Abstract
blaOXA-134 genes and variants were sought in 21 species of Acinetobacter and found in A. lwoffii, genomic species 9 (regarded as synonyms), and A. schindleri. Sequencing revealed a 9-bp deletion in the gene in the type strain of genomic species 9 (ATCC 9957) relative to the gene in the type strain of A. lwoffii (ATCC 15309). Primers based on the gene without the deletion gave specific amplification of 29 of 30 clinical isolates of A. lwoffii/genomic species 9.
TEXT
Species within the Acinetobacter genus are difficult to distinguish except by molecular methods. Hospital isolates are dominated by A. baumannii, which is usually resistant to most antibiotics and can relatively readily be identified by detection of the blaOXA-51-like intrinsic carbapenemase gene or by other PCR methods (3, 5, 14). However, it is clear that other species are also clinically important, with rpoB sequencing having been instrumental in providing a means of species identification facilitating assessment of the prevalence and clinical relevance of some of the less common species (6, 8, 15, 16). Among these, A. lwoffii/genomic species 9 (now considered synonyms for the same species) (10) is relatively commonly found from blood, sometimes as a result of contamination from the skin but also associated with bacteremia (12, 13), and a rapid method of detection would be useful. Recently, a carbapenem-hydrolyzing class D β-lactamase, OXA-134, has been described in A. lwoffii, providing a potential target for species-specific identification (4). In the original study, the gene and its variants (referred to as blaOXA-134-like) were sought using flanking primers, but we chose to amplify a smaller, internal fragment so that detection could be combined with amplification of variable regions of the rpoB gene. This provides an internal control for Acinetobacter species in the PCR and affords the possibility of identifying the non-lwoffii isolates by sequencing the rpoB amplicon, if required.
We have used a similar approach for detecting A. baumannii for some time, with primers for amplification of the intrinsic blaOXA-51-like being combined with those for amplification of rpoB; primers for detection of acquired OXA and class 1 integrase genes are also included (Table 1) (14, 17). The PCR is carried out using the Qiagen multiplex PCR kit as recommended by the manufacturers, with an annealing temperature of 57°C, in a reaction mixture volume of 25 μl. However, a substantial proportion of blood isolates of this genus identify as A. lwoffii, and for these, screening for this species is often more relevant.
Table 1.
Primers for characterization of isolates of Acinetobacter spp.
| Target | Primer | Sequence (5′ to 3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| blaOXA-134-like/rpoB PCRa | ||||
| rpoB | Ac696F | TAYCGYAAAGAYTTGAAAGAAG | 858 | 8 |
| Ac1598R | CGBGCRTGCATYTTGTCRT | |||
| A. lwoffii/g sp. 9 blaOXA-134-like | OXA-134F3 | ACTCAATCSACYCAAGCCA | 223 | This study |
| OXA-134_307R2 | GTTTCTTGCCATCCCATTTA | |||
| Alternative blaOXA-134-like primer pairsa | ||||
| blaOXA-134-like | OXA-134_29F2 | TGAGTTGCTTGGGCCTGA | 281/290 | This study |
| OXA-134_309R | GCGTTTCTTGCCATCCC | |||
| blaOXA-134a | OXA-134_74F | TCCCATCTCAAAGCATTTC | 234 | This study |
| OXA-134_307R2 | GTTTCTTGCCATCCCATTTA | |||
| blaOXA-58-like,23-like,51-like,40-like, -143/class 1 integrase gene/rpoB multiplex PCR | ||||
| rpoB | Ac696F | TAYCGYAAAGAYTTGAAAGAAG | 858 | 8 |
| Ac1598R | CGBGCRTGCATYTTGTCRT | |||
| blaOXA-58-like | mpOXA-58-likeF | AAGTATTGGGGCTTGTGCTG | 599 | 17 |
| mpOXA-58-likeR | CCCCTCTGCGCTCTACATAC | |||
| blaOXA-23-like | mpOXA-23-likeF | GATCGGATTGGAGAACCAGA | 501 | 17 |
| mpOXA-23-likeR | ATTTCTGACCGCATTTCCAT | |||
| blaOXA-51-like | mpOXA-51-likeF | TAATGCTTTGATCGGCCTTG | 353 | 17 |
| mpOXA-51-likeR | TGGATTGCACTTCATCTTGG | |||
| blaOXA-40-like | mpOXA-24-likeF | GGTTAGTTGGCCCCCTTAAA | 246 | 17 |
| mpOXA-24-likeR | AGTTGAGCGAAAAGGGGATT | |||
| blaOXA-143 | OXA-143 303F2 | GATTTTCAAATGGGACGGT | 90 | This study |
| OXA-143 392R2 | GGAACTGCTGAAAGTGCC | |||
| Class 1 integrase gene | Int1F | CAGTGGACATAAGCCTGTTC | 160 | 7 |
| Int1R | CCCGAGGCATAGACTGTA | |||
The OXA-134F3/OXA-134_307R2 primer pair (in bold) specifically detected most clinical isolates (29/30) of A. lwoffii/genomic species 9; those with the blaOXA-134a allele (which are missed) were detected using the OXA-134_74F/OXA-134_307R2 primer pair. The OXA-134_29F2/OXA-134_309R primers amplified blaOXA-134-like in A. lwoffii, genomic species 9 and A. schindleri.
Primers OXA-134_29F2 and OXA-134_309R (Table 1) were designed from consensus regions of the blaOXA-134a sequence of isolate AL3 (GenBank accession number HQ122933) and the corresponding sequence of the type strain of A. lwoffii (ATCC 15309), kindly provided by Jacqueline Chan following whole-genome sequencing. PCRs were carried out in 25-μl volumes containing 10 pmol of each primer (OXA-134_29F2, OXA-134_309R, and rpoB primers Ac696F and Ac1598R), 1× Qiagen CoralLoad buffer, deoxynucleoside triphosphates (200 μM each), 2 μl cell lysate, and Taq DNA polymerase (1.5 U). Reaction conditions were 94°C for 3 min, followed by 35 cycles of 94°C for 45 s, 50°C for 45 s, and 72°C for 1 min, followed by a 5-min final extension at 72°C.
This first version of the PCR was tested on type strains of the former genomic species 1 to 17, most of which are now named species (http://www.bacterio.cict.fr/a/acinetobacter.html) (Table 2) and 80 clinical isolates received from United Kingdom hospitals, previously subjected to the blaOXA-58-like, -23-like, -51-like, -40-like, -143/class 1 integrase gene PCR and rpoB sequencing. These included 30 isolates that had been identified as A. lwoffii/genomic species 9 and representatives of other clinically relevant species, as follows: A. ursingii (n = 4), A. parvus (n = 5), A. schindleri (n = 4), A. gyllenbergii (n = 1), A. soli (n = 1) (species not otherwise represented), A. calcoaceticus (n = 2), A. baumannii (n = 10), A. pittii (n = 3), A. nosocomialis (n = 2), A. junii (n = 1), A. haemolyticus (n = 1), A. radioresistens (n = 4), A. johnsonii (n = 3), genomic species 13BJ (n = 1), genomic species 16 (n = 1), A. bereziniae (n = 3), and A. baylyi or A. guillouiae (n = 3). Reaction products were separated by agarose gel electrophoresis followed by staining with GelRed (3× in 0.1 M NaCl) and photography under UV illumination or on a QIAxcel instrument (Qiagen) using the high-resolution cartridge and 3-kb/15-bp alignment marker.
Table 2.
Reference strains used in this study
| Genomic speciesc | Species name | Isolate |
|---|---|---|
| 1 | A. calcoaceticus | ATCC 23055 |
| 2 | A. baumannii | ATCC 19606 |
| 3 | A. pittii | ATCC 19004 |
| 4 | A. haemolyticus | ATCC 17906 |
| 5 | A. junii | ATCC 17908 |
| 6 | Acinetobacter genomic species 6 | ATCC 17979 |
| 7 | A. johnsonii | ATCC 17909 |
| 8a | A. lwoffii | ATCC 15309 |
| 9a | Acinetobacter genomic species 9 | ATCC 9957 |
| 10 | A. bereziniae | ATCC 17924 |
| 11 | A. guillouiae | ATCC 11171 |
| 12 | A. radioresistens | ATCC 43998 |
| 13BJ | Acinetobacter genomic species 13 | ATCC 17905 |
| 13TUb | A. nosocomialis | ATCC 17903 |
| 14BJb | Acinetobacter genomic species 14 | Bouvet 382 |
| 15 | Acinetobacter genomic species 15 | Bouvet 79 |
| 16 | Acinetobacter genomic species 16 | ATCC 17988 |
| 17 | Acinetobacter genomic species 17 | Bouvet 942 |
Probably synonyms for the same species (10).
Probably synonyms for the same species (10).
Genomic species numbering is according to Bouvet and Grimont (1) and Bouvet and Jeanjean (BJ) (2), unless indicated otherwise (TU; Tjernberg and Ursing [13a]).
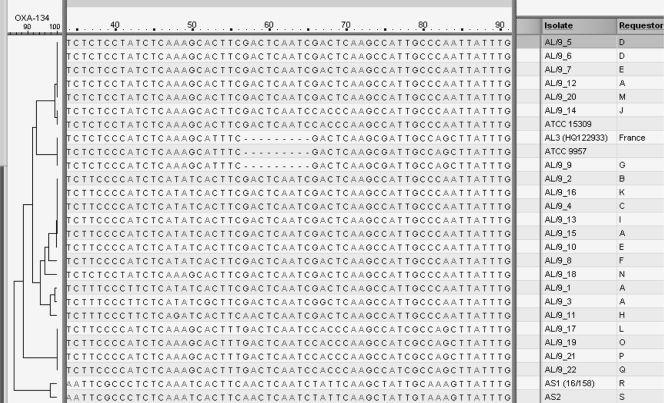
While the type strains of A. lwoffii (ATCC 15309) and genomic species 9 (ATCC 9957) and field isolates of A. lwoffii/genomic species 9 gave the blaOXA-134-like band in this PCR, so too did our isolates of A. schindleri (4/4). Two isolates (ATCC 9957 and a clinical isolate of A. lwoffii/genomic species 9) gave slightly smaller amplicons than the other isolates. The blaOXA-134-like products from representatives of each of these groups were sequenced and compared in a BioNumerics (Applied Maths) database (Fig. 1). The gene in the type strain of genomic species 9 (ATCC 9957) has a deletion of 9 bp relative to the gene in the type strain of A. lwoffii and in most of the field isolates of A. lwoffii/genomic species 9 that we sequenced (21/22); one field isolate (isolate AL/9_9) also had this deletion, as does the AL3 isolate in the original description of blaOXA-134a (HQ122933). The A. schindleri sequences showed less than 90% (∼85%) homology to those of the other species and clearly clustered apart from them.
Fig 1.
Screenshot of comparison of partial blaOXA-134-like sequences corresponding to nucleotides 37 to 310 of the coding sequence of blaOXA-134a (GenBank accession number HQ122933) showing the region with the 9-bp deletion in ATCC 9957, AL3, and clinical isolate AL/9_9, using BioNumerics (Applied Maths) software. Isolates were identified as A. lwoffii/genomic species 9 (AL/9_1 to AL/9_22) or A. schindleri (AS1 and AS2) by rpoB sequence cluster analysis. Each isolate was from a different patient, from United Kingdom hospitals A to S, collected between 2009 and 2011. Comparison was by the unweighted-pair group method with arithmetic mean (UPGMA).
Among the A. lwoffii/genomic species 9 isolates, there were four clusters of blaOXA-134-like sequences, that with the deletion being one of them (Fig. 1). On the basis of the type strains, one might anticipate that the allele with the deletion may distinguish genomic species 9 from A. lwoffii, but that is not supported by rpoB sequence cluster analysis, which shows that the clinical isolate with the deletion shared the same rpoB sequence as isolates without it and clustered more closely with ATCC 15309 than ATCC 9957, supporting a single species. In view of the extent of sequence diversity in blaOXA-134-like, two primer pairs were designed to specifically detect A. lwoffii/genomic species 9, one with a forward primer (OXA-134F3) to detect alleles without the deletion and the second with a forward primer (OXA-134_74F) to detect the allele with the deletion; a slightly modified reverse primer (OXA-134_307R2) was used, since this was found to give cleaner PCR products. As before, each pair was used in combination with the rpoB primers (Ac696F and Ac1598R), and similar conditions were used (94°C for 3 min followed by 32 cycles of 94°C for 45 s, 52°C for 45 s, and 72°C for 1 min, followed by a 5-min final extension at 72°C); each primer was present at 8 pmol per reaction mixture. The same panel of isolates was used. All the isolates of A. lwoffii/genomic species 9 gave a blaOXA-134-like band of the expected size (223 bp) with the OXA-134F3/OXA-134_307R2 primer pair, with the exception of that with the deletion, while none of the other species, including A. schindleri and the type strain of genomic species 9, gave this band. All isolates gave the rpoB band. When the OXA-134_74F/OXA-134_307R2 primer pair was used, the type strain of genomic species 9 and the isolate with the deletion gave the blaOXA-134-like band; some of the other A. lwoffii/genomic species 9 isolates gave slightly larger, fainter bands. We recommend screening with the OXA-134F3/OXA-134_307R2 primer pair to identify A. lwoffii/genomic species 9, which will detect most isolates. Those with the deletion, which will be missed, can be identified either by sequencing the rpoB amplicon or by screening with the OXA-134_74F/OXA_307R2 primer pair.
The finding of highly similar alleles of blaOXA-134-like in A. lwoffii and genomic species 9 supports their being assigned to a single species. Our results on isolates from 30 patients from 22 hospitals indicate that most field isolates of A. lwoffii/genomic species 9 do not have the deletion associated with the type strain of genomic species 9. Although there is some potential for intrinsic OXA genes to move between species (e.g., blaOXA-51-like from A. baumannii to A. nosocomialis [formerly genomic species 13TU] [9] and blaOXA-23-like from A. radioresistens to A. baumannii [11]), blaOXA-134-like was only found in the species described here among our panel. The PCR using the OXA-134F3/OXA-134_307R2 primer pair provides a rapid method of specific detection of A. lwoffii/genomic species 9. Should confirmation of the identification be required, the rpoB amplicon can additionally be sequenced.
Nucleotide sequence accession numbers.
The partial blaOXA-134-like sequences of ATCC 9957, two clinical isolates of A. lwoffii/genomic species 9 (AL/9_7 and AL/9_10), and a clinical isolate of A. schindleri (AS1) are deposited in GenBank under accession numbers JN203134, JN804564, JN804565, and JN203135, respectively.
ACKNOWLEDGMENTS
We are grateful to Jacqueline Chan and Mark Pallen at the University of Birmingham for providing the blaOXA-134-like sequence of the type strain of A. lwoffii, to the Genomic Services Unit at Centre for Infections, Colindale, for sequencing services, and to colleagues in hospital laboratories for sending these isolates.
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Bouvet PJ, Grimont PA. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov., and emended description of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36:228–240 [Google Scholar]
- 2. Bouvet PJ, Jeanjean S. 1989. Delineation of new proteolytic genomic species in the genus Acinetobacter. Res. Microbiol. 140:291–299 [DOI] [PubMed] [Google Scholar]
- 3. Chen TL, et al. 2007. Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:801–806 [DOI] [PubMed] [Google Scholar]
- 4. Figueiredo S, et al. 2010. OXA-134, a naturally occurring carbapenem-hydrolyzing class D beta-lactamase from Acinetobacter lwoffii. Antimicrob. Agents Chemother. 54:5372–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Higgins PG, Wisplinghoff H, Krut O, Seifert H. 2007. A PCR-based method to differentiate between Acinetobacter baumannii and Acinetobacter genomic species 13TU. Clin. Microbiol. Infect. 13:1199–1201 [DOI] [PubMed] [Google Scholar]
- 6. Karah N, et al. 2011. Species identification and molecular characterization of Acinetobacter spp. blood culture isolates from Norway. J. Antimicrob. Chemother. 66:738–744 [DOI] [PubMed] [Google Scholar]
- 7. Koeleman JGM, Stoof J, van der Bijl MW, Vandenbroucke-Grauls CMJE, Savelkoul PHM. 2001. Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J. Clin. Microbiol. 39:8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. La Scola B, Gundi VA, Khamis A, Raoult D. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 44:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee YT, et al. 2009. First identification of blaOXA-51-like in non-baumannii Acinetobacter spp. J. Chemother. 21:514–520 [DOI] [PubMed] [Google Scholar]
- 10. Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poirel L, Figueiredo S, Cattoir V, Carattoli A, Nordmann P. 2008. Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 52:1252–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Regalado NG, Martin G, Antony SJ. 2009. Acinetobacter lwoffii: bacteremia associated with acute gastroenteritis. Travel Med. Infect. Dis. 7:316–317 [DOI] [PubMed] [Google Scholar]
- 13. Tega L, et al. 2007. Catheter-related bacteremia and multidrug-resistant Acinetobacter lwoffii. Emerg. Infect. Dis. 13:355–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a. Tjernberg I, Ursing J. 1989. Clinical strains of Acinetobacter classified by DNA-DNA hybridization. APMIS 97:595–605 [DOI] [PubMed] [Google Scholar]
- 14. Turton JF, et al. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44:2974–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turton JF, Shah J, Ozongwu C, Pike R. 2010. Incidence of Acinetobacter species other than A. baumannii among clinical isolates of Acinetobacter: evidence for emerging species. J. Clin. Microbiol. 48:1445–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van den Broek PJ, et al. 2009. Endemic and epidemic Acinetobacter species in a university hospital, an eight years' survey. J. Clin. Microbiol. 47:3593–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Woodford N, et al. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351–353 [DOI] [PubMed] [Google Scholar]