Abstract
We describe a novel HPV 9G DNA chip test for the accurate and reliable genotyping of human papillomavirus (HPV). The HPV 9G DNA chip test established its efficiency in terms of a signal-to-background ratio (SBR) of 200, which is 50 times superior to commercial HPV DNA chips, and 100% target-specific hybridization at 25°C. We compared the genotyping results for the 439 clinical samples by the HPV 9G DNA chip test with the sequencing results for the MY11/GP6+ (M2) primer set-mediated PCR products. The discrimination of HPV genotypes in the 151 HPV-positive clinical samples by the HPV 9G DNA chip test were 100% identical with the sequencing analysis. The clinical sensitivities of HPV genotyping by the HPV 9G DNA chip test and a commercial HPV DNA chip test were 100% and 88%, respectively. However, the clinical specificities of HPV genotyping by the HPV 9G DNA chip test and the commercial HPV DNA chip test were 100% and 94%, respectively. The 100% clinical sensitivity and specificity of the HPV 9G DNA chip test make it a promising diagnostic tool for HPV genotyping.
INTRODUCTION
Cervical cancer is the second most common cancer in terms of both incidence and mortality worldwide (6). Molecular epidemiologic evidence clearly indicates that certain types of human papillomavirus (HPV) are the principal causes of invasive cervical cancer (19) and cervical intraepithelial neoplasia (11, 16). Genital HPV types are subdivided into low-risk types (HPV-6, HPV-11, HPV-34, HPV-40, and HPV-42), found mainly in genital warts (14), and high-risk types (HPV-16, HPV-18, HPV-31, HPV-33, HPV-35, HPV-39, HPV-45, HPV-51, HPV-52, HPV-56, HPV-58, HPV-59, HPV-66, and HPV-68), associated with invasive cervical cancer. Several studies indicate that persistent infection with a high-risk HPV type is strongly associated with cervical dysplasia and carries a greater risk of subsequent progression to cervical cancer (3, 8). Therefore, highly proficient HPV-genotyping tests that can screen for specific high-risk HPV types are essential. In addition, the importance of HPV testing has been increasing because genetic typing is required to predict clinical progression (2).
However, the traditional methods of HPV detection, such as morphological and immunological methods, lack the ability to detect the specific HPV type (13). The Southern blot hybridization test is considered a highly sensitive test for HPV detection. However, it is unsuitable for clinical use because it is labor-intensive and requires fresh samples (5). Sequencing analysis is the gold standard for the detection of genotypes in clinical samples, but the major disadvantage of this method is that it cannot differentiate specific HPV genotypes in multiply infected clinical samples.
Although several working systems based on oligonucleotide arrays have been established, their signal-to-background ratios (SBR) and 100% target-specific hybridization, leading to results identical to those of sequencing analysis in singly infected clinical samples, are major issues (13). The high background signals are due to the use of high photomultiplier tube (PMT) gains (80 to 90%) of reported microarrays (1, 9). Considering the drawbacks of current HPV detection and the discrimination tests, a highly proficient HPV detection test is essential.
Recently reported 9G (GGGGGGGGG) DNA chips based on 9G DNA chip technology show high SBR (PMT gain = 48%) and 100% target-specific hybridization, with more than 90% hybridization efficiency at 25°C in 30 min (17, 18). Thus, based on the 9G DNA chip technology, we developed a novel HPV 9G DNA chip and an HPV 9G DNA chip test for the detection and discrimination of HPV genotypes in clinical samples. The HPV 9G DNA chip test is based on the detection of the PCR products of HPV in clinical samples. Moreover, single and multiple infections can be detected by 25°C hybridization for 30 min on the HPV 9G DNA chip. The overall detection time of the HPV 9G DNA chip test is 40 min. In this study, we evaluated the accuracy of the HPV 9G DNA chip test for the detection and typing of HPV in 439 clinical samples. The results of the HPV 9G DNA chip test were compared with the results of cervical cytology, HPV DNA sequencing analysis, and a commercial HPV DNA chip test.
MATERIALS AND METHODS
Materials.
All chemicals were purchased from Sigma-Aldrich Chemicals, South Korea. All oligonucleotides were purchased from Bioneer, South Korea. Glass slides (2.5 by 7.5 cm) were purchased from Paul Marienfeld GmbH & Co. KG, Germany. All washing solvents for the substrates were of high-performance liquid chromatography (HPLC) grade and were from SK Chemicals, South Korea. Ultrapure water (18 MΩ/cm) was obtained with a Milli-Q purification system (Millipore). The commercial HPV DNA chip was purchased from Biomedlab Inc., Seoul, South Korea.
Instruments.
Oligonucleotides were spotted using a Qarray2 microarrayer (Genetix Technologies, Inc.). Hybridization was done at 25°C using a commercial incubator, and then the slides were dried using a commercial centrifuge (1,000 rpm). The fluorescence signal of the microarray was measured on a ScanArrayLite (GSI Lumonics), and the images were analyzed with Quant Array software (Packard Bioscience).
Compositions of the solutions used.
The compositions of the different solutions used were as follows: immobilization solution (pH 7.4), 15% glycerol, 50 mM butyl amine, 600 mM NH4Cl; blocking buffer solution (pH 7.4), 0.5% milk casein in 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate); hybridization buffers (pH 7.4), 25% formamide, 0.1% Triton X-100, 6× SSC; washing buffer solution A (pH 7.4), 0.1% SDS in 4× SSC; washing buffer solution B (pH 7.4), 4× SSC.
Generalized probe selection method.
Probes were selected according to our recent report on the generalized probe selection method (15). In brief, the probes were designed to hybridize with the MY11/GP6+ primer set-mediated PCR products of the HPV genotypes. The generalized probe selection method uses a single artificial mutation in the immobilized probe (with one artificial mutation), which eliminates the problem of cross-hybridization shown by the original probes (without artificial mutation). Therefore, we chose probes (with one artificial mutation [Table 1]) that are highly specific for the particular HPV genotype and do not show any cross-hybridization with other HPV genotypes. The absence of cross-hybridization indicates that there are no chances of false-positive results. The other, minor details of the probe selection method can be found in reference 15.
Table 1.
Sequences of probes immobilized on the HPV 9G DNA chip for HPV genotyping
| Probe or primer name | HPV type, probe, or primera | Sequenceb |
|---|---|---|
| 1 | 16 | 5′-GGGGGGGGGCTTTATCCTACGACTTGGGGAGG |
| 2 | 18 | 5′-GGGGGGGGGCTTTATTAGCAGACTTGTTGAGG |
| 3 | 6 | 5′-GGGGGGGGGCTTTATCATGCGTCATATGGAAG |
| 4 | 66 | 5′-GGGGGGGGGCTTTATCCTTCGCCATATGGAGG |
| 5 | 39 | 5′-GGGGGGGGGCTTTATTACCAGGCACGTGTAGG |
| 6 | 42 | 5′-GGGGGGGGGCTTTATTTTAAGACTTGCTGAAG |
| 7 | 31 | 5′-GGGGGGGGGCTTTATTTTAAGACATAGTGAGG |
| 8 | 45 | 5′-GGGGGGGGGCTTTATTAGTAGACATATGGAGG |
| 9 | 11 | 5′-GGGGGGGGGCTTTATCATGCGCCATATGGAGG |
| 10 | 59 | 5′-GGGGGGGGGCTTTATTGCCAGACATATGGAGG |
| 11 | 51 | 5′-GGGGGGGGGCTTTATTATTAGGCATAGGGAAG |
| 12 | 34 | 5′-GGGGGGGGGCTTTATCCTCAGACTTGCAGAAG |
| 13 | 33 | 5′-GGGGGGGGGCTTTATTATAAGACATGTTGAAG |
| 14 | 52 | 5′-GGGGGGGGGCTTTATCCTTCGTCAAGGCGAGG |
| 15 | 40 | 5′-GGGGGGGGGCTTTATTTTGCGTCATAGGGAGG |
| 16 | 68 | 5′-GGGGGGGGGCTTTATTGTTAGGCATGTTGAGG |
| 17 | 56 | 5′-GGGGGGGGGCTTTATCCTTAGACATGTGTAGG |
| 18 | 35 | 5′-GGGGGGGGGCTTTATTTTAAGGCAAGGTGAAG |
| 19 | 58 | 5′-GGGGGGGGGCTTTATTGTACGTCATATTGAAG |
| 20 | HC | 5′-GGGGGGGGGTTTCCTAGTGGCTCTATGGTAAC |
| 21 | PC | 5′-GGGGGGGGGTGATTTACAGTTTATDTTTC |
| 22 | PCR | 5′-GGGGGGGGGATTGGCATGBKGARGARTWTGA |
| 23 | NC | 5′-GGGGGGGGGAAAGCTGCTGCTCGTCGTCGTCGT |
| Target1 | HC-Cy5-T1 | 3′-GGATCACCGAGATACCATTGGAGACTGCG-Cy5-5′ |
| Forward | FP | 3′-GCMCAGGGWCATAAYAATGG-5′ |
| Reverse | RP-Cy5 | 3′-GAAAHATAAACTGTAAATCATAYTC-Cy5-5′ |
HC, probe for hybridization control; PC, probe for primer control (positive control); PCR, probe for PCR control; HC-Cy5-T1, target oligonucleotide for HC probe.
GGGGGGGGG, 9G for immobilization of probes on AMCA slides; CTTTAT, vertical spacer group.
Typical method for preparation of the HPV 9G DNA chip.
The HPV 9G DNA chip was prepared according to our previous report (17) using the probes obtained by our recently reported generalized probe selection method for DNA chips (15). The immobilization solutions containing the probes (probe 1 to probe 23) corresponding to the 19 HPV genotypes—the 14 high-risk groups (HPV-16, HPV-18, HPV-31, HPV-33, HPV-35, HPV-39, HPV-45, HPV-51, HPV-52, HPV-56, HPV-58, HPV-59, HPV-66, and HPV-68) and the 5 low-risk groups (HPV-6, HPV-11, HPV-34, HPV-40, and HPV-42), along with the hybridization control (HC), positive control (PC), and PCR control (PCR) probes (Table 1), were spotted on the AMCA (aminocalix[4]arene) slide using a microarrayer. The spots were arranged to make 8 by 7 pixels. The microarrayed AMCA slide was kept in an incubator (25°C; 50% humidity) for 4 h to immobilize the oligonucleotides. The slide was then suspended in blocking buffer solution at 25°C for 30 min in order to remove the excess oligonucleotides and to deactivate the nonspotted area. Then, the slide was rinsed with washing buffer solutions A and B for 5 min each and then dried with a commercial centrifuge to obtain the HPV 9G DNA chip. Before hybridization, the 9G HPV DNA chips were covered with Secure-Seal hybridization chambers. An immobilization density of 6.2 pmol/cm2 was calculated for the probes immobilized on the HPV 9G DNA chip according to the method in our previous report (17).
General hybridization procedure.
Each hybridization chamber of the HPV 9G DNA chip was covered with a mixture of 35 μl of hybridization buffer solution, 10 μl of Cy5-HC-T1 cys-labeled hybridization control target probe (660 fmol/μl), and 5 μl of Cy5-labeled PCR product of an HPV genotype (e.g., HPV-16 or HPV-18) and incubated at 25°C for 30 min. For the washing step, the HPV 9G DNA chip was rinsed with washing buffer solutions A and B successively at 25°C for 2 min each in order to remove the excess target DNA and dried with a commercial centrifuge (1,000 rpm). The fluorescence signal of the microarray was measured on a ScanArrayLite, and the images were analyzed with Quant Array software. Each experiment was done more than three times.
Clinical samples.
Samples were collected consecutively from 439 Korean women who visited the Department of Obstetrics and Gynecology, CHA Clinic, South Korea. The samples were collected by scraping the uterine cervical canal with a small cytobrush after a Pap smear, and the brush was put into a 15-ml centrifuge tube containing phosphate-buffered saline. The specimens were collected as part of an informed consent protocol approved by the clinical studies committee of CHA Bundang Hospital, South Korea.
Study subjects.
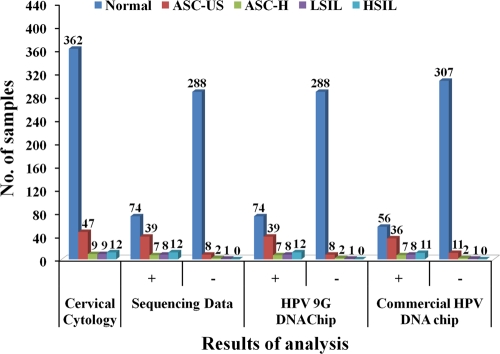
Cervical cytology was performed on all 439 cervical samples. Out of 439 samples, 77 produced HPV-positive results, while the remaining 362 samples were found to be normal (no HPV was detected) (Table 1). The 77 HPV-positive samples were categorized as atypical squamous cells of undetermined significance (ASC-US) (47 [61.0%]); atypical squamous cells, cannot exclude high-grade squamous intraepithelial lesions (HSIL) (ASC-US-H) (9 [11.7%]), low-grade squamous intraepithelial lesions (LSIL) (9 [11.7%]), and HSIL (12 [15.6%]). The results of cervical cytology were compared with the results of sequencing analysis, the HPV 9G DNA chip test, and the commercial HPV DNA chip test (Table 2).
Table 2.
Comparison of the results of HPV genotyping by cervical cytology with the results of sequencing analysis, the HPV 9G DNA chip test, and a commercial HPV DNA chip test (439 clinical samples)
| Cervical cytology result | No. of samples | No. (%) of samples |
|||||
|---|---|---|---|---|---|---|---|
| Sequencing |
HPV 9G DNA chip |
Commercial HPV DNA chip |
|||||
| HPV+ | HPV− | HPV+ | HPV− | HPV+ | HPV− | ||
| Normal | 362 | 74 (20.8) | 288 | 74 (20.8) | 288 | 56 (15.0) | 307 |
| ASC-US | 47 | 39 (83.0) | 8 | 39 (83.0) | 8 | 36 (76.6) | 11 |
| ASC-US-H | 9 | 7 (77.8) | 2 | 7 (77.8) | 2 | 7 (77.8) | 2 |
| LSIL | 9 | 8 (88.9) | 1 | 8 (88.9) | 1 | 8 (88.9) | 1 |
| HSIL | 12 | 12 (100) | 12 (100) | 11 (92.3) | |||
DNA extraction and PCR amplification.
MY09/MY11 primer set-mediated PCR (MY-PCR) and GP5+/GP6+ primer set-mediated PCR (GP+-PCR) are the most frequently used amplification systems for the detection of HPV DNA in clinical samples, amplifying DNA fragments in the conserved L1 region of approximately 450 bp and 150 bp, respectively. Further, type-specific PCR primer sets allow the identification of individual genotypes. The MY11/GP6+ primer set consists of fixed nucleotide sequences for the forward and reverse primers and detects a wide range of HPV types by using a lower annealing temperature during PCR (4, 7).
The whole HPV genomic DNA extracted from clinical samples was amplified by duplex PCR to generate amplicons. The HPV DNA was amplified with primers RP and FP (Table 1). The PCR mixture consisted of 10 μl of the extracted DNA, 10 μl of each primer (RP and FP), PCR premix (catalog number K-2016V1; Bioneer Inc., Daejan, South Korea) containing deoxyribonucleotide triphosphate, 2 U of Fast Start Taq DNA polymerase in an amplification buffer containing 2 mM MgCl2, and a tracking dye (Cy5). All tubes were incubated for 2 min at 50°C before PCR was started. Amplification was performed with the following steps: predenaturation for 5 min at 94°C, 45 cycles of 30 s each for denaturation at 94°C, 45 cycles of 30 s each for annealing at 65°C, 45 cycles of 30 s each for elongation at 72°C, and a final elongation step of 7 min at 72°C. Five microliters of PCR product was subjected to agarose gel electrophoresis using a 2% agarose standard run in 1× Tris-borate EDTA. Five microliters of this Cy5-labeled PCR product was used for further hybridization experiments on the HPV 9G DNA chip for HPV detection and genotyping. The whole procedure was followed for all 439 clinical samples.
RESULTS
HPV detection and genotyping by sequencing.
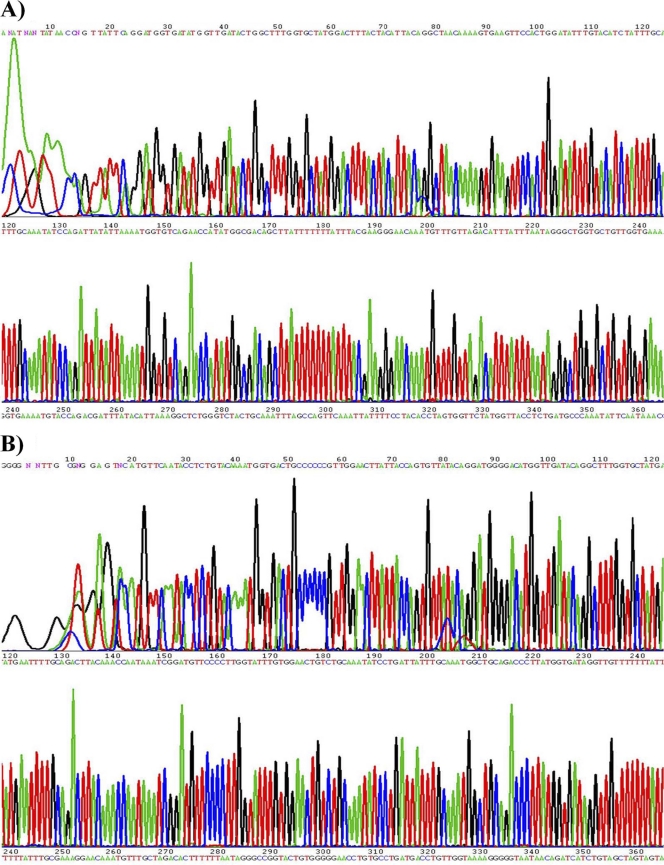
The primed PCR product was added to the sequencing reaction mixture. Sequencing was performed bidirectionally with the BigDye3 terminator cycle-sequencing kit (PE Applied Biosystems) using an ABI Prism 310 genomic analyzer (PE Applied Biosystems) at a dispensing pressure of 60,000 Pa with 8-ms open times and 65-s cycle times. The sequencing procedure was carried out by stepwise elongation of the primer strand upon cyclic dispensation of the different deoxynucleoside triphosphates (Amersham Pharmacia Biotech). A charge-coupled device (CCD) camera detected the light output resulting from nucleotide incorporation. The data were obtained in a graphic format (Fig. 1). Out of the 439 clinical samples, 151 were found to be HPV positive in the sequencing analysis. Six out of 151 samples were infected with multiple HPV genotypes. It is well known that HPV genotyping by sequencing analysis of samples containing multiple infections is difficult due to the presence of a mixed peak in sequencing data. Therefore, the samples with multiple infections were first tested with the HPV 9G DNA chip. Based on the types found on the HPV 9G DNA chip, type-specific primers sets were used to obtain specific PCR products, which were analyzed by sequencing to identify the HPV genotypes in the same sample. The samples with more than one HPV genotype were also detected on the HPV 9G DNA chip. HPV-16/HPV-58, HPV-16/HPV-68, and HPV-18/HPV-11 are examples of the samples with multiple infections found by the HPV 9G DNA chip, as well as by sequencing of the type-specific PCR products.
Fig 1.
Examples of HPV DNA sequencing. (A) HPV-16. (B) HPV-11.
HPV 9G DNA chip test.
We used the HPV 9G DNA chip (provided by Biometrix Technology Inc., Chuncheon, South Korea) as a method for HPV genotyping in clinical samples. The HPV 9G DNA chip consists of 19 type-specific probes—14 high-risk types (HPV-16, HPV-18, HPV-31, HPV-33, HPV-35, HPV-39, HPV-45, HPV-51, HPV-52, HPV-56, HPV-58, HPV-59, HPV-66, and HPV-68) and 5 low-risk types (HPV-6, HPV-11, HPV-34, HPV-40, and HPV-42). The 5 μl of PCR product was subjected to agarose gel electrophoresis, using a 2% agarose gel, and the product size of the HPV DNA was found to be 250 bp.
Each hybridization chamber of the HPV 9G DNA chip was covered with a mixture of 35 μl of the hybridization buffer solution, 10 μl of Cy5-HC-T1 (660 fmol/μl), and 5 μl of Cy5-labeled HPV genotype PCR product (e.g., HPV-16 or HPV-18) and incubated at 25°C for 30 min. The DNA chip was then rinsed with washing buffer solutions A and B successively for 2 min each in order to remove the excess target DNA and dried with a commercial centrifuge (1,000 rpm). The fluorescence signal of the microarray was measured on a ScanArrayLite, and the images were analyzed with Quant Array software. Each experiment was done more than three times.
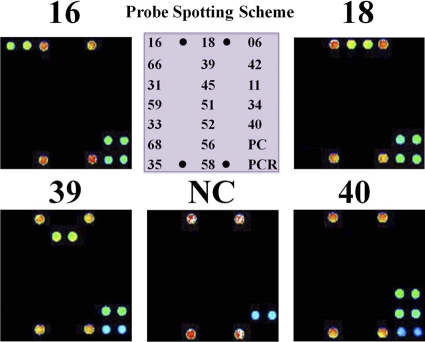
HPV amplicons can be hybridized with type-specific oligonucleotide probes and visualized on HPV 9G DNA chips as double-positive spots (Fig. 2) when HPV DNA is present in the amplified PCR product. However, the samples that showed a positive band of 250 bp on gel electrophoresis but were absent on the spots for 19 HPV genotypes on the HPV 9G DNA chip were designated HPV-other, as they can be visualized on the spots for the PC. None of the negative controls (without DNA) revealed HPV positivity. Interestingly, the excellent specificity of the probes immobilized using the new 9G DNA chip technology ensured an SBR of more than 200 (Fig. 2), which is 50 times better than the commercial HPV DNA chips. The target hybridization provides 150 to 300 times stronger signal intensity than nontarget hybridization. The HPV 9G DNA chip allowed the genotyping of a number of HPVs in one reaction. This is advantageous for diagnostic purposes when genotyping HPV in clinical samples.
Fig 2.
HPV genotyping by the HPV 9G DNA chip probe-spotting scheme. Shown are fluorescence images after hybridization of the immobilized probes on HPV 9G DNA chips with Cy5-labeled PCR products of the HPV genotypes. NC, negative control. PMT gain = 48.
The results of HPV genotyping by the HPV 9G DNA chip were compared with the results of HPV genotyping by sequencing analysis and the commercial HPV DNA chip in the 439 clinical samples in which specific HPV genotypes were detected (Table 2). Out of the 439 clinical samples, 151 were found to be HPV positive in sequencing analysis. Six out of 151 samples were infected with multiple HPV genotypes. These results were 100% identical with those of the sequencing analysis (Tables 2 and 3 and Fig. 3). However, it is well known that the detection of multiple infections in a sample by sequencing is not easy and is often unreliable.
Table 3.
Comparison of the results of HPV genotyping by sequencing analysis, HPV 9G DNA chip, and commercial HPV DNA chip in the 439 clinical samples in which specific HPV genotypes were detected
| HPV type | No. of samplesa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Normal |
ASC-US |
ASC-US-H |
LISL |
HSIL |
||||||
| A/B | C | A/B | C | A/B | C | A/B | C | A/B | C | |
| 16 | 11 | 9 | 7 | 7 | 3 | 3 | 1 | 1 | 6 | 6 |
| 18 | 4 | 4 | ||||||||
| 45 | 2 | 1 | ||||||||
| 31 | 2 | 2 | 2 | 1 | 1 | 1 | ||||
| 6 | 2 | 1 | 1 | 1 | ||||||
| 11 | 1 | |||||||||
| 34 | 1 | 1 | 1 | 1 | ||||||
| 35 | 1 | 1 | ||||||||
| 39 | 7 | 5 | 1 | 1 | 1 | 1 | ||||
| 40 | 7 | 7 | ||||||||
| 42 | 1 | 1 | 2 | 2 | ||||||
| 51 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| 56 | 2 | 2 | 4 | 4 | 2 | 2 | ||||
| 59 | ||||||||||
| 58 | 5 | 4 | 6 | 4 | 1 | 1 | 4 | 4 | ||
| 66 | 2 | 2 | 1 | 1 | ||||||
| 68 | 5 | 1 | 1 | 1 | ||||||
| 52 | 11 | 10 | 7 | 7 | 2 | 2 | ||||
| 33 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| 16/58 | 1 | 1 | ||||||||
| 16/68 | 1 | 1 | ||||||||
| 31/39/51 | 1 | |||||||||
| 18/11 | 1 | 1 | ||||||||
| 18/6 | 1 | 1 | ||||||||
| 66/11 | 1 | 1 | ||||||||
| 26 | 2 | 2 | ||||||||
| 44 | 1 | 1 | ||||||||
| 70 | 2 | |||||||||
| 81 | 1 | |||||||||
| 84 | 1 | 1 | ||||||||
| 86 | 1 | |||||||||
| Negative | 288 | 307 | 8 | 11 | 2 | 2 | 1 | 1 | ||
A, results of sequencing analysis; B, results of HPV 9G DNA chip analysis; C, results of the commercial HPV DNA chip analysis.
Fig 3.
Comparison of the results of HPV testing by cervical cytology, sequencing, the HPV 9G DNA chip test, and a commercial HPV DNA chip test.
However, it is interesting that the HPV 9G DNA chip test can detect multiple infections. Thus, based on the detection of multiple infections by the HPV 9G DNA chip, type-specific primers were used for PCR amplification and the PCR products were analyzed by sequencing. The results were in accordance with the types detected in the HPV 9G DNA chip. The multiply infected samples were found to have only two genotypes in the HPV 9G DNA chip test, which is quite unusual and is probably due to the primer set used in this study, which is well known to express two major genotypes in multiply infected samples. The results of the sequencing test for the HPV genotypes HPV-16 (18.5%), HPV-52 (13.2%), HPV-58 (10.6%), HPV-39 (5.9%), HPV-56 (5.2%), and HPV-40 (4.6%) were 100% identical with those of the HPV 9G DNA chip test.
Commercial HPV DNA chip test.
HPV genotyping of 439 clinical samples was performed using a commercial HPV DNA chip test following the manufacturer's protocol. The results of the genotyping test by commercial HPV DNA chip were compared with the results of the HPV-genotyping test by sequencing and the HPV 9G DNA chip test (Table 3 and Fig. 3). For 19 (12.6%) of the 151 HPV-positive samples, the commercial HPV DNA chip was not in agreement with sequencing and the HPV 9G DNA chip test.
It is interesting that in 151 (100%) of the 151 cases, the HPV types from the HPV DNA-sequencing test were in agreement with the types from the HPV 9G DNA chip test. The HPV 9G DNA chip test detected 20% more HPV-16 genotypes in the normal samples than the commercial HPV DNA chip. An additional 12 cases of HPV high-risk genotypes (HPV-45, HPV-58, HPV-31, and HPV-68) were also detected by the HPV 9G DNA chip test.
It is interesting that the 439 clinical samples showed 100% identical results in sequencing and the HPV 9G DNA chip assay. However, there were 22 discrepant samples among the results from the HPV 9G DNA chip test or sequencing analysis and the commercial HPV DNA chip test. Therefore, to cross check the results obtained with the HPV 9G DNA chip test and the commercial HPV DNA chip test in these 22 discrepant samples, we performed sequencing analysis of type-specific PCR products of the HPV genotypes detected by the HPV 9G DNA chip test. The results indicated that the HPV genotypes detected by the HPV 9G DNA chip test were in accordance with the HPV genotypes detected in the sequencing analysis of type-specific PCR products.
Statistical analysis.
The accuracy of the HPV 9G DNA chip test (Table 4) for the detection of HPV genotypes in the 439 clinical samples was calculated from the sensitivity (the true-positive rate) and specificity (the true-negative rate) and 95% confidence intervals (CI). Considering that the HPV 9G DNA chip test showed results that were 100% identical with those of sequencing analysis, the samples that were positive by sequencing analysis and the HPV 9G DNA chip test were defined as true positive and the specimens that were negative by both methods were defined as true negative.
Table 4.
Sensitivity and specificity of the HPV 9G DNA chip in HPV genotyping
| HPV-genotyping test | Sensitivity (95% CI)a | Specificity (95% CI)a |
|---|---|---|
| HPV 9G DNA chip | 1.0 (0.98–1.0) | 1.0 (0.99–1.0) |
| Commercial HPV DNA chip | 0.88 (0.83–0.93) | 0.94 (0.90–0.96) |
The sensitivity and specificity were calculated by assuming that the sequencing analysis of the clinical samples was correct.
DISCUSSION
The HPV 9G DNA chips obtained by the 9G DNA chip technology showed excellent performance in terms of the SBR of 200 and 100% target-specific hybridization. The target hybridization provides 150 to 300 times stronger signal intensity than nontarget hybridization, which serves as a key to the proficient genotyping by HPV 9G DNA chips. The clinical sensitivities of HPV genotyping by the HPV 9G DNA chip test and the commercial HPV DNA chip test were 100% and 88%, respectively. However, the clinical specificities of HPV genotyping by the HPV 9G DNA chip test and the commercial HPV DNA chip test were 100% and 94%, respectively. The sensitivity and specificity shown by the commercial HPV DNA chip test in these experiments are comparable to those of reported DNA chip tests for HPV detection (10, 12). The clinical sensitivity and specificity of cervical cytology were 67% and 80%, respectively. It is evident from the results of the HPV 9G DNA chip test that many clinical samples designated normal by cervical cytology were found to be high-risk-type HPV genotypes (HPV-16, 11cases; HPV-53, 11 cases; and HPV-68, 5 cases). The low sensitivity and specificity of cervical cytology may lead to improper diagnosis. On the other hand, the HPV 9G DNA chip test shows great accuracy in the genotyping of HPVs in clinical samples.
The 100% sensitivity and specificity of the HPV 9G DNA chip test in the detection and discrimination of the 19 HPV genotypes establish the accuracy of the HPV 9G DNA chip test in the clinical diagnosis of HPV infection. The following factors probably contribute to proficient genotyping by the HPV 9G DNA chip test: (i) 30-min hybridization and 2 rounds of 2-min washing at 25°C, (ii) an SBR for the HPV 9G DNA chip of 200 compared to 2.5 to 5 for the other DNA chips, and (iii) 100% target specificity of the HPV 9G DNA chip (results that were 100% identical with those of sequencing analysis).
The results for the sequencing of the HPV genotypes HPV-16 (18.5%), HPV-52 (13.2%), HPV-58 (10.6%), HPV-39 (5.9%), HPV-56 (5.2%), and HPV-40 (4.6%) were 100% identical with those of the HPV 9G DNA chip test. The HPV-genotyping test based on the HPV 9G DNA chip is an accurate method for detection and genotyping of HPV. The genotypes from the HPV 9G DNA chip test were in 100% agreement with the genotypes from HPV DNA sequencing. It is interesting that the six samples containing multiple HPV genotypes were identified in the HPV 9G DNA chip test. It is well known that the detection of multiple infections in a sample by sequencing is not easy and is often unreliable. However, it is interesting that the HPV 9G DNA chip test can detect multiple infections. Hence, based on the detection of multiple infections by the HPV 9G DNA chip test, type-specific primers were used for PCR amplification, and the PCR products were analyzed by sequencing. The results were in accordance with the types detected in the HPV 9G DNA chip test.
The multiply infected samples were found to have only two genotypes in the HPV 9G DNA chip test, which is quite unusual, but it is due to the primer set used in this study, which is well known to express two major genotypes in multiply infected samples (18). The HPV 9G DNA chip test has proven that the preliminary findings of cervical cytology of HPV-infected samples may lead to the false detection of HPV. Out of 362 clinical samples that were designated normal (HPV negative) by cervical cytology, 72 (20.4%) were identified as HPV positive in the HPV 9G DNA chip test.
TaqMan PCR or other PCR techniques are also sensitive methods for the detection of HPV DNA. Currently, standard nested HPV PCR can be performed using degenerative primers followed by direct sequencing of the PCR product. Alternatively, PCR products can be hybridized to DNAs of known HPV types to determine the type of HPV present in the PCR-amplified sample (e.g., the Roche HPV Amplicor system). Both assays are time-consuming and are often associated with false-positive results.
However, the HPV 9G DNA chip test takes only 60 min, including 30 min of hybridization time, for the highly specific detection of the HPV genotypes. Moreover, the advantage of the HPV 9G DNA chip test is that it uses simple hybridization and washing steps, which allows it to be used by small laboratories and does not require highly trained professionals. The HPV 9G DNA chip test can efficiently detect the 19 known HPV genotypes.
In conclusion, the excellent sensitivity and the specificity of the HPV 9G DNA chip were demonstrated by the signal-to-noise ratio of more than 200 and the target hybridization/nontarget hybridization ratio of 150 to 300. The newly developed HPV 9G DNA chip test has been successfully used as a diagnostic tool, since it discriminates many HPV genotypes in 30 min and also identifies multiple infections. Furthermore, in this study, the accuracy of the HPV 9G DNA chip test for HPV genotyping could be certified by comparison with the sequencing data, which were found to be 100% identical in all cases. The HPV 9G DNA chip test demonstrates clinical value in decision making, as it can identify and discriminate the 19 different HPV genotypes with 100% accuracy. The efficient detection and discrimination of the 439 clinical samples make the HPV 9G DNA chip test a promising diagnostic tool for accurate HPV genotyping.
ACKNOWLEDGMENT
This work was supported by the Ministry of Knowledge and Economy of Korea.
We declare that there are no conflicts of interest.
Footnotes
Published ahead of print 14 December 2011
REFERENCES
- 1. Choi Y-D, Jung W-W, Nam J-H, Choi H-S, Park C-S. 2005. Detection of HPV genotypes in cervical lesions by the HPV DNA chip and sequencing. Gynecol. Oncol. 98:369–375 [DOI] [PubMed] [Google Scholar]
- 2. Choi BS, et al. 2003. Genital human papillo-mavirus genotyping by HPV oligonucleotide microarray in Korean commercial sex workers. J. Med. Virol. 71:440–445 [DOI] [PubMed] [Google Scholar]
- 3. Chua K-L, Hjerpe A. 1996. Persistence of HPV infections preceding cervical carcinoma. Cancer 77:121–127 [DOI] [PubMed] [Google Scholar]
- 4. de Roda Husman AM, Walboomers JMM, van den Brule AJC, Meijer CJLM, Snijders PJF. 1995. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 76:1057–1062 [DOI] [PubMed] [Google Scholar]
- 5. Ferenczy A. 1995. Viral testing for genital human papillomavirus infections: recent progress and clinical potentials. Int. J. Gynecol. Cancer 5:321–328 [DOI] [PubMed] [Google Scholar]
- 6. Ferlay J, Bray F, Pisani P, Parkin DM. 2001. Parkin Globocan 2000: cancer incidence, mortality and prevalence worldwide, version 1.0. IARC CancerBase no. 5. IARC Press, Lyons, France [Google Scholar]
- 7. Fuessel Haws AL, et al. 2004. Nested PCR with the PGMY09/11 and GP5+/6+ primer sets improves detection of HPV DNA in cervical samples. J. Virol. Methods 122:87–93 [DOI] [PubMed] [Google Scholar]
- 8. Ho GYF, et al. 1995. Persistent genital HPV infection as a risk factor for persistent cervical dysplasia. J. Natl. Cancer Inst. 87:1365–1371 [DOI] [PubMed] [Google Scholar]
- 9. Hwang TS, et al. 2003. Detection and typing of HPV genotypes in various cervical lesions by HPV oligonucleotide microarray. Gynecol. Oncol. 90:51–56 [DOI] [PubMed] [Google Scholar]
- 10. Kim CJ, et al. 2003. HPV oligonucleotide microarray-based detection of HPV genotypes in cervical neoplastic lesions. Gynecol. Oncol. 89:210–217 [DOI] [PubMed] [Google Scholar]
- 11. Kjaer SK, et al. 1996. Human papillomavirus—the most significant risk determinant of cervical intraepithelial neoplasia. Int. J. Cancer 65:601–606 [DOI] [PubMed] [Google Scholar]
- 12. Lee G-Y, et al. 2005. Human papillomavirus (HPV) genotyping by HPV DNA chip in cervical cancer and precancerous lesions. Int. J. Gynecol. Cancer 15:81–87 [DOI] [PubMed] [Google Scholar]
- 13. Liu CH, et al. 2003. Possibility of using DNA chip technology for diagnosis of human papillomavirus. J. Biochem. Mol. Biol. 36:349–353 [DOI] [PubMed] [Google Scholar]
- 14. Munoz N, et al. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518–527 [DOI] [PubMed] [Google Scholar]
- 15. Nimse SB, et al. 2011. A generalized probe selection method for DNA chips. Chem. Commun. 47:12444–12446 [DOI] [PubMed] [Google Scholar]
- 16. Schiffman MH, et al. 1993. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 85:958–964 [DOI] [PubMed] [Google Scholar]
- 17. Song K, et al. 2011. 9G DNAChip: microarray based on the multiple interactions of 9 consecutive guanines. Chem. Commun 47:7101–7103 [DOI] [PubMed] [Google Scholar]
- 18. Song K, et al. 2011. 9G DNAChip: a platform for the efficient detection of the proteins. Chem. Commun. 47:7616–7618 [DOI] [PubMed] [Google Scholar]
- 19. Walboomers JMM, et al. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12–19 [DOI] [PubMed] [Google Scholar]