Iron, copper, and zinc are all essential nutrients. The electron transfer properties of iron and copper are fundamental to processes such as respiration and photosynthesis. Zinc forms the catalytic center in numerous enzymes and has an important structural role in a wide range of proteins. However, all these metals can be toxic if their levels and distribution are not carefully regulated, as their inappropriate binding may compromise cellular function. The uncontrolled redox activity of iron and copper can also lead to the generation of damaging oxygen radicals. Therefore, organisms maintain cytoplasmic metal concentrations at a nontoxic level that is sufficient for growth. A variety of homeostatic mechanisms have been identified, which include the control of translation and RNA stability by iron-regulatory proteins and the metal-dependent trafficking or degradation of metal transporters (39, 109, 138). This review focuses on the role that metal-responsive transcription factors have in regulating trace metal metabolism. These factors are able to sense changes in metal concentrations and coordinate the expression of genes that are involved in the acquisition, distribution, sequestration, and use of metals. Consequently, the ability to mediate metal-responsive gene expression is an important aspect of metal homeostasis in those organisms that contain these factors.
IRON
Iron is extremely insoluble in the presence of oxygen at physiological pH. Organisms that live in an oxygen environment have therefore evolved specific mechanisms to acquire what would otherwise be an unavailable nutrient. These systems of iron acquisition in many fungi are regulated at the transcriptional level by iron availability. The proteins that mediate this control are, to date, the only known iron-responsive transcription factors within eukaryotes. In the budding yeast Saccharomyces cerevisiae, genes that are involved in the compartmentalization and use of iron are regulated in a similar way to those genes that are involved in iron acquisition. This global regulation of iron metabolism may be established to be the norm in other eukaryotic microorganisms.
The iron metabolism of S. cerevisiae has been the most intensively studied of all the fungi (reviewed in references 83, 114, and 146). This organism can grow in both aerobic and anaerobic environments and can utilize a variety of carbon sources by using both fermentative and respiratory metabolism. This range of growth conditions influences iron availability and the cell requirements for iron. Under anaerobic conditions, iron is in the ferrous form and therefore more readily available. Conversely, cells that are respiring require additional iron for the various iron-containing proteins of the mitochondrial respiratory chain at a time when iron is less soluble. Therefore, mutations that are detrimental to iron metabolism often result in a more severe phenotype when this yeast grows by using a respiratory carbon source.
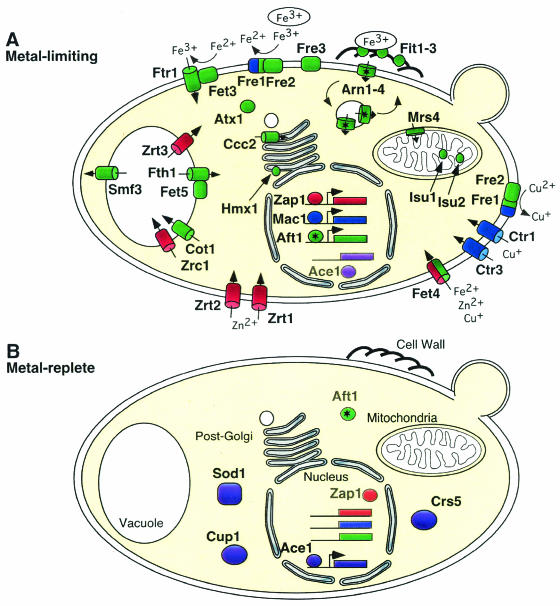
S. cerevisiae contains a variety of genes that are transcriptionally induced in response to low iron and which encode proteins that are involved in iron acquisition at the cell surface (Table 1; Fig. 1A). Free iron is taken into the cell by both high- and low-affinity transport systems (Fet3, Ftr1, and Fet4) (36, 136). The high-affinity complex contains a ferroxidase (Fet3) that requires copper as a cofactor. Consequently, genes that are involved in the trafficking and transport of copper to this protein (ATX1 and CCC2) are also regulated at the transcriptional level by iron (93, 156, 160). High-affinity iron uptake is therefore compromised when cells experience low copper levels. A cell surface ferric reductase activity is also required for high-affinity iron uptake (33, 34). The majority of this activity is provided by two flavocytochromes (Fre1 and Fre2) that reduce ferric iron to provide ferrous iron as a substrate for the high-affinity transport system (48, 49). S. cerevisiae can also acquire iron through siderophores, which are low-molecular-weight organic molecules that specifically chelate iron. S. cerevisiae does not synthesize its own siderophores, but it is able to utilize those that are produced by other microorganisms (114). A family of transporters (Arn1 to Arn4) that cycle between the cell surface and an endosomal compartment mediate siderophore uptake (162). As an alternative to siderophore uptake, the siderophore iron can be reduced by the cell surface reductases (Fre1 to Fre4) to provide ferrous iron as a substrate for the Fet3/Ftr1 high-affinity uptake system (162). A group of mannoproteins (Fit1 to Fit3) facilitates siderophore iron uptake by sequestering this iron chelate within the cell wall (117). In addition to those genes that are involved in cell surface iron acquisition, a number of genes involved in other aspects of iron metabolism are transcriptionally induced under low-iron conditions (Table 1). These include genes that encode vacuolar transport systems (Fet5, Fth1, and Smf3), a mitochondrial transporter (Mrs4), and proteins involved in the biosynthesis of iron-sulfur clusters (Isu1 and Isu2) (43, 47, 115, 116, 129, 145).
TABLE 1.
Genes that are regulated by metal-responsive transcription factorsa
| Transcription factorb | Description | Gene name(s) | Reference(s) |
|---|---|---|---|
| Aft1 | Transporters | FET4, FET5, FTR1, FTH1, SMF3, MRS4, CCC2, COT1 | 72, 115, 124, 125, 149, 156 |
| Cu chaperone | ATX1 | 93 | |
| Ferroxidase | FET3, FET5 | 124, 154 | |
| Metalloreductases | FRE1, FRE2, FRE3, FRE4, FRE5, FRE6 | 98, 154 | |
| Cell wall proteins | FIT1, FIT2, FIT3 | 117 | |
| Siderophore transport | ARN1, ARN2, ARN3, ARN4 | 162 | |
| Fe-S biosynthesis | ISU1, ISU2 | 47 | |
| Otherd | TIS11, HMX1, AKR1, PCL5, YOR387c, YHL035c, YMR034c, ICY2, PRY1, YDL124w | 44, 118, 125, 135 | |
| Aft2 | Transporters | SMF3, MRS4, FTR1, COT1 | 115, 125 |
| Cu chaperone | ATX1 | 14 | |
| Ferroxidase | FET3, FET5 | 14, 124 | |
| Metalloreductase | FRE1 | 125 | |
| Cell wall protein | FIT1, FIT3, FIT2 | 124, 125 | |
| Fe-S biosynthesis | ISU1 | 125 | |
| Otherd | BNA2, ECM4, LAP4, TIS11, YOL083w, YGR146c, YHL035c | 125 | |
| Fep1 | Transporter | fip1+ | 107 |
| Ferroxidase | fio+ | 107 | |
| Siderophore transport | str1+, str2+, str3+ | 108 | |
| SREA | Siderophore biosynthesis | sidA, sidB, sidC, amcA, atrH, estA | 103 |
| Siderophore transport | mirA, mirB, mirC | 103 | |
| Otherd | cycA, acoA, lysF | 102 | |
| Urbs1 | Siderophore biosynthesis | sid1, sid2 | 2, 161 |
| Ace1 | Cu metallothioneins | CUP1, CRS5 | 29, 139, 150 |
| Cellular stress response | SOD1 | 55 | |
| Amt1 | Cu metallothioneins | MT-I, MT-IIa, MT-IIb | 141, 172 |
| Gene regulation | AMT1 | 174 | |
| Crf1 | Cu metallothioneins | MTPI, MTPI1 | 46 |
| Mac1 | Cu transporters | CTR1, CTR3 | 85, 155 |
| Metalloreductases | FRE1, FRE7 | 49, 98 | |
| Otherd | YFR055w, YJL217w, YLR213c | 57 | |
| Cuf1 | Cu transporters | ctr4+, ctr5+, ctr6+ | 7, 9, 84 |
| Fe transport | fip+c | 84 | |
| Metalloreductase | frp1+c | 84 | |
| Multicopper oxidase | fio1+c | 84 | |
| GRISEA | Cu transport | PaCTR3 | 16 |
| Cellular stress response | PaSOD2 | 18 | |
| Crr1 | Heme biosynthesis | CPX1 | 68 |
| Photosystem I maintanence | CRD1, CTH1c | 100, 101 | |
| Electron transfer | CYC6 | 67, 99 | |
| Zap1 | Zn transporters | ZRT1, ZRT2, ZRT3, ZRC1, FET4, ZRG17 | 94, 95, 96, 149, 159, 166, 167 |
| Gene regulation | ZAP1, NRG2 | 94, 168 | |
| Phosphate/lipid metabolism | YOL002c | 94 | |
| Metabolic enzymes | DPP1, ADH4, MNT2, ADE17, TKL2, URA10 | 94 | |
| Vacuolar proteases | PRC1, PEP4 | 94 | |
| Otherd | MCD4, ZPS1, RAD27, ZIP1, GRE2, BAG7, FLO1, YNL254c, YLL020c, YGL258w, YOR387c, YJR061w, YMR086w, YOL131w, GPG1, COS1, COS2, COS3, COS4, COS6, COS8, YJL132w, ICY2, PST1, YBL048w, YBL049w, YNL234w, YDR492w, YKL174c, PHM7 | 94, 159 | |
| Mammalian MTF-1 | Zn transporters | ZNT-1, ZTL1 | 27, 87 |
| Metallothioneins | MT-I, MT-II | 66 | |
| Cellular stress response | AFP, LCN1, PIGF, γ-GCS | 56, 59, 92 | |
| Other | AHSG, CBG, PMP22, XIST, ACVR2b | 92 | |
| Drosophila MTF-1 | Metallothioneins | MTNA, MTNB, MTNC, MTND | 38, 163 |
The genes that are listed are (i) metal regulated and (ii) have a consensus sequence or sequence that resembles the consensus binding site for the relevant transcription factor within the promoter region and/or encode for a protein that is involved in metal metabolism. Genes that have been analyzed in such a way to demonstrate a more direct interaction with the factor in question are shown in bold. Examples of experimental evidence that is consistent with a direct interaction include analysis of expression in the presence and absence of the wild-type or constitutive alleles of the relevant factor and/or the use of reporter constructs containing the metal-responsive element(s) of the target gene in question.
No target gene has been identified for the SRE and SreP transcription factors.
Genes that are repressed, rather than activated, by the named transcription factor.
Genes which do not fall into any of the listed categories.
FIG. 1.
Protein products of metalloregulated genes involved in metal homeostasis in S. cerevisiae. Products of genes that are activated under metal-limiting conditions (A) and metal-replete conditions (B) by Aft1 (green), Mac1 (blue), Zap1 (red), and Ace1 (purple) are shown. Iron that is bound to siderophores has been circled, and stars indicate proteins that undergo iron-dependent cellular trafficking. The metal ion specificities of proteins required for metal uptake are indicated. See the text for further details of the functional roles of each protein.
Iron-dependent gene regulation in S. cerevisiae is mediated by two transcription factors. Aft1 and Aft2 (for “activator of ferrous transport”) activate gene expression when iron is scarce. Consequently, strains that lack both these factors exhibit reduced expression of the iron regulon (14, 24, 124, 154, 156). The genes that code for these factors are thought to have arisen from a genome duplication event (130). As with many other paralogous genes within S. cerevisiae, AFT1 and AFT2 code for proteins that have significant regions of identity and overlapping functions. The DNA-binding domain of each protein is in a highly conserved N-terminal region, and a conserved cysteine-to-phenylalanine mutation in both proteins generates a factor that activates the high expression of the iron regulon irrespective of iron concentrations (124, 154). There are clear phenotypic differences in strains that separately lack Aft1 and Aft2. An aft1 null strain exhibits low ferrous iron uptake and grows poorly under low-iron conditions or on a respiratory carbon source (24, 154). No phenotype has been attributed to an aft2 null strain. An aft1 aft2 double null strain is, however, more sensitive to low-iron growth than a single aft1 null strain, which is consistent with the functional similarity of these factors (14, 124). The partial redundancy of these factors allows Aft2 to complement an aft1 null strain when it is overexpressed from a plasmid (124).
The properties of Aft1 and Aft2 that distinguish them from each other have not been fully identified. Both factors mediate gene regulation via an iron-responsive element that contains the core sequence 5′-CACCC-3′ (125, 156). It is likely that sequences adjacent to this element influence the ability of each factor to mediate regulation via a particular iron-responsive element (125). The differential regulation of individual genes by Aft1 and Aft2 results in each factor generating a distinct global transcriptional profile (125). Aft1 autoregulation, which is consistent with the in vivo binding of Aft1 to its own promoter, may influence Aft1 control of the iron regulon (89). Critical to the function of Aft1 is its ability to shuttle between the nucleus and cytoplasm in response to iron levels. Nuclear export is dependent on a leucine rich N-terminal nuclear export signal (NES), mutation of which results in the nuclear retention of Aft1 and constitutive expression of Aft1-regulated genes (157). Aft1 nuclear import is mediated by a direct interaction with the nuclear import factor Pse1 (144). Aft1 contains two distinct basic nuclear localization signals, which together are sufficient and necessary to direct Aft1 to the nucleus (144).
Aft1 function is regulated in response to glucose levels independently of iron. Certain genes in the iron regulon are induced immediately following entry into the diauxic shift when cells adapt from fermentative to respiratory metabolism. This control is dependent on both the global regulatory complex Snf1/Snf4 and on Aft1 (64). The iron regulon also responds to glucose levels via the cyclic AMP-dependent protein kinase A. TPK2 encodes a protein kinase A catalytic subunit, and a tpk2 null strain shows derepressed expression of the iron regulon (123). Tpk2-dependent and Snf1/Snf4-dependent regulation of the iron regulon is consistent with an increased requirement for iron during respiratory metabolism (64, 123). Direct phosphorylation of Aft1 that is independent of Snf1 and cyclic AMP levels occurs when cells undergo a transient or permanent cell cycle arrest. Conditions that result in this modification of Aft1 include the change or removal of a carbon source, traversal of the diauxic shift, and the shifting of temperature-sensitive cdc28 or cdc25 mutants to a nonpermissive temperature (24, 64). The functional consequences of this cell cycle-dependent phosphorylation of Aft1 are not clear.
The mechanisms(s) by which the Aft1 and Aft2 factors sense iron are not fully understood. Nucleocytoplasmic shuttling of Aft1 in response to iron is fundamental to the ability of a cell to sense iron. However, the signal that shifts the equilibrium of Aft1 localization that results in nuclear translocation is not known. It is also not clear if that signal acts at the level of Aft1 nuclear export or import. The phosphorylation of Aft1 and the effect of the various phosphorylation pathways on the iron regulon described above are not involved in iron sensing per se. These regulatory pathways integrate Aft1 function with other aspects of cellular metabolism such as carbon source utilization. However, phosphorylation by an unknown signaling pathway could be the triggering event for Aft1 nuclear localization. Alternatively, Aft1 may bind iron directly and the loss of iron binding could initiate the Aft1 response. Direct metal binding of iron as a signal of iron status has been demonstrated with various prokaryotic iron-responsive transcription factors (reviewed in reference 6). Consistent with this hypothesis, the iron regulon in S. cerevisiae is sensitive to the intracellular chelation of iron and mutants that accumulate iron in the mitochondria exhibit enhanced expression of the iron regulon (44, 63). In addition, iron-responsive gene regulation is significantly compromised in cells that are unable to synthesize heme (28). The Fe(II)/Fe(III) redox equlibrium in the cell may also influence iron metabolism (63). This is supported by the phenotype of a sod1 null strain that lacks superoxide dismutase activity. This mutant is sensitive to oxidative stress and has higher than wild-type levels of Fe(III) with a concurrent increase in the expression of an Aft1/2-regulated gene (35, 134).
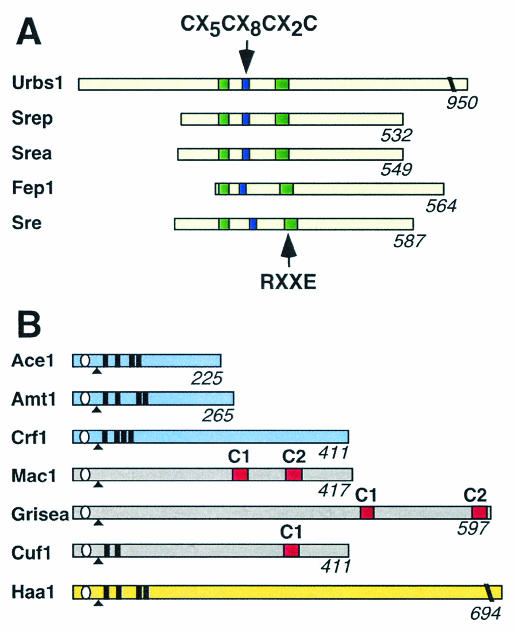
Iron-regulated gene expression in fungi other than S. cerevisiae is mediated by a group of GATA-type transcription factors. These include Fep1 from Schizosaccharomyces pombe, SREA from Aspergillus nidulans, SRE from Neurospora crassa, SreP from Penicillium chrysogenum, and Urbs1 from Ustilago maydis (60, 61, 107, 147, 171). These factors regulate, or are predicted to regulate, the expression of genes involved in siderophore production, siderophore transport, and free iron transport (Table 1). GATA factors are a group of transcription factors that are characterized by conserved zinc finger motifs and their ability to bind to a core 5′-GATA-3′ element. The fungal iron-responsive GATA factors contain two zinc finger Cys-X2-Cys-X17-Cys-X2-Cys motifs that flank a region that contains four conserved cysteine residues. Adjacent to the C-terminal zinc finger of each factor is a basic region that is conserved in other eukaryotic GATA factors (Fig. 2A). The iron-responsive GATA-type factors repress the transcription of their target genes in response to high iron. Therefore, although the iron-responsive GATA factors are transcriptional repressors and the Aft factors are transcriptional activators, both classes of factors ensure that their target genes are induced when the relevant organism senses that iron is limiting.
FIG. 2.
Schematic representation of the fungal iron-responsive GATA factors (A) and copper-responsive transcription factors (B). Shown are Mac1, Ace1, and Haa1 from S. cerevisiae, Fep1 and Cuf1 from S. pombe, SREA from A. nidulans, SRE from N. crassa, SreP from P. chrysogenum, Urbs1 from U. maydis, GRISEA from P. anserina, Amt1 from C. glabrata, and Crf1 from Y. lipolytica. In panel A, the conserved zinc finger motifs (green rectangles), the cysteine-rich region (blue rectangle), and the RXXE motif of the iron-responsive GATA factors are indicated. In panel B, the following motifs are shown: conserved zinc modules (white ovals), conserved (R/K)GRP motifs (black triangles), positions of N-terminal Cys-X-Cys or Cys-X2-Cys motifs outside the zinc motif (black rectangles), and positions of the C1 and C2 motifs (red rectangles). Proteins that are active under copper-limiting or copper-replete conditions are shown in grey and blue, respectively. Proteins that are not copper-responsive are shown in yellow.
The phenotypes of strains that lack an iron-responsive GATA factor are consistent with deregulation of iron metabolism. Wild-type siderophore production is repressed under high iron conditions but derepressed in strains that lack Urbs1, SREA, and SRE (61, 147, 170). Uptake of 59Fe(III) is higher in an sreA null strain than in a wild-type strain, and this confers sensitivity to the iron-dependent antibiotics phleomycin and streptonigrin (61). A fep1 null strain exhibits constitutive cell surface metalloreductase activity and is also sensitive to phleomycin (107). Consistent with these phenotypes, mutant strains that lack the relevant GATA factor exhibit constitutive expression of genes involved in the acquisition of iron from the environment (2, 103, 107, 108, 161). Fep1-dependent repression also requires Tup11 and Tup12 that may act as corepressors in a complex with Fep1 (108). A similar role in the regulation of the iron regulon in Candida albicans has also been proposed for the homologous Tup1, although the factor that recruits it to the relevant promoters has not been identified (80).
The number of functional 5′-GATA-3′ elements differs in the target promoters of the iron-responsive GATA factors. Fep1 regulates gene expression via two adjacent sites or a single site depending on the gene in question (107, 108). The full Urbs1-dependent regulation of a gene involved in siderophore synthesis requires two adjacent 5′-GATA-3′ elements, although these are not in themselves sufficient to confer repression of a reporter gene. Therefore, Urbs1 may interact with other GATA sites in this particular promoter region (2). The in vitro affinity of SRE for two adjacent 5′-GATA-3′ elements is dependent on the spacing between those two sites (62). Urbs1 and Fep1 have a higher in vitro affinity for one site when the target DNA contains two adjacent 5′-GATA-3′ elements. The loss of this high-affinity site results in a greater loss of in vivo repression of a reporter gene than does loss of the low-affinity site (2, 107). In addition, the phenotypes of various Urbs1 mutants suggest that, of the two zinc fingers, the C-terminal finger is more important for DNA binding (3).
Evidence to date suggests that the iron-responsive GATA factors bind iron. Recombinant SRE is reddish brown in color and gives a spectrum that is characteristic of iron-binding proteins, which is lost when the protein is reduced (62). The diagnostic spectrum of the wild-type protein is lost with proteins that contain cysteine-to-serine mutations in the conserved region between the zinc fingers. In vivo, these substitutions result in a constitutive repressor that does not respond to iron (62). Furthermore, the in vitro DNA-binding ability and stability of recombinant Fep1 is dependent on the protein being expressed in cells that are grown in high-iron medium before the protein is purified (107). In addition to the conserved cysteines between the zinc fingers, it has also been proposed that a conserved RXXE motif in the C-terminal zinc finger is a potential iron-binding site (107). An arginine-to-leucine mutation in the same motif in Urbs1 renders it unable to respond to iron (3). As with the iron-responsive factors from S. cerevisiae, further work is required to determine the precise mechanism of iron sensing by these factors.
COPPER
A number of homeostatic mechanisms have been identified which ensure that copper is maintained at a level sufficient for, but not toxic to, cell growth. In mammals, posttranslational mechanisms, such as the intracellular trafficking of copper transporters and the copper-stimulated endocytosis and degradation of proteins involved in copper uptake, play a major role in copper homeostasis (109, 112, 113). Although posttranslational control of transporters exists in fungi, copper homeostasis in these organisms is also mediated by the transcriptional regulation of genes involved in copper acquisition, mobilization, and sequestration (105). To date, six copper-responsive fungal factors have been characterized in detail. Ace1 (also known as Cup2), Amt1, and Crf1 activate gene expression in response to elevated copper while Mac1, GRISEA, and Cuf1 activate gene expression in response to copper deficiency. Homeostasis through copper-responsive transcriptional regulation has been observed in insects and plants, as well as in fungi, suggesting that this mechanism of copper control is widespread in nature (69, 126, 169).
Factors that are activated in response to copper regulate the expression of genes encoding proteins involved in copper sequestration and/or protection against copper toxicity (Table 1). In S. cerevisiae, resistance to copper is primarily mediated by the Ace1-dependent induction of the CUP1 gene (Fig. 1B) (139, 150). CUP1 encodes a small, cysteine-rich, copper-binding metallothionein that protects cells by sequestering copper and thereby preventing its toxicity (23, 42, 77, 153). Ace1 also regulates the expression of a second metallothionein gene (CRS5) and the copper and zinc superoxide dismutase gene (SOD1) (29, 55). Functional orthologs of Ace1 confer copper resistance in Candida glabrata (Amt1) and Yarrowia lipolytica (Crf1) (46, 173). Although Amt1 protects cells from copper by regulating the expression of three metallothionein genes, metallothionein expression is still copper responsive in a crf1 mutant strain (46, 141, 172). This latter result suggests that Crf1 guards against copper overload by regulating the expression of a yet-unidentified target gene(s) (46).
Regulatory factors that are active during copper deficiency regulate the expression of genes encoding proteins involved in increasing cytosolic copper (Table 1). In S. cerevisiae, Mac1 protects cells from copper starvation by activating the expression of the high-affinity copper uptake systems encoded by CTR1 and CTR3 (Fig. 1A) (85, 155). Mac1 also regulates the expression of a cell surface Fe3+/Cu2+ reductase (FRE1) and a putative reductase of unknown cellular localization (FRE7) (49, 98). The posttranslational degradation of Ctr1 under conditions of excess copper also requires Mac1. It is possible that an uncharacterized Mac1 target gene (Table 1) encodes a protein that is essential for this regulation or that Mac1 itself functions as a protease or protease-recruiting factor under copper-replete conditions (158).
High-affinity copper uptake is regulated at the transcriptional level in S. pombe and Podospora anserina by the factors Cuf1 and GRISEA, respectively (7, 16, 84). In addition to directly regulating copper uptake, Cuf1 stimulates mobilization of copper from vacuolar stores by regulating the expression of the CTR6 vacuolar efflux system (9). GRISEA activates the expression of P. anserina SOD2, a gene that is not copper regulated in S. pombe or S. cerevisiae (17). P. anserina SOD2 encodes mitochondrial manganese superoxide dismutase. In P. anserina, low intracellular copper levels lead to reduced activity of the copper-requiring enzyme, cytochrome c oxidase. This in turn results in the induction of an iron-dependent pathway that utilizes an alternative terminal oxidase. Induction of P. anserina SOD2 under copper-limiting conditions may therefore be important for protection from oxidative stress inherent in the utilization of the alternate oxidase (16-18).
In addition to activating copper transporter genes, Cuf1 represses the expression of the iron-regulated fip1+, fio1+, and frp1+ genes that encode proteins required for iron uptake (84). Similar to S. cerevisiae, iron uptake in S. pombe requires copper. Cuf1-dependent repression of these genes therefore ensures that iron uptake is inhibited under copper-limiting conditions. As copper levels increase (through Cuf1-dependent copper uptake and copper mobilization), Cuf1 is inactivated, which leads to the loss of Cuf1-mediated repression of fip1+, fio1+, and frp1+ (84). It is currently unknown whether Cuf1 mediates this repression by recruiting a corepressor to these promoters or whether Cuf1 inhibits binding of a transcriptional activator. A similar regulatory pathway may also exist in S. cerevisiae, since the iron-regulated FET3 gene also appears to show Mac1-dependent repression (84). However, it is not clear whether this is a direct result of Mac1 acting as a repressor at the FET3 promoter or whether this is a pleiotropic affect of altered iron homeostasis in strains that lack or express a constitutive allele of MAC1. Thus, the ability of Cuf1, and possibly Mac1, to act as both a repressor and activator allows the coordinated expression of genes involved in both copper and iron homeostasis.
A number of structural domains are conserved between the six known copper regulatory factors (Fig. 2B). Ace1, Amt1, and Crf1 all contain a zinc-binding domain, a conserved (R/K)GRP sequence motif, and eight cysteine residues that are arranged in Cys-X-Cys or Cys-X2-Cys motifs (45, 137, 142). The cysteine-rich motifs form a polycopper cluster that binds 4 Cu(I) ions cooperatively while the zinc-binding domain and (R/K)GRP motif are essential for minor groove site-specific binding (32, 41, 53, 81, 142, 143). Mac1, GRISEA, and Cuf1 share regions of homology to Ace1 but lack all the cysteine-rich motifs required in forming the Ace1-Amt1 N-terminal polycopper cluster. The C terminus of Mac1 contains two Cys-X-Cys-X4-Cys-X-Cys-X2-Cys-X2-His motifs that have been designated C1 and C2 (or REPI and REPII, respectively) (Fig. 2B) (54, 78, 175). The C1 and C2 motifs lie within transactivation domains and bind four Cu(I) ions in a polycopper cluster (20, 73). Similar to Mac1, GRISEA contains two cysteine-rich domains (15). Cuf1 contains only one of these motifs, designated C1 (84).
An important facet of copper homeostasis is that copper regulates the activity of each transcription factor. Evidence to date indicates that the conserved structural domains within each class of copper regulatory factors are important for copper sensing. Copper-dependent DNA-binding activity primarily regulates Ace1 and Amt1 activity. Both factors bind as a monomer, in a copper-dependent manner, to upstream activating sequences (22, 45, 70, 140, 172). The copper-induced gene activation is thought to be mediated by the formation of the N-terminal polycopper cluster in response to copper, which converts Ace1 and Amt1 from a nonactive form to an active DNA-binding form (reviewed in reference 152). Haa1, a transcriptional activator in S. cerevisiae, contains the eight conserved cysteine residues that are required for polycopper cluster formation in Ace1, yet is not regulated by copper. Amino acids present in the Haa1 N-terminal region but not present in Ace1 or Amt1 may disrupt polycopper cluster formation and prevent this domain from being used as a copper regulatory domain (79). Additional regulatory mechanisms control both Amt1 and Crf1 activity. Amt1 autoregulates its own expression, a critical factor in copper resistance, since cells that are unable to autoactivate AMT1 are sensitive to growth on copper (174). Crf1 activity is subject to copper-dependent nuclear translocation (46).
Mac1 mediates the response to copper limitation by binding as a homodimer, in a site-specific manner, to copper-responsive cis-acting elements (CuREs) that are located in the promoter regions of target genes (71, 76, 85, 131, 155). In vivo, activation of gene expression requires multiple CuREs, which are arranged in tandem or as inverted repeats and have a synergistic effect on gene expression (74, 85, 155). On exposure to copper, repression of Mac1 is primarily mediated by a copper-dependent interaction between the C1 domain and the DNA-binding domain. This interaction inhibits both transactivation domain function and DNA-binding activity (54, 74, 85). In support of this model, mutations that substitute single cysteine residues in the Mac1 C1 motif lead to a total loss of copper-responsive regulation (78, 155, 175).
Cuf1 activity is similarly controlled by a copper-dependent interaction between the DNA-binding domain and the C1 domain (8). However, a number of differences between Mac1 and Cuf1 regulation have been observed. The N-terminal region of Cuf1 exhibits a higher percentage of sequence identity with the corresponding Ace1 domain than with the N-terminal domain of its functional homolog Mac1. Indeed, Cuf1 can activate Ace1 target gene expression when introduced into an ace1 null S. cerevisiae strain (7, 8). A second difference between Mac1 and Cuf1 is that the substitution of cysteine residues in the Cuf1 C1 domain only leads to a partial loss of regulation. This result suggests that the cysteine residues of Cuf1 are not equally involved in copper coordination (8). Similarities and differences between the regulation of Mac1 and GRISEA by copper are also observed. For example, copper-responsive repression of GRISEA is mediated by an interaction between the DNA-binding domain and the C2 domain rather than the C1 domain (15).
While the copper-dependent control of Mac1 activity via an intramolecular interaction remains undisputed, more-recent studies of Mac1 have superimposed a number of additional regulatory mechanisms. Mac1 must be phosphorylated to bind to CuREs, and homodimerization of Mac1 is essential for maximal in vivo activity (65, 76, 131). It is noteworthy that overexpression of Mac1 constructs that lack the homodimerization domain are fully functional proteins (74, 148). Homodimerization of Mac1 may therefore allow a protein that is normally expressed at a low level to achieve maximal activation of target gene expression. A third additional level of regulation is that in the presence of excess copper, C-terminally tagged Mac1 undergoes a rapid copper-dependent degradation. However, this effect is lost when Mac1 is overexpressed (74, 175). Finally, the C2 transactivation domain may have a role in modulating the activity of Mac1 (78, 148). Further studies that identify other proteins required for Mac1 regulation may reveal the finer details of how Mac1 responds to copper.
In addition to Mac1 and its functional homologs, novel copper regulatory factors may also exist in the plant kingdom. Copper is an essential cofactor of a number of the enzymes required for photosynthesis. During copper deficiency, the photosynthetic algae Chlamydomonas reinhardtii can bypass this copper requirement by decreasing its reliance on copper-requiring enzymes and using alternative enzymes that utilize heme cofactors. At the transcriptional level, this transition is mediated by the Crr1-dependent induction of CPX1 and CYC6 (120, 121). These genes encode the enzyme coproporphyrinogen oxidase that is required for heme biosynthesis and cytochrome c6, a heme-containing electron transfer catalyst. Crr1 also reciprocally regulates the expression of the partially redundant genes CRD1 and CTH1, which are required for the maintenance of photosystem I under copper-limiting and -replete conditions, respectively (100, 101). Although the precise genetic locus of Crr1 has yet to be identified, a number of observations suggest that Crr1 is not simply an ortholog of Mac1. First, one CuRE is both necessary and sufficient to mediate copper-responsive regulation (119). Second, the consensus sequence of GTAC, rather than the Mac1 consensus 5′-TTTGC(T/G)C(A/G)-3′, is found in all known Crr1 target genes and is essential for copper-responsive transcription (119). Finally, Crr1 possibly responds to Cu2+ not Cu+, since Hg2+ and not Ag+ will also repress Crr1 target gene transcription (119).
ZINC
To a major extent, zinc homeostasis generally parallels copper homeostasis in that both posttranslational and transcriptional homeostatic regulatory mechanisms function together to maintain zinc at an optimal level under conditions of either zinc limitation or zinc excess. For example, in S. cerevisiae, expression of the high-affinity zinc uptake gene ZRT1 increases in response to zinc limitation, whereas under zinc-replete conditions, Zrt1 undergoes zinc-induced endocytosis and is degraded in the vacuole (39). However, unlike iron and copper, zinc-responsive transcription factors are found in fungi, mammals, fish, and possibly plants, suggesting that the transcriptional control of genes involved in zinc homeostasis is of universal importance (31, 58, 110, 151, 168). To date, two factors that control gene expression in response to zinc have been characterized in detail. These are Zap1 from S. cerevisiae, which activates gene expression in response to zinc deficiency, and mammalian MTF-1, which is activated by zinc (151, 168).
Under zinc-limiting conditions, Zap1 increases the expression of three zinc uptake systems encoded by the ZRT1, ZRT2, and FET4 genes (Fig. 1A) (149, 166, 167). Zap1 also stimulates the release of zinc from the vacuolar zinc store by activating the expression of the ZRT3 vacuolar efflux system (95). A fifth target of Zap1 is ZRC1, a gene that encodes a vacuolar zinc influx system (96). Although it seems counterintuitive that Zap1 up-regulates the expression of a gene associated with lowering cytoplasmic zinc levels, recent studies have revealed that the increased expression of ZRC1 in response to zinc limitation is a proactive mechanism to protect zinc-limited cells from possible exposure to high zinc levels (97). In addition to the five zinc transporter genes, Zap1 regulates the expression of 42 other genes, some of which may have additional roles in zinc homeostasis (Table 1) (94).
In mammals, MTF-1 plays a central role in protecting cells against zinc toxicity. This is partly achieved by increasing the expression of MT-1 and MT-2, two genes that encode zinc-binding metallothioneins (66). MTF-1 also lowers cytoplasmic zinc levels by regulating the expression of a zinc efflux system encoded by the ZnT-1 gene (87). A further putative target gene of MTF-1 is hZTL1, a gene that encodes a zinc uptake transporter that is localized to the enterocyte apical membrane (27). While increased hZTL1 expression may assist efficient uptake of zinc from a zinc-rich diet, this apparent regulation counteracts other homeostatic mechanisms. Perhaps this unusual regulation ensures that zinc is effectively absorbed from the intestine while other transcriptional and posttranslational homeostatic mechanisms maintain cellular zinc at an optimal level under these conditions.
In addition to regulating genes involved in zinc homeostasis, MTF-1 regulates the expression of a number of other genes (Table 1) (92). In mice, MTF-1 is an essential gene, with knockout mice dying in utero at approximately day 14 of gestation due to degeneration of hepatocytes (59). Although the exact reason for the lethality of the MTF-1 knockout is currently unknown, a number of candidate target genes of MTF-1 that are essential for embryonic development (C/EBPα and α-fetoprotein) could provide clues to the lethality phenotype (92). Contrary to the lethality of MTF-1 in mice, an MTF-1 knockout in Drosophila melanogaster is viable (38). In Drosophila, however, copper is a more potent inducer of MTF-1 activity than zinc and MTF-1 plays a dual role in regulating genes involved in resistance to copper toxicity and in preventing copper deficiency (38, 163). Thus, MTF-1 can have species-specific cellular roles in addition to zinc homeostasis.
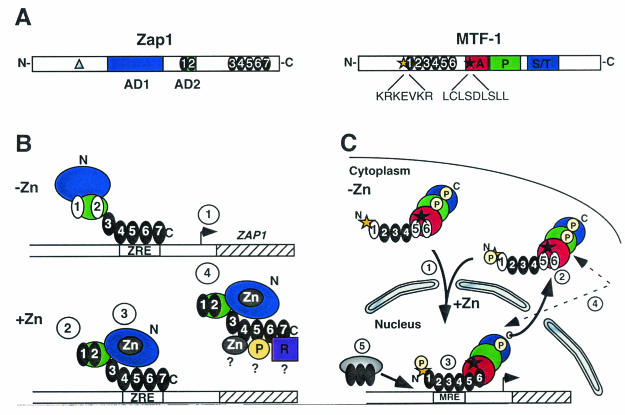
Zap1 and MTF-1 have a number of features that are common to transcriptional activators, which include transactivation domains and DNA-binding domains containing C2H2-type zinc finger motifs (Fig. 3A). Zap1 contains two acidic activation domains (168). The first activation domain is located at the N terminus in a region rich in cysteine and histidine residues, and the second activation domain maps to a region containing two C2H2-type zinc finger domains (11). A further five C-terminal zinc finger domains are all essential for Zap1 DNA-binding activity (10, 40, 165). MTF-1 encodes a 72.5-kDa protein that contains six C2H2-type zinc finger domains and three transactivation domains (Fig. 3A) (21, 122).
FIG. 3.
(A) Schematic diagram of S. cerevisiae Zap1 and mouse MTF-1. The following are shown: zinc finger domains (numbered black ovals), position of the cysteine-to-serine mutation in Zap1-1up (grey triangle), MTF-1 nuclear localization signal (gold star), MTF-1 NES (black star), and MTF-1 acid-, proline-, and serine/threonine-rich activation domains (red, green, and blue boxes labeled A, P, and S/T, respectively). (B) The multiple levels of Zap1 regulation: 1, autoregulation; 2, AD2 repression; 3, AD1 repression; 4, DNA-binding control. (C) The multiple levels of MTF-1 regulation: 1, nuclear translocation; 2, nucleocytoplasmic shuttling; 3, DNA-binding control; 4, posttranslational modification by phosphorylation; 5, interactions and binding inhibition by other factors. The absence of zinc in a metalloregulatory finger is indicated by a white numbered oval. Possible phosphorylation events (yellow circle labeled P), a putative repressor (purple square), and target genes (hatched boxes) are shown.
A critical feature in understanding zinc homeostasis is determining how these factors sense zinc. Multiple regulatory mechanisms contribute to the inactivation of Zap1 by zinc (Fig. 3B). At the transcriptional level, Zap1 binds to a zinc-responsive element located within its own promoter and autoregulates its own expression. Zap1 activity is also regulated by up to three posttranslational mechanisms (12, 165, 168). The most understood regulatory mechanism is the autonomous repression of activation domain 2 (AD2) by zinc. Both of the zinc finger domains that are located in AD2 (Znf1 and Znf2) are required for zinc-responsive repression. In vitro, zinc binds with a slightly lower affinity and, notably, with a much higher lability to the Znf1-Znf2 pair relative to a control pair of zinc finger domains (11). Moreover, in vivo, residues that form the packing interface between the two fingers are an essential component of zinc-responsive repression. Thus, as zinc increases, the zinc occupancy of Znf1 and Znf2 may result in an interfinger, protein-protein interaction that masks critical residues that are essential for transactivation domain function (11).
In the absence of AD2 regulation, both AD1 and the Zap1 DNA-binding domain are negatively regulated by zinc by independent mechanisms (12). Repression of AD1 most likely involves a zinc-dependent intramolecular interaction with the Zap1 DNA-binding domain that masks activation domain function. In support of this model, AD1 is rich in potential zinc-coordinating ligands and repression of AD1 by zinc requires the Zap1 DNA-binding domain. In addition, a less-zinc-responsive mutant allele of ZAP1 (ZAP1-1up) encodes a cysteine-to-serine mutation in a region that is immediately upstream from AD1 (Fig. 3A) (12, 168). Zap1 DNA-binding activity may also be regulated by zinc, since the Zap1 DNA-binding domain is able to confer zinc-responsiveness onto a heterologous activation domain (12). At present, the precise mechanism by which zinc inhibits Zap1 DNA-binding activity is unknown.
Unlike Zap1, which is primarily regulated by zinc, MTF-1 activity can be regulated by zinc, other divalent metal ions, and various stress conditions in vivo (4, 51, 91). In vitro, zinc stimulates transcriptional induction by MTF-1, whereas induction in response to cadmium, copper, or H2O2 additionally requires the presence of zinc-saturated metallothionein (164). Moreover, in Drosophila, MTF-1 is primarily regulated by copper, whereas in transfected mammalian cells, Drosophila MTF-1 responds to zinc like mammalian MTF-1 (163). Thus, other aspects of metal ion homeostasis, such as metal release from metallothionein, may influence the primary metal specificity of MTF-1.
MTF-1 is regulated at multiple levels by zinc (Fig. 3C) (4, 51, 91). The first level of regulation of MTF-1 involves its cellular localization. Under noninducing conditions, MTF-1 predominantly resides in the cytoplasm. Upon addition of zinc or cadmium, MTF-1 rapidly translocates to the nucleus (128, 133). Mutation of the MTF-1 NES results in the retention of MTF-1 in the nucleus in a form that is able to bind to a metal response element (MRE) in response to zinc but is unable to activate transcription. Thus, a second level of regulation involving the MTF-1 cellular position could potentially be nucleocytoplasmic shuttling, i.e., MTF-1 might undergo a constant cycle of activation, nuclear import, deactivation, nuclear export, and then reactivation (128). The third level of regulation is the control of DNA-binding activity (13, 30, 66, 82). MTF-1 DNA-binding activity increases upon the addition of zinc. Consequently, one model is that one or more of the zinc finger domains bind zinc with a low affinity or low stability; thus, full DNA-binding activity is only achieved upon full metallation of the regulatory zinc finger(s) (66, 151).
Recent in vitro studies have clearly demonstrated that there is conformational heterogeneity in the MTF-1 DNA-binding zinc fingers such that the zinc bound to Znf5, Znf6, and to a lesser extent, Znf1 is less stable than the zinc bound to the remaining finger domains (Znf2, Znf3, and Znf4) (25, 26, 52). While Znf2, Znf3, and Znf4 form the core DNA-binding domain in vitro, deletion of Znf1 or Znf5 and Znf6 leads to attenuation of zinc-induced MTF-1 DNA binding at the endogenous MT-1 promoter in vivo (75). Thus, Znf1, Znf5, and Znf6 are required for maximal binding under zinc-replete conditions (75). Similar effects are not seen in mutants containing deletions of Znf5 or Znf6, whether examined in vitro or with an MRE-reporter construct, suggesting that binding zinc in these regulatory fingers may stabilize an MTF-1-chromatin complex (13, 75, 82).
MTF-1 activity is also controlled by phosphorylation. MTF-1 is phosphorylated in both an uninduced and an induced state. However, the level of phosphorylation is stimulated two- to fourfold by the addition of zinc. Kinase inhibitor studies have revealed that this phosphorylation is an essential component of zinc-responsive MTF-1 activation. Since kinase inhibitors have little effect on the nuclear import or DNA-binding activity of MTF-1, signal transduction pathways may use phosphorylation or dephosphorylation to control activation domain function (88, 127). Finally, under specific conditions, other factors can influence MTF-1 activity. The precise activity of MTF-1 may also be dependent on the MRE sequence, the chromatin architecture of the target loci, cell type, developmental stage of growth, and growth conditions (1, 5, 50, 104).
Both Zap1 and MTF-1 use regulatory zinc finger domains to sense zinc; however, it is currently unknown what properties of a zinc finger make it regulatory. The regulatory zinc fingers all match the consensus zinc finger sequence (Phe/Tyr-X-CysX2-4-Cys-X3-[Phe]-X5-Leu-X2-His-X2-3-His), with the exception of the conserved central phenylalanine. In Znf1 and Znf2 from Zap1, this residue is a cysteine and glycine, respectively. Attempts to convert the Zap1, Znf1, and Znf2 fingers back to the consensus sequence (by converting the cysteine and glycine residues at the finger tip to phenylalanine) have no effect on AD2 activity, suggesting that the high lability of the zinc bound to these fingers is not simply caused by these substitutions (11). Importantly, these zinc fingers function together as a pair, suggesting that other residues that stabilize pair formation may have important regulatory functions. In MTF-1, Znf5 fluctuates between the canonical ββα structure and another structure or structures upon addition of excess zinc (52). Znf5 contains five additional potential zinc-coordinating ligands that are located in the Cys-X4-Cys loop and the α-helix. The high conservation of these residues and the properties of Znf5 have led to the speculation that these residues may bind an additional zinc ion that destabilizes Znf5 (52). Thus, a precise knowledge of what makes these particular fingers bind zinc with a higher lability than other fingers will help us to understand their regulatory function.
Another unanswered question is what zinc pools do MTF-1 and Zap1 sense. In Escherichia coli, the zinc sensors Zur and ZntR respond to femtomolar levels of zinc (less than 1 atom of zinc per cell) (106). These data suggest that the majority of cellular zinc is bound in a yet-unidentified bioavailable zinc pool that could consist of proteins, small molecules, or other macromolecules that either strongly or weakly chelate zinc. Because of the predicted low levels of free zinc, it has been proposed that zinc-trafficking factors may be required to deliver zinc to proteins (106). Although the precise level of free zinc in eukaryotic cells is unknown, it is interesting that both MTF-1 and Zap1 sense zinc in the nanomolar to subnanomolar range (11, 52). If free intracellular zinc levels are maintained at much less than nanomolar concentrations in eukaryotic cells, then the eukaryotic zinc sensors must be detecting fluctuations in a bioavailable zinc pool. Studies with eukaryotic sensors may therefore help us to answer questions such as what molecule(s) or protein(s) potentially delivers zinc to Zap1 and MTF-1. These studies also raise many other interesting questions, such as whether all zinc-sensing factors rely on regulatory zinc fingers or whether other types of zinc domains, such as the GATA or LIM domain, can be used.
CONCLUSIONS
The transcription factors described here have provided insight into the molecular mechanisms involved in the control of eukaryotic metal ion homeostasis. The regulation of these proteins at multiple levels is a recurring theme and may be characteristic of all eukaryotic metalloregulatory factors. There is also compelling evidence that these factors directly sense metal, although, to date, there is no direct in vivo evidence that their metal occupancy changes with cellular metal ion status. Therefore, alternative mechanisms of sensing, such as signal transduction pathways, that involve protein kinases cannot be discounted at present.
It is likely that other eukaryotic metal-responsive transcription factors exist. There are many fungal genes that are regulated in response to metal ions independent of the factors described in this review (19, 90, 102). In other eukaryotes, metal-regulated genes are continuously being identified, suggesting that other metalloregulators exist (for examples, see references 37, 58, 69, 86, 110, 111, 126, and 132). The influence of metals on human health has generated considerable interest in the understanding of the principles involved in metal homeostasis. As more metal-responsive transcription factors are discovered in both plants and mammals, it should become apparent whether or not there are general principles of metal ion sensing that are conserved throughout the eukaryotes. This will be indispensable to our understanding of intracellular metal homeostasis.
Acknowledgments
We thank Dennis Winge, Heather Carr, and Paul Cobine for helpful discussion and comments on the manuscript.
J.R. and A.B are both members of the Winge lab, whose work is supported by grant CA 61286 from the National Cancer Institute, National Institutes of Health.
REFERENCES
- 1.Adilakshmi, T., and R. O. Laine. 2002. Ribosomal protein S25 mRNA partners with MTF-1 and La to provide a p53-mediated mechanism for survival or death. J. Biol. Chem. 277:4147-4151. [DOI] [PubMed] [Google Scholar]
- 2.An, Z., B. Mei, W. M. Yuan, and S. A. Leong. 1997. The distal GATA sequences of the sid1 promoter of Ustilago maydis mediate iron repression of siderophore production and interact directly with Urbs1, a GATA family transcription factor. EMBO J. 16:1742-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, Z., Q. Zhao, J. McEvoy, W. M. Yuan, J. L. Markley, and S. A. Leong. 1997. The second finger of Urbs1 is required for iron-mediated repression of sid1 in Ustilago maydis. Proc. Natl. Acad. Sci. USA 94:5882-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews, G. K. 2001. Cellular zinc sensors: MTF-1 regulation of gene expression. Biometals 14:223-237. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, G. K., D. K. Lee, R. Ravindra, P. Lichtlen, M. Sirito, M. Sawadogo, and W. Schaffner. 2001. The transcription factors MTF-1 and USF1 cooperate to regulate mouse metallothionein-I expression in response to the essential metal zinc in visceral endoderm cells during early development. EMBO J. 20:1114-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 7.Beaudoin, J., and S. Labbé. 2001. The fission yeast copper-sensing transcription factor Cuf1 regulates the copper transporter gene expression through an AceI/Amt1-like recognition sequence. J. Biol. Chem. 276:15472-15480. [DOI] [PubMed] [Google Scholar]
- 8.Beaudoin, J., A. Mercier, R. Langlois, and S. Labbé. 2003. The Schizosaccharomyces pombe Cuf1 is composed of functional modules from two distinct classes of copper metalloregulatory transcription factors. J. Biol. Chem. 278:14565-14577. [DOI] [PubMed] [Google Scholar]
- 9.Bellemare, D. R., L. Shaner, K. A. Morano, J. Beaudoin, R. Langlois, and S. Labbé. 2002. Ctr6, a vacuolar membrane copper transporter in Schizosaccharomyces pombe. J. Biol. Chem. 277:46676-46686. [DOI] [PubMed] [Google Scholar]
- 10.Bird, A., M. V. Evans-Galea, E. Blankman, H. Zhao, H. Luo, D. R. Winge, and D. J. Eide. 2000. Mapping the DNA binding domain of the Zap1 zinc-responsive transcriptional activator. J. Biol. Chem. 275:16160-16166. [DOI] [PubMed] [Google Scholar]
- 11.Bird, A. J., K. McCall, M. Kramer, E. Blankman, D. R. Winge, and D. J. Eide. 2003. Zinc fingers can act as Zn2+ sensors to regulate transcriptional activation domain function. EMBO J. 22:5137-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bird, A. J., H. Zhao, H. Luo, L. T. Jensen, C. Srinivasan, M. Evans-Galea, D. R. Winge, and D. J. Eide. 2000. A dual role for zinc fingers in both DNA binding and zinc sensing by the Zap1 transcriptional activator. EMBO J. 19:3704-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bittel, D. C., I. V. Smirnova, and G. K. Andrews. 2000. Functional heterogeneity in the zinc fingers of metalloregulatory protein metal response element-binding transcription factor-1. J. Biol. Chem. 275:37194-37201. [DOI] [PubMed] [Google Scholar]
- 14.Blaiseau, P.-L., E. Lesuisse, and J.-M. Camadro. 2001. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J. Biol. Chem. 276:34221-34226. [DOI] [PubMed] [Google Scholar]
- 15.Borghouts, C., and H. D. Osiewacz. 1998. GRISEA, a copper-modulated transcription factor from Podospora anserina involved in senescence and morphogenesis, is an ortholog of MAC1 in Saccharomyces cerevisiae. Mol. Gen. Genet. 260:492-502. [DOI] [PubMed] [Google Scholar]
- 16.Borghouts, C., C. Q. Scheckhuber, O. Stephan, and H. D. Osiewacz. 2002. Copper homeostasis and aging in the fungal model system Podospora anserina: differential expression of PaCtr3 encoding a copper transporter. Int. J. Biochem. Cell. Biol. 34:1355-1371. [DOI] [PubMed] [Google Scholar]
- 17.Borghouts, C., C. Q. Scheckhuber, A. Werner, and H. D. Osiewacz. 2002. Respiration, copper availability and SOD activity in P. anserina strains with different lifespan. Biogerontology 3:143-153. [DOI] [PubMed] [Google Scholar]
- 18.Borghouts, C., A. Werner, T. Elthon, and H. D. Osiewacz. 2001. Copper-modulated gene expression and senescence in the filamentous fungus Podospora anserina. Mol. Cell. Biol. 21:390-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borrelly, G. P., M. D. Harrison, A. K. Robinson, S. G. Cox, N. J. Robinson, and S. K. Whitehall. 2002. Surplus zinc is handled by Zym1 metallothionein and Zhf endoplasmic reticulum transporter in Schizosaccharomyces pombe. J. Biol. Chem. 277:30394-30400. [DOI] [PubMed] [Google Scholar]
- 20.Brown, K. R., G. L. Keller, I. J. Pickering, H. H. Harris, G. N. George, and D. R. Winge. 2002. Structures of the cuprous-thiolate clusters of the mac1 and ace1 transcriptional activators. Biochemistry 41:6469-6476. [DOI] [PubMed] [Google Scholar]
- 21.Brugnera, E., O. Georgiev, F. Radtke, R. Heuchel, E. Baker, G. R. Sutherland, and W. Schaffner. 1994. Cloning, chromosomal mapping, and characterization of the human metal-regulatory transcription factor MTF-1. Nucleic Acids Res. 22:3167-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchman, C., P. Skroch, J. Welch, S. Fogel, and M. Karin. 1989. The CUP2 gene product, regulator of yeast metallothionein expression, is a copper-activated DNA-binding protein. Mol. Cell. Biol. 9:4091-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butt, T. R., E. J. Sternberg, J. A. Gorman, P. Clark, D. Hamer, M. Rosenberg, and S. T. Crooke. 1984. Copper metallothionein of yeast, structure of the gene, and regulation of expression. Proc. Natl. Acad. Sci. USA 81:3332-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casas, C., M. Aldea, C. Espinet, C. Gallego, R. Gil, and E. Herrero. 1997. The AFT1 transcriptional factor is differentially required for expression of high-affinity iron uptake genes in Saccharomyces cerevisiae. Yeast 13:621-637. [DOI] [PubMed] [Google Scholar]
- 25.Chen, X., A. Agarwal, and D. P. Giedroc. 1998. Structural and functional heterogeneity among the zinc fingers of human MRE-binding transcription factor-1. Biochemistry 37:11152-11161. [DOI] [PubMed] [Google Scholar]
- 26.Chen, X., M. Chu, and D. P. Giedroc. 1999. MRE-binding transcription factor-1: weak zinc-binding finger domains 5 and 6 modulate the structure, affinity, and specificity of the metal-response element complex. Biochemistry 38:12915-12925. [DOI] [PubMed] [Google Scholar]
- 27.Cragg, R. A., G. R. Christie, S. R. Phillips, R. M. Russi, S. Kury, J. C. Mathers, P. M. Taylor, and D. Ford. 2002. A novel zinc-regulated human zinc transporter, hZTL1, is localized to the enterocyte apical membrane. J. Biol. Chem. 277:22789-22797. [DOI] [PubMed] [Google Scholar]
- 28.Crisp, R. J., A. Pollington, C. Galea, S. Jaron, Y. Yamiguchi-Iwai, and J. Kaplan. 2003. Inhibition of heme biosynthesis prevents transcription of iron uptake genes in yeast. J. Biol. Chem. 278:45499-45506. [DOI] [PubMed] [Google Scholar]
- 29.Culotta, V. C., W. R. Howard, and X. F. Liu. 1994. CRS5 encodes a metallothionein-like protein in Saccharomyces cerevisiae. J. Biol. Chem. 269:25295-252302. [PubMed] [Google Scholar]
- 30.Dalton, T. P., D. Bittel, and G. K. Andrews. 1997. Reversible activation of mouse metal response element-binding transcription factor 1 DNA binding involves zinc interaction with the zinc finger domain. Mol. Cell. Biol. 17:2781-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalton, T. P., W. A. Solis, D. W. Nebert, and M. J. Carvan III. 2000. Characterization of the MTF-1 transcription factor from zebrafish and trout cells. Comp. Biochem. Physiol. B 126:325-335. [DOI] [PubMed] [Google Scholar]
- 32.Dameron, C. T., D. R. Winge, G. N. George, M. Sansone, S. Hu, and D. Hamer. 1991. A copper-thiolate polynuclear cluster in the ACE1 transcription factor. Proc. Natl. Acad. Sci. USA 88:6127-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dancis, A., R. D. Klausner, A. G. Hinnebusch, and J. G. Barriocanal. 1990. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2294-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dancis, A., D. G. Roman, G. J. Anderson, A. G. Hinnebusch, and R. D. Klausner. 1992. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc. Natl. Acad. Sci. USA 89:3869-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Freitas, J. M., A. Liba, R. Meneghini, J. S. Valentine, and E. B. Gralla. 2000. Yeast lacking Cu-Zn superoxide dismutase show altered iron homeostasis. Role of oxidative stress in iron metabolism. J. Biol. Chem. 275:11645-11649. [DOI] [PubMed] [Google Scholar]
- 36.Dix, D. R., J. T. Bridgham, M. A. Broderius, C. A. Byersdorfer, and D. J. Eide. 1994. The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J. Biol. Chem. 269:26092-26099. [PubMed] [Google Scholar]
- 37.Dufner-Beattie, J., F. Wang, Y. M. Kuo, J. Gitschier, D. Eide, and G. K. Andrews. 2003. The acrodermatitis enteropathica gene ZIP4 encodes a tissue-specific, zinc-regulated zinc transporter in mice. J. Biol. Chem. 278:33474-33481. [DOI] [PubMed] [Google Scholar]
- 38.Egli, D., A. Selvaraj, H. Yepiskoposyan, B. Zhang, E. Hafen, O. Georgiev, and W. Schaffner. 2003. Knockout of 'metal-responsive transcription factor' MTF-1 in Drosophila by homologous recombination reveals its central role in heavy metal homeostasis. EMBO J. 22:100-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eide, D. J. 2003. Multiple regulatory mechanisms maintain zinc homeostasis in Saccharomyces cerevisiae. J. Nutr. 133:1532S-1535S. [DOI] [PubMed] [Google Scholar]
- 40.Evans-Galea, M. V., E. Blankman, D. G. Myszka, A. J. Bird, D. J. Eide, and D. R. Winge. 2003. Two of the five zinc fingers in the Zap1 transcription factor DNA binding domain dominate site-specific DNA binding. Biochemistry 42:1053-1061. [DOI] [PubMed] [Google Scholar]
- 41.Farrell, R. A., J. L. Thorvaldsen, and D. R. Winge. 1996. Identification of the Zn(II) site in the copper-responsive yeast transcription factor, AMT1: a conserved Zn module. Biochemistry 35:1571-1580. [DOI] [PubMed] [Google Scholar]
- 42.Fogel, S., and J. W. Welch. 1982. Tandem gene amplification mediates copper resistance in yeast. Proc. Natl. Acad. Sci. USA 79:5342-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foury, F., and T. Roganti. 2002. Deletion of the mitochondrial carrier genes MRS3 and MRS4 suppresses mitochondrial iron accumulation in a yeast frataxin-deficient strain. J. Biol. Chem. 277:24475-24483. [DOI] [PubMed] [Google Scholar]
- 44.Foury, F., and D. Talibi. 2001. Mitochondrial control of iron homeostasis. A genome wide analysis of gene expression in a yeast frataxin-deficient strain. J. Biol. Chem. 276:7762-7768. [DOI] [PubMed] [Google Scholar]
- 45.Fürst, P., S. Hu, R. Hackett, and D. Hamer. 1988. Copper activates metallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell 55:705-717. [DOI] [PubMed] [Google Scholar]
- 46.García, S., M. Prado, R. Dégano, and A. Domínguez. 2002. A copper-responsive transcription factor, CRF1, mediates copper and cadmium resistance in Yarrowia lipolytica. J. Biol. Chem. 277:37359-37368. [DOI] [PubMed] [Google Scholar]
- 47.Garland, S. A., K. Hoff, L. E. Vickery, and V. C. Culotta. 1999. Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron-sulfur cluster assembly. J. Mol. Biol. 294:897-907. [DOI] [PubMed] [Google Scholar]
- 48.Georgatsou, E., and D. Alexandraki. 1994. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Georgatsou, E., L. A. Mavrogiannis, G. S. Fragiadakis, and D. Alexandraki. 1997. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 272:13786-13792. [DOI] [PubMed] [Google Scholar]
- 50.Ghoshal, K., J. Datta, S. Majumder, S. Bai, X. Dong, M. Parthun, and S. T. Jacob. 2002. Inhibitors of histone deacetylase and DNA methyltransferase synergistically activate the methylated metallothionein I promoter by activating the transcription factor MTF-1 and forming an open chromatin structure. Mol. Cell. Biol. 22:8302-8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giedroc, D. P., X. Chen, and J. L. Apuy. 2001. Metal response element (MRE)-binding transcription factor-1 (MTF-1): structure, function, and regulation. Antioxid. Redox Signal. 3:577-596. [DOI] [PubMed] [Google Scholar]
- 52.Giedroc, D. P., X. Chen, M. A. Pennella, and A. C. LiWang. 2001. Conformational heterogeneity in the C-terminal zinc fingers of human MTF-1: an NMR and zinc-binding study. J. Biol. Chem. 276:42322-42332. [DOI] [PubMed] [Google Scholar]
- 53.Graden, J. A., M. C. Posewitz, J. R. Simon, G. N. George, I. J. Pickering, and D. R. Winge. 1996. Presence of a copper(I)-thiolate regulatory domain in the copper-activated transcription factor Amt1. Biochemistry 35:14583-14589. [DOI] [PubMed] [Google Scholar]
- 54.Graden, J. A., and D. R. Winge. 1997. Copper-mediated repression of the activation domain in the yeast Mac1p transcription factor. Proc. Natl. Acad. Sci. USA 94:5550-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gralla, E. B., D. J. Thiele, P. Silar, and J. S. Valentine. 1991. ACE1, a copper-dependent transcription factor, activates expression of the yeast copper, zinc superoxide dismutase gene. Proc. Natl. Acad. Sci. USA 88:8558-8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Green, C. J., P. Lichtlen, N. T. Huynh, M. Yanovsky, K. R. Laderoute, W. Schaffner, and B. J. Murphy. 2001. Placenta growth factor gene expression is induced by hypoxia in fibroblasts: a central role for metal transcription factor-1. Cancer Res. 61:2696-2703. [PubMed] [Google Scholar]
- 57.Gross, C., M. Kelleher, V. R. Iyer, P. O. Brown, and D. R. Winge. 2000. Identification of the copper regulon in Saccharomyces cerevisiae by DNA microarrays. J. Biol. Chem. 275:32310-32316. [DOI] [PubMed] [Google Scholar]
- 58.Grotz, N., T. Fox, E. Connolly, W. Park, M. L. Guerinot, and D. Eide. 1998. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc. Natl. Acad. Sci. USA 95:7220-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Günes, C., R. Heuchel, O. Georgiev, K. H. Müller, P. Lichtlen, H. Blüthmann, S. Marino, A. Aguzzi, and W. Schaffner. 1998. Embryonic lethality and liver degeneration in mice lacking the metal-responsive transcriptional activator MTF-1. EMBO J. 17:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haas, H., K. Angermayr, and G. Stöffler. 1997. Molecular analysis of a Penicillium chrysogenum GATA factor encoding gene (sreP) exhibiting significant homology to the Ustilago maydis urbs1 gene. Gene 184:33-37. [DOI] [PubMed] [Google Scholar]
- 61.Haas, H., I. Zadra, G. Stöffler, and K. Angermayr. 1999. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J. Biol. Chem. 274:4613-4619. [DOI] [PubMed] [Google Scholar]
- 62.Harrison, K. A., and G. A. Marzluf. 2002. Characterization of DNA binding and the cysteine rich region of SRE, a GATA factor in Neurospora crassa involved in siderophore synthesis. Biochemistry 41:15288-15295. [DOI] [PubMed] [Google Scholar]
- 63.Hassett, R. F., A. M. Romeo, and D. J. Kosman. 1998. Regulation of high affinity iron uptake in the yeast Saccharomyces cerevisiae. Role of dioxygen and Fe(II). J. Biol. Chem. 273:7628-7636. [DOI] [PubMed] [Google Scholar]
- 64.Haurie, V., H. Boucherie, and F. Sagliocco. 2003. The Snf1 protein kinase controls the induction of genes of the iron uptake pathway at the diauxic shift in Saccharomyces cerevisiae. J. Biol. Chem. 278:45391-45396. [DOI] [PubMed] [Google Scholar]
- 65.Heredia, J., M. Crooks, and Z. Zhu. 2001. Phosphorylation and Cu+ coordination-dependent DNA binding of the transcription factor Mac1p in the regulation of copper transport. J. Biol. Chem. 276:8793-8797. [DOI] [PubMed] [Google Scholar]
- 66.Heuchel, R., F. Radtke, O. Georgiev, G. Stark, M. Aguet, and W. Schaffner. 1994. The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J. 13:2870-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hill, K. L., H. H. Li, J. Singer, and S. Merchant. 1991. Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6. Analysis of the kinetics and metal specificity of its copper-responsive expression. J. Biol. Chem. 266:15060-15067. [PubMed] [Google Scholar]
- 68.Hill, K. L., and S. Merchant. 1995. Coordinate expression of coproporphyrinogen oxidase and cytochrome c6 in the green alga Chlamydomonas reinhardtii in response to changes in copper availability. EMBO J. 14:857-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Himelblau, E., H. Mira, S. J. Lin, V. C. Culotta, L. Peñarrubia, and R. M. Amasino. 1998. Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiol. 117:1227-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huibregtse, J. M., D. R. Engelke, and D. J. Thiele. 1989. Copper-induced binding of cellular factors to yeast metallothionein upstream activation sequences. Proc. Natl. Acad. Sci. USA 86:65-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jamison McDaniels, C. P., L. T. Jensen, C. Srinivasan, D. R. Winge, and T. D. Tullius. 1999. The yeast transcription factor Mac1 binds to DNA in a modular fashion. J. Biol. Chem. 274:26962-26967. [DOI] [PubMed] [Google Scholar]
- 72.Jensen, L. T., and V. C. Culotta. 2002. Regulation of Saccharomyces cerevisiae FET4 by oxygen and iron. J. Mol. Biol. 318:251-260. [DOI] [PubMed] [Google Scholar]
- 73.Jensen, L. T., M. C. Posewitz, C. Srinivasan, and D. R. Winge. 1998. Mapping of the DNA binding domain of the copper-responsive transcription factor Mac1 from Saccharomyces cerevisiae. J. Biol. Chem. 273:23805-23811. [DOI] [PubMed] [Google Scholar]
- 74.Jensen, L. T., and D. R. Winge. 1998. Identification of a copper-induced intramolecular interaction in the transcription factor Mac1 from Saccharomyces cerevisiae. EMBO J. 17:5400-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang, H., P. J. Daniels, and G. K. Andrews. 2003. Putative zinc-sensing zinc fingers of metal-response element-binding transcription factor-1 stabilize a metal-dependent chromatin complex on the endogenous metallothionein-I promoter. J. Biol. Chem. 278:30394-30402. [DOI] [PubMed] [Google Scholar]
- 76.Joshi, A., M. Serpe, and D. J. Kosman. 1999. Evidence for (Mac1p)2. DNA ternary complex formation in Mac1p-dependent transactivation at the CTR1 promoter. J. Biol. Chem. 274:218-226. [DOI] [PubMed] [Google Scholar]
- 77.Karin, M., R. Najarian, A. Haslinger, P. Valenzuela, J. Welch, and S. Fogel. 1984. Primary structure and transcription of an amplified genetic locus: the CUP1 locus of yeast. Proc. Natl. Acad. Sci. USA 81:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keller, G., C. Gross, M. Kelleher, and D. R. Winge. 2000. Functional independence of the two cysteine-rich activation domains in the yeast Mac1 transcription factor. J. Biol. Chem. 275:29193-29199. [DOI] [PubMed] [Google Scholar]
- 79.Keller, G., E. Ray, P. O. Brown, and D. R. Winge. 2001. Haa1, a protein homologous to the copper-regulated transcription factor AceI, is a novel transcriptional activator. J. Biol. Chem. 276:38697-38702. [DOI] [PubMed] [Google Scholar]
- 80.Knight, S. A., E. Lesuisse, R. Stearman, R. D. Klausner, and A. Dancis. 2002. Reductive iron uptake by Candida albicans: role of copper, iron and the TUP1 regulator. Microbiology 148:29-40. [DOI] [PubMed] [Google Scholar]
- 81.Koch, K. A., and D. J. Thiele. 1996. Autoactivation by a Candida glabrata copper metalloregulatory transcription factor requires critical minor groove interactions. Mol. Cell. Biol. 16:724-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koizumi, S., K. Suzuki, Y. Ogra, P. Gong, and F. Otuska. 2000. Roles of zinc fingers and other regions of the transcription factor human MTF-1 in zinc-regulated DNA binding. J. Cell. Physiol. 185:464-472. [DOI] [PubMed] [Google Scholar]
- 83.Kosman, D. J. 2003. Molecular mechanisms of iron uptake in fungi. Mol. Microbiol. 47:1185-1197. [DOI] [PubMed] [Google Scholar]
- 84.Labbé, S., M. M. Peña, A. R. Fernandes, and D. J. Thiele. 1999. A copper-sensing transcription factor regulates iron uptake genes in Schizosaccharomyces pombe. J. Biol. Chem. 274:36252-36260. [DOI] [PubMed] [Google Scholar]
- 85.Labbé, S., Z. Zhu, and D. J. Thiele. 1997. Copper-specific transcriptional repression of yeast genes encoding critical components in the copper transport pathway. J. Biol. Chem. 272:15951-15958. [DOI] [PubMed] [Google Scholar]
- 86.La Fontaine, S., J. M. Quinn, S. S. Nakamoto, M. D. Page, V. Gohre, J. L. Moseley, J. Kropat, and S. Merchant. 2002. Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot. Cell 1:736-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langmade, S. J., R. Ravindra, P. J. Daniels, and G. K. Andrews. 2000. The transcription factor MTF-1 mediates metal regulation of the mouse ZnT1 gene. J. Biol. Chem. 275:34803-34809. [DOI] [PubMed] [Google Scholar]
- 88.LaRochelle, O., V. Gagné, J. Charron, J. W. Soh, and C. Séguin. 2001. Phosphorylation is involved in the activation of metal-regulatory transcription factor 1 in response to metal ions. J. Biol. Chem. 276:41879-41888. [DOI] [PubMed] [Google Scholar]
- 89.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 90.Lesuisse, E., R. Santos, B. F. Matzanke, S. A. Knight, J. M. Camadro, and A. Dancis. 2003. Iron use for haeme synthesis is under control of the yeast frataxin homologue (Yfh1). Hum. Mol. Genet. 12:879-889. [DOI] [PubMed] [Google Scholar]
- 91.Lichtlen, P., and W. Schaffner. 2001. Putting its fingers on stressful situations: the heavy metal-regulatory transcription factor MTF-1. Bioessays 23:1010-1017. [DOI] [PubMed] [Google Scholar]
- 92.Lichtlen, P., Y. Wang, T. Belser, O. Georgiev, U. Certa, R. Sack, and W. Schaffner. 2001. Target gene search for the metal-responsive transcription factor MTF-1. Nucleic Acids Res. 29:1514-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin, S. J., R. A. Pufahl, A. Dancis, T. V. O'Halloran, and V. C. Culotta. 1997. A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J. Biol. Chem. 272:9215-9220. [PubMed] [Google Scholar]
- 94.Lyons, T. J., A. P. Gasch, L. A. Gaither, D. Botstein, P. O. Brown, and D. J. Eide. 2000. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 97:7957-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.MacDiarmid, C. W., L. A. Gaither, and D. Eide. 2000. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19:2845-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.MacDiarmid, C. W., M. A. Milanick, and D. J. Eide. 2002. Biochemical properties of vacuolar zinc transport systems of Saccharomyces cerevisiae. J. Biol. Chem. 277:39187-39194. [DOI] [PubMed] [Google Scholar]
- 97.MacDiarmid, C. W., M. A. Milanick, and D. J. Eide. 2003. Induction of the ZRC1 metal tolerance gene in zinc-limited yeast confers resistance to zinc shock. J. Biol. Chem. 278:15065-15072. [DOI] [PubMed] [Google Scholar]
- 98.Martins, L. J., L. T. Jensen, J. R. Simon, G. L. Keller, D. R. Winge, and J. R. Simons. 1998. Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J. Biol. Chem. 273:23716-23721. [DOI] [PubMed] [Google Scholar]
- 99.Merchant, S., K. Hill, and G. Howe. 1991. Dynamic interplay between two copper-titrating components in the transcriptional regulation of cyt c6. EMBO J. 10:1383-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moseley, J., J. Quinn, M. Eriksson, and S. Merchant. 2000. The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19:2139-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moseley, J. L., M. D. Page, N. P. Alder, M. Eriksson, J. Quinn, F. Soto, S. M. Theg, M. Hippler, and S. Merchant. 2002. Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14:673-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oberegger, H., M. Schoeser, I. Zadra, M. Schrettl, W. Parson, and H. Haas. 2002. Regulation of freA, acoA, lysF, and cycA expression by iron availability in Aspergillus nidulans. Appl. Environ. Microbiol. 68:5769-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oberegger, H., I. Zadra, M. Schoeser, B. Abt, W. Parson, and H. Haas. 2002. Identification of members of the Aspergillus nidulans SREA regulon: genes involved in siderophore biosynthesis and utilization. Biochem. Soc. Trans. 30:781-783. [DOI] [PubMed] [Google Scholar]
- 104.Ogra, Y., K. Suzuki, P. Gong, F. Otsuka, and S. Koizumi. 2001. Negative regulatory role of Sp1 in metal responsive element-mediated transcriptional activation. J. Biol. Chem. 276:16534-16539. [DOI] [PubMed] [Google Scholar]
- 105.Ooi, C. E., E. Rabinovich, A. Dancis, J. S. Bonifacino, and R. D. Klausner. 1996. Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane copper transporter Ctr1p in the apparent absence of endocytosis. EMBO J. 15:3515-3523. [PMC free article] [PubMed] [Google Scholar]
- 106.Outten, C. E., and T. V. O'Halloran. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488-2492. [DOI] [PubMed] [Google Scholar]
- 107.Pelletier, B., J. Beaudoin, Y. Mukai, and S. Labbé. 2002. Fep1, an iron sensor regulating iron transporter gene expression in Schizosaccharomyces pombe. J. Biol. Chem. 277:22950-22958. [DOI] [PubMed] [Google Scholar]
- 108.Pelletier, B., J. Beaudoin, C. C. Philpott, and S. Labbé. 2003. Fep1 represses expression of the fission yeast Schizosaccharomyces pombe siderophore-iron transport system. Nucleic Acids Res. 31:4332-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Peña, M. M., J. Lee, and D. J. Thiele. 1999. A delicate balance: homeostatic control of copper uptake and distribution. J. Nutr. 129:1251-1260. [DOI] [PubMed] [Google Scholar]
- 110.Pence, N. S., P. B. Larsen, S. D. Ebbs, D. L. Letham, M. M. Lasat, D. F. Garvin, D. Eide, and L. V. Kochian. 2000. The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc. Natl. Acad. Sci. USA 97:4956-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Petit, J.-M., O. van Wuytswinkel, J.-F. Briat, and S. Lobréaux. 2001. Characterization of an iron-dependent regulatory sequence involved in the transcriptional control of AtFer1 and ZmFer1 plant ferritin genes by iron. J. Biol. Chem. 276:5584-5590. [DOI] [PubMed] [Google Scholar]
- 112.Petris, M. J., J. F. Mercer, J. G. Culvenor, P. Lockhart, P. A. Gleeson, and J. Camakaris. 1996. Ligand-regulated transport of the Menkes copper P-type ATPase efflux pump from the Golgi apparatus to the plasma membrane: a novel mechanism of regulated trafficking. EMBO J. 15:6084-6095. [PMC free article] [PubMed] [Google Scholar]
- 113.Petris, M. J., K. Smith, J. Lee, and D. J. Thiele. 2003. Copper-stimulated endocytosis and degradation of the human copper transporter, hCtr1. J. Biol. Chem. 278:9639-9646. [DOI] [PubMed] [Google Scholar]
- 114.Philpott, C. C., O. Protchenko, Y. W. Kim, Y. Boretsky, and M. Shakoury-Elizeh. 2002. The response to iron deprivation in Saccharomyces cerevisiae: expression of siderophore-based systems of iron uptake. Biochem. Soc. Trans. 30:698-702. [DOI] [PubMed] [Google Scholar]
- 115.Portnoy, M. E., L. T. Jensen, and V. C. Culotta. 2002. The distinct methods by which manganese and iron regulate the Nramp transporters in yeast. Biochem. J. 362:119-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Portnoy, M. E., X. F. Liu, and V. C. Culotta. 2000. Saccharomyces cerevisiae expresses three functionally distinct homologues of the nramp family of metal transporters. Mol. Cell. Biol. 20:7893-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Protchenko, O., T. Ferea, J. Rashford, J. Tiedeman, P. O. Brown, D. Botstein, and C. C. Philpott. 2001. Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:49244-49250. [DOI] [PubMed] [Google Scholar]
- 118.Protchenko, O., and C. C. Philpott. 2003. Regulation of intracellular heme levels by HMX1, a homologue of heme oxygenase, in Saccharomyces cerevisiae. J. Biol. Chem. 278:36582-36587. [DOI] [PubMed] [Google Scholar]
- 119.Quinn, J. M., P. Barraco, M. Eriksson, and S. Merchant. 2000. Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J. Biol. Chem. 275:6080-6089. [DOI] [PubMed] [Google Scholar]
- 120.Quinn, J. M., and S. Merchant. 1995. Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7:623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Quinn, J. M., S. S. Nakamoto, and S. Merchant. 1999. Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper response elements and increased translation from a copper deficiency-specific form of the transcript. J. Biol. Chem. 274:14444-14454. [DOI] [PubMed] [Google Scholar]
- 122.Radtke, F., O. Georgiev, H. Müller, E. Brugnera, and W. Schaffner. 1995. Functional domains of the heavy metal-responsive transcription regulator MTF-1. Nucleic Acids Res. 23:2277-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Robertson, L. S., H. C. Causton, R. A. Young, and G. R. Fink. 2000. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. USA 97:5984-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rutherford, J. C., S. Jaron, E. Ray, P. O. Brown, and D. R. Winge. 2001. A second iron-regulatory system in yeast independent of Aft1p. Proc. Natl. Acad. Sci. USA 98:14322-14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rutherford, J. C., S. Jaron, and D. R. Winge. 2003. Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 278:27636-27643. [DOI] [PubMed] [Google Scholar]
- 126.Ruzsa, S. M., and J. G. Scandalios. 2003. Altered Cu metabolism and differential transcription of Cu/ZnSod genes in a Cu/ZnSOD-deficient mutant of maize: evidence for a Cu-responsive transcription factor. Biochemistry 42:1508-1516. [DOI] [PubMed] [Google Scholar]
- 127.Saydam, N., T. K. Adams, F. Steiner, W. Schaffner, and J. H. Freedman. 2002. Regulation of metallothionein transcription by the metal-responsive transcription factor MTF-1: identification of signal transduction cascades that control metal-inducible transcription. J. Biol. Chem. 277:20438-20445. [DOI] [PubMed] [Google Scholar]
- 128.Saydam, N., O. Georgiev, M. Y. Nakano, U. F. Greber, and W. Schaffner. 2001. Nucleo-cytoplasmic trafficking of metal-regulatory transcription factor 1 is regulated by diverse stress signals. J. Biol. Chem. 276:25487-25495. [DOI] [PubMed] [Google Scholar]
- 129.Schilke, B., C. Voisine, H. Beinert, and E. Craig. 1999. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:10206-10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Seoighe, C., and K. H. Wolfe. 1999. Updated map of duplicated regions in the yeast genome. Gene 238:253-261. [DOI] [PubMed] [Google Scholar]
- 131.Serpe, M., A. Joshi, and D. J. Kosman. 1999. Structure-function analysis of the protein-binding domains of Mac1p, a copper-dependent transcriptional activator of copper uptake in Saccharomyces cerevisiae. J. Biol. Chem. 274:29211-29219. [DOI] [PubMed] [Google Scholar]
- 132.Skelton, A. P. F., N. J. Robinson, and P. B. Goldsbrough. 1998. Metallothionein-like genes and phytochelatins in higher plants, p. 398-430. In S. Silver and W. Walden (ed.), Metal ions in gene regulation. Chapman & Hall, London, England.
- 133.Smirnova, I. V., D. C. Bittel, R. Ravindra, H. Jiang, and G. K. Andrews. 2000. Zinc and cadmium can promote rapid nuclear translocation of metal response element-binding transcription factor-1. J. Biol. Chem. 275:9377-9384. [DOI] [PubMed] [Google Scholar]
- 134.Srinivasan, C., A. Liba, J. A. Imlay, J. S. Valentine, and E. B. Gralla. 2000. Yeast lacking superoxide dismutase(s) show elevated levels of “free iron” as measured by whole cell electron paramagnetic resonance. J. Biol. Chem. 275:29187-29192. [DOI] [PubMed] [Google Scholar]
- 135.Stadler, J. A., and R. J. Schweyen. 2002. The yeast iron regulon is induced upon cobalt stress and crucial for cobalt tolerance. J. Biol. Chem. 277:39649-39654. [DOI] [PubMed] [Google Scholar]
- 136.Stearman, R., D. S. Yuan, Y. Yamaguchi-Iwai, R. D. Klausner, and A. Dancis. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552-1557. [DOI] [PubMed] [Google Scholar]
- 137.Szczypka, M. S., and D. J. Thiele. 1989. A cysteine-rich nuclear protein activates yeast metallothionein gene transcription. Mol. Cell. Biol. 9:421-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Theil, E. C., and R. S. Eisenstein. 2000. Combinatorial mRNA regulation: iron regulatory proteins and iso-iron-responsive elements (Iso-IREs). J. Biol. Chem. 275:40659-40662. [DOI] [PubMed] [Google Scholar]
- 139.Thiele, D. J. 1988. ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene. Mol. Cell. Biol. 8:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thiele, D. J., and D. H. Hamer. 1986. Tandemly duplicated upstream control sequences mediate copper-induced transcription of the Saccharomyces cerevisiae copper-metallothionein gene. Mol. Cell. Biol. 6:1158-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Thorvaldsen, J. L., A. K. Sewell, C. L. McCowen, and D. R. Winge. 1993. Regulation of metallothionein genes by the ACE1 and AMT1 transcription factors. J. Biol. Chem. 268:12512-12518. [PubMed] [Google Scholar]
- 142.Thorvaldsen, J. L., A. K. Sewell, A. M. Tanner, J. M. Peltier, I. J. Pickering, G. N. George, and D. R. Winge. 1994. Mixed Cu+ and Zn2+ coordination in the DNA-binding domain of the AMT1 transcription factor from Candida glabrata. Biochemistry 33:9566-9577. [DOI] [PubMed] [Google Scholar]
- 143.Turner, R. B., D. L. Smith, M. E. Zawrotny, M. F. Summers, M. C. Posewitz, and D. R. Winge. 1998. Solution structure of a zinc domain conserved in yeast copper-regulated transcription factors. Nat. Struct. Biol. 5:551-555. [DOI] [PubMed] [Google Scholar]
- 144.Ueta, R., A. Fukunaka, and Y. Yamaguchi-Iwai. 2003. Pse1p mediates the nuclear import of the iron-responsive transcription factor Aft1p in Saccharomyces cerevisiae. J. Biol. Chem. 278:50120-50127. [DOI] [PubMed] [Google Scholar]
- 145.Urbanowski, J. L., and R. C. Piper. 1999. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 274:38061-38070. [DOI] [PubMed] [Google Scholar]
- 146.Van Ho, A., D. M. Ward, and J. Kaplan. 2002. Transition metal transport in yeast. Annu. Rev. Microbiol. 56:237-261. [DOI] [PubMed] [Google Scholar]
- 147.Voisard, C., J. Wang, J. L. McEvoy, P. Xu, and S. A. Leong. 1993. urbs1, a gene regulating siderophore biosynthesis in Ustilago maydis, encodes a protein similar to the erythroid transcription factor GATA-1. Mol. Cell. Biol. 13:7091-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Voutsina, A., G. S. Fragiadakis, A. Boutla, and D. Alexandraki. 2001. The second cysteine-rich domain of Mac1p is a potent transactivator that modulates DNA binding efficiency and functionality of the protein. FEBS Lett. 494:38-43. [DOI] [PubMed] [Google Scholar]
- 149.Waters, B. M., and D. J. Eide. 2002. Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J. Biol. Chem. 277:33749-33757. [DOI] [PubMed] [Google Scholar]
- 150.Welch, J., S. Fogel, C. Buchman, and M. Karin. 1989. The CUP2 gene product regulates the expression of the CUP1 gene, coding for yeast metallothionein. EMBO J. 8:255-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Westin, G., and W. Schaffner. 1988. A zinc-responsive factor interacts with a metal-regulated enhancer element (MRE) of the mouse metallothionein-1 gene. EMBO J. 7:3763-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Winge, D. R., J. A. Graden, M. C. Posewitz, L. J. Martins, L. T. Jensen, and J. R. Simon. 1997. Sensors that mediate copper-specific activation and repression of gene expression. J. Biol. Inorg. Chem. 2:2-10. [Google Scholar]
- 153.Winge, D. R., K. B. Nielson, W. R. Gray, and D. H. Hamer. 1985. Yeast metallothionein: sequence and metal-binding properties. J. Biol. Chem. 260:14464-14470. [PubMed] [Google Scholar]
- 154.Yamaguchi-Iwai, Y., A. Dancis, and R. D. Klausner. 1995. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 14:1231-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yamaguchi-Iwai, Y., M. Serpe, D. Haile, W. Yang, D. J. Kosman, R. D. Klausner, and A. Dancis. 1997. Homeostatic regulation of copper uptake in yeast via direct binding of MAC1 protein to upstream regulatory sequences of FRE1 and CTR1. J. Biol. Chem. 272:17711-17718. [DOI] [PubMed] [Google Scholar]
- 156.Yamaguchi-Iwai, Y., R. Stearman, A. Dancis, and R. D. Klausner. 1996. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 15:3377-3384. [PMC free article] [PubMed] [Google Scholar]
- 157.Yamaguchi-Iwai, Y., R. Ueta, A. Fukunaka, and R. Sasaki. 2002. Subcellular localization of Aft1 transcription factor responds to iron status in Saccharomyces cerevisiae. J. Biol. Chem. 277:18914-18918. [DOI] [PubMed] [Google Scholar]
- 158.Yonkovich, J., R. McKenndry, X. Shi, and Z. Zhu. 2002. Copper ion-sensing transcription factor Mac1p post-translationally controls the degradation of its target gene product Ctr1p. J. Biol. Chem. 277:23981-23984. [DOI] [PubMed] [Google Scholar]
- 159.Yuan, D. S. 2000. Zinc-regulated genes in Saccharomyces cerevisiae revealed by transposon tagging. Genetics 156:45-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yuan, D. S., R. Stearman, A. Dancis, T. Dunn, T. Beeler, and R. D. Klausner. 1995. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin-like oxidase required for iron uptake. Proc. Natl. Acad. Sci. USA 92:2632-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yuan, W. M., G. D. Gentil, A. D. Budde, and S. A. Leong. 2001. Characterization of the Ustilago maydis sid2 gene, encoding a multidomain peptide synthetase in the ferrichrome biosynthetic gene cluster. J. Bacteriol. 183:4040-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yun, C. W., J. S. Tiedeman, R. E. Moore, and C. C. Philpott. 2000. Siderophore-iron uptake in Saccharomyces cerevisiae. Identification of ferrichrome and fusarinine transporters. J. Biol. Chem. 275:16354-16359. [DOI] [PubMed] [Google Scholar]
- 163.Zhang, B., D. Egli, O. Georgiev, and W. Schaffner. 2001. The Drosophila homolog of mammalian zinc finger factor MTF-1 activates transcription in response to heavy metals. Mol. Cell. Biol. 21:4505-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang, B., O. Georgiev, M. Hagmann, C. Gunes, M. Cramer, P. Faller, M. Vasák, and W. Schaffner. 2003. Activity of metal responsive transcription factor 1 by toxic heavy metals and H2O2 in vitro is modulated by metallothionein. Mol. Cell. Biol. 23:8471-8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhao, H., E. Butler, J. Rodgers, T. Spizzo, S. Duesterhoeft, and D. Eide. 1998. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J. Biol. Chem. 273:28713-28720. [DOI] [PubMed] [Google Scholar]
- 166.Zhao, H., and D. Eide. 1996. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 93:2454-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhao, H., and D. Eide. 1996. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271:23203-23210. [DOI] [PubMed] [Google Scholar]
- 168.Zhao, H., and D. J. Eide. 1997. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:5044-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhou, H., K. M. Cadigan, and D. J. Thiele. 2003. A copper regulated transporter required for copper acquisition, pigmentation and specific stages of development in Drosophila melanogaster. J. Biol. Chem. 278:48210-48218. [DOI] [PubMed] [Google Scholar]
- 170.Zhou, L., and G. A. Marzluf. 1999. Functional analysis of the two zinc fingers of SRE, a GATA-type factor that negatively regulates siderophore synthesis in Neurospora crassa. Biochemistry 38:4335-4341. [DOI] [PubMed] [Google Scholar]
- 171.Zhou, L. W., H. Haas, and G. A. Marzluf. 1998. Isolation and characterization of a new gene, sre, which encodes a GATA-type regulatory protein that controls iron transport in Neurospora crassa. Mol. Gen. Genet. 259:532-540. [DOI] [PubMed] [Google Scholar]
- 172.Zhou, P., M. S. Szczypka, T. Sosinowski, and D. J. Thiele. 1992. Expression of a yeast metallothionein gene family is activated by a single metalloregulatory transcription factor. Mol. Cell. Biol. 12:3766-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhou, P., and D. J. Thiele. 1991. Isolation of a metal-activated transcription factor gene from Candida glabrata by complementation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88:6112-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhou, P., and D. J. Thiele. 1993. Rapid transcriptional autoregulation of a yeast metalloregulatory transcription factor is essential for high-level copper detoxification. Genes Dev. 7:1824-1835. [DOI] [PubMed] [Google Scholar]
- 175.Zhu, Z., S. Labbe, M. M. Pena, and D. J. Thiele. 1998. Copper differentially regulates the activity and degradation of yeast Mac1 transcription factor. J. Biol. Chem. 273:1277-1280. [DOI] [PubMed] [Google Scholar]