: the analysis of method comparison studies. Statistician 32:307–317 [Google Scholar]
- 2. Baughman AL, et al. 2008. Utility of composite reference standards and latent class analysis in evaluating the clinical accuracy of diagnostic tests for pertussis. Clin. Vaccine Immunol. 15:106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bland JM, Altman DG. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307–310 [PubMed] [Google Scholar]
- 4. Carvalho MDGS, et al. 2007. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J. Clin. Microbiol. 45:2460–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cavrini F, Liguori G, Andreoli A, Sambri V. 2010. Multiple nucleotide substitutions in the Neisseria meningitidis serogroup C ctrA gene cause false-negative detection by real-time PCR. J. Clin. Microbiol. 48:3016–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. CDC 2010. Streptococcus laboratory protocols. http://www.cdc.gov/ncidod/biotech/strep/protocols.htm
- 7. Claus H, Maiden MC, Maag R, Frosch M, Vogel U. 2002. Many carried meningococci lack the genes required for capsule synthesis and transport. Microbiology 148:1813–1819 [DOI] [PubMed] [Google Scholar]
- 8. Cohn AC, et al. 2010. Changes in Neisseria meningitidis disease epidemiology in the United States, 1998-2007: implications for prevention of meningococcal disease. Clin. Infect. Dis. 50:184–191 [DOI] [PubMed] [Google Scholar]
- 9. Corless CE, et al. 2001. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J. Clin. Microbiol. 39:1553–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. da Gloria Carvalho M, et al. 2010. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes. J. Clin. Microbiol. 48:1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dolan-Livengood JM, Miller YK, Martin LE, Urwin R, Stephens DS. 2003. Genetic basis for nongroupable Neisseria meningitidis. J. Infect. Dis. 187:1616–1628 [DOI] [PubMed] [Google Scholar]
- 12. Dolan Thomas J, et al. 2011. sodC-based real-time PCR for detection of Neisseria meningitidis. PLoS One 6:e19361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geslin P, Legrand P, Lemoine JL, Squinazi F, Borderon JC. 1977. The detection of bacterial antigens by counter-immunoelectrophoresis in N. meningitidis, H. influenzae serotype b, S. pneumoniae infections. Diagnostic value and evolutive aspect (in 216 cases). Pathol. Biol. (Paris) 25:711–721 (In French.) [PubMed] [Google Scholar]
- 14. Harrison LH. 2010. Epidemiological profile of meningococcal disease in the United States. Clin. Infect. Dis. 50(Suppl 2):S37–S44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jin P, et al. 2009. First report of putative Streptococcus pneumoniae serotype 6D among nasopharyngeal isolates from Fijian children. J. Infect. Dis. 200:1375–1380 [DOI] [PubMed] [Google Scholar]
- 16. Khatami A, Pollard AJ. 2010. The epidemiology of meningococcal disease and the impact of vaccines. Expert Rev. Vaccines 9:285–298 [DOI] [PubMed] [Google Scholar]
- 17. La Scolea LJ, Jr, Dryja D. 1984. Quantitation of bacteria in cerebrospinal fluid and blood of children with meningitis and its diagnostic significance. J. Clin. Microbiol. 19:187–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levine OS, et al. 2010. Global status of Haemophilus influenzae type b and pneumococcal conjugate vaccines: evidence, policies, and introductions. Curr. Opin. Infect. Dis. 23:236–241 [DOI] [PubMed] [Google Scholar]
- 19. Lynch JP, III, Zhanel GG. 2010. Streptococcus pneumoniae: epidemiology and risk factors, evolution of antimicrobial resistance, and impact of vaccines. Curr. Opin. Pulm. Med. 16:217–225 [DOI] [PubMed] [Google Scholar]
- 20. McIntosh ED, Reinert RR. 2011. Global prevailing and emerging pediatric pneumococcal serotypes. Expert Rev. Vaccines 10:109–129 [DOI] [PubMed] [Google Scholar]
- 21. Metzgar D, et al. 2009. Evaluation and validation of a real-time PCR assay for detection and quantitation of human adenovirus 14 from clinical samples. PLoS One 4:e7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mothershed EA, et al. 2004. Use of real-time PCR to resolve slide agglutination discrepancies in serogroup identification of Neisseria meningitidis. J. Clin. Microbiol. 42:320–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Norskov-Lauritsen N. 2009. Detection of cryptic genospecies misidentified as Haemophilus influenzae in routine clinical samples by assessment of marker genes fucK, hap, and sodC. J. Clin. Microbiol. 47:2590–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Brien KL, et al. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902 [DOI] [PubMed] [Google Scholar]
- 25. Peltola H. 2000. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 13:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poland GA. 2010. Prevention of meningococcal disease: current use of polysaccharide and conjugate vaccines. Clin. Infect. Dis. 50(Suppl 2):S45–S53 [DOI] [PubMed] [Google Scholar]
- 27. Stephens DS, Greenwood B, Brandtzaeg P. 2007. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369:2196–2210 [DOI] [PubMed] [Google Scholar]
- 28. Wang X, et al. 2011. Detection of bacterial pathogens in Mongolia meningitis surveillance with a new real-time PCR assay to detect Haemophilus influenzae. Int. J. Med. Microbiol. 301:303–309 [DOI] [PubMed] [Google Scholar]
- 29. Watt JP, et al. 2009. Burden of disease caused by Haemophilus influenzae type b in children younger than 5 years: global estimates. Lancet 374:903–911 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.