Abstract
Campylobacter jejuni is a major zoonotic pathogen. A highly virulent, tetracycline-resistant C. jejuni clone (clone SA) has recently emerged in ruminant reservoirs and has become the predominant cause of sheep abortion in the United States. To determine whether clone SA is associated with human disease, we compared the clinical isolates of clone SA from sheep abortions with the human isolates of the PulseNet National Campylobacter databases at the CDC and the FDA using pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and serotyping. The combined SmaI and KpnI PFGE pattern designations of clone SA from sheep were indistinguishable from those of 123 (9.03%) human C. jejuni isolates (total, 1,361) in the CDC database, among which 56 were associated with sporadic infections and 67 were associated with outbreaks that occurred in multiple states from 2003 to 2010. Most of the outbreaks were attributed to raw milk, while the sources for most of the sporadic cases were unknown. All clone SA isolates examined, including PFGE-matched human isolates, belong to sequence type 8 (ST-8) by MLST and serotype HS:1,8, further indicating the clonality of the related isolates from different host species. Additionally, C. jejuni clone SA was identified in raw milk, cattle feces, the feces and bile of healthy sheep, and abortion cases of cattle and goats, indicating the broad distribution of this pathogenic clone in ruminants. These results provide strong molecular and epidemiological evidence for zoonotic transmission of this emergent clone from ruminants to humans and indicate that C. jejuni clone SA is an important threat to public health.
INTRODUCTION
Campylobacter jejuni is a leading cause of bacterial food-borne gastroenteritis worldwide and is a major public health problem (1, 2, 57). As reported by the Centers for Disease Control and Prevention (CDC) FoodNet surveillance program in 2009, Campylobacter ranked second (13.02 per 100,000 population) only to Salmonella (15.19 per 100,000 population) among the causes of laboratory-confirmed bacterial food-borne illnesses in the United States (9). A very recent estimate by the CDC further indicates that Campylobacter is not only among the most common causes of domestically acquired food-borne illnesses in humans (over 800,000 cases per year), but also is among the leading causes of hospitalization (over 8,000 annually) in the United States (49). Campylobacter infections in humans are usually characterized by self-limiting watery/bloody diarrhea, abdominal cramps, nausea, and fever; however, severe neurological sequelae, bacteremia, and other extraintestinal complications may develop infrequently (4, 42). Antibiotic therapy with fluoroquinolones or macrolides may be required for patients with severe or chronic infections (4).
As a zoonotic pathogen, Campylobacter is widespread in food-producing animals, and thus, foods of animal origin are frequently contaminated with the organism. The majority of Campylobacter infections in humans are sporadic and predominantly associated with poor handling of raw chicken or consumption of undercooked chicken (17, 19, 23, 24). Other risk factors include contact with house pets, exposure to farm animals, and the consumption of raw milk, untreated water, and undercooked beef, pork, and shellfish (3, 19). Outbreaks due to Campylobacter are most commonly associated with consumption of raw milk, contaminated surface water, and chicken meat (3, 24, 27, 28). Despite the prominent role of poultry in the transmission of campylobacteriosis, recent molecular epidemiological studies indicate that ruminants also contribute significantly to both sporadic cases and the outbreak of human infections via contaminated milk, water, and produce (11, 14, 30, 32, 41).
Pathogens constantly evolve in response to selective pressures, and adaptation leads to the emergence and spread of new pathogenic variants and clones (34, 58). Recently, we discovered that a highly virulent C. jejuni clone (clone SA, for sheep abortion) has emerged as the predominant cause of Campylobacter-associated abortion in sheep in the United States (48). This finding represents a paradigm shift, because diverse Campylobacter species and strains were responsible for sheep abortion in the United States prior to the predominance of clone SA (15). All isolates of clone SA were resistant to tetracycline (48), the only class of antibiotics that is approved for use in treating sheep abortion and which is commonly used for prevention and control of the disease in the United States. These findings suggest that tetracycline use might have facilitated, at least partially, the emergence and spread of clone SA. The high virulence of clone SA in causing abortion was confirmed using a pregnant-guinea-pig model, in which the distinct abortifacient ability of the clone was shown compared with other C. jejuni strains (7).
In a previous study (48), we noticed that the multilocus sequence typing (MLST) type (ST-8) of clone SA was identical to that of several human C. jejuni isolates that were in the Campylobacter MLST database (http://pubmlst.org/campylobacter/). This circumstantial evidence suggested that clone SA may also cause disease in the human host. To evaluate this possibility, we compared the pulsed-field gel electrophoresis (PFGE) profiles of clone SA with those of human C. jejuni isolates deposited in the PulseNet National Campylobacter database at the CDC (www.cdc.gov/pulsenet/pathogens_pages/campylobacter_jejuni.htm). In addition, the presence of clone SA in different types of retail meats was investigated by comparison with the PFGE database at the Food and Drug Administration (FDA) (http://www.fda.gov/cvm/default.html). Finally, we determined the prevalence of clone SA in animal reservoirs by surveying a lamb slaughterhouse.
MATERIALS AND METHODS
Sources of Campylobacter isolates for typing.
The source of C. jejuni isolates obtained from ovine abortion cases was described previously (48). Among the PFGE-matched isolates, two human isolates (FDA 17848 and FDA 17817) and two chicken isolates (FDA N337 and FDA N342) were obtained from the Center for Veterinary Medicine at the FDA for analysis, and 27 human C. jejuni isolates were obtained by the National Campylobacter and Helicobacter Reference Laboratory at the CDC from the Wyoming State Public Health Laboratory (n = 4), Colorado State Public Health Laboratory (n = 1), South Dakota State Public Health Laboratory (n = 19), and Kansas State Public Health Laboratory (n = 3) for molecular typing by MLST. Also, C. jejuni isolates associated with cattle abortions from California (n = 9; between 2003 and 2005), Iowa (n = 3; between 2006 and 2009), and North Dakota (n = 1; 2005) and goat abortions from Iowa (n = 14; between 2004 and 2010) were included for molecular typing. Additionally, 48 C. jejuni isolates from healthy sheep were analyzed by MLST and PFGE.
PFGE.
PFGE using SmaI and KpnI restriction enzymes was performed following the CDC's standardized PulseNet protocol for C. jejuni (46). The PFGE patterns of clone SA were compared with those of the human C. jejuni isolates deposited in the PulseNet National Campylobacter database at the CDC to determine the genetic relatedness between the isolates from animals and humans. The FDA's PFGE database was interrogated for possible matches to retail meat C. jejuni isolates. PFGE was also performed on the slaughterhouse isolates to determine the prevalence of clone SA in healthy animals. The gel patterns were analyzed with the BioNumerics v.4.01 software program (Applied Maths, Kortrijk, Belgium) using the Dice similarity coefficient with 0.5% optimization and 1.0 to 1.25% position tolerance. The strains were clustered by the unweighted-pair group method using average linkages (UPGMA) to reflect similarities. Isolates whose restriction profiles had a similarity coefficient of ≥90% were considered a PFGE cluster having closely related (clonal) genotypes, as described elsewhere (45).
MLST.
MLST for C. jejuni (http://pubmlst.org/campylobacter/) was performed as described elsewhere (16, 48) on representative C. jejuni isolates from different sources to further confirm the strain relatedness. Of the 123 human isolates in the PulseNet National Campylobacter database that were indistinguishable by PFGE from clone SA subtypes I and II, 27 (obtained from the four State Public Health Laboratories mentioned above) were available for MLST analysis. MLST was also performed to determine the genetic diversity among C. jejuni isolates obtained from the lamb slaughterhouse. The standardized index of association (IAs) between different MLST loci and the mean genetic diversity (H, which reflects allelic diversity across all loci and ranges from 0 to 1) were calculated using the LIAN program version 3.5 (31). If there is complete linkage equilibrium (statistical independence of alleles at all loci), the IAs is equal to zero (as it is within a freely recombining population); on the other hand, this value is significantly different from zero for populations with a clonal structure, where significant linkage disequilibrium exists (a value of 1 indicates absolute linkage disequilibrium).
The eBURST program (version 3) with the default settings (where STs within the same group share identical alleles at ≥6 of the 7 MLST loci with at least one other ST in the group) was used to depict the evolutionary relationships within all the ST-21 complex isolates, which includes ST-8, present in the Campylobacter MLST database (20).
Serotyping.
Penner heat-stable (HS) serotyping was performed using a panel of antisera, as described elsewhere (21), on four representative C. jejuni isolates of clone SA. The four isolates were IA5908 and IA3902 (from sheep abortions; subtypes I and II based on the KpnI restriction pattern, respectively), FDA 17848 (from human gastroenteritis), and 1E2B2a (from bile of healthy sheep). These C. jejuni isolates were of the same MLST genotype (ST-8) and had nearly indistinguishable PFGE patterns (shown in Fig. 1 and 2). They were chosen for serotyping because they have the same MLST sequence type (ST-8) and very close (but not identical) PFGE patterns but were isolated from diverse sources.
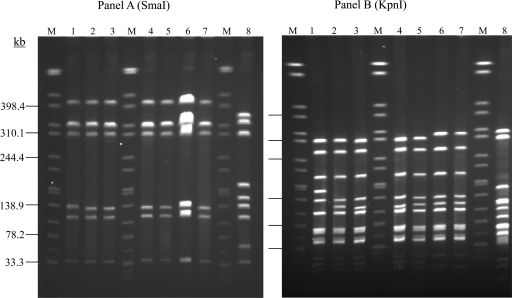
Fig 1.
PFGE analysis of C. jejuni isolates from different sources using SmaI (A) or KpnI (B). The isolates were FDA 17848 and FDA 17817 (lanes 1 and 2; from human), FDA N337 and FDA N342 (lanes 3 and 4; from chicken), clone SA subtype I (IA5908; lane 5) and subtype II (IA3902; lane 6), a goat abortion isolate (IA625; lane 7), and C. jejuni NCTC 11168 (lane 8). M, size marker (Salmonella sp. strain Braenderup H9812 genomic DNA digested with XbaI).
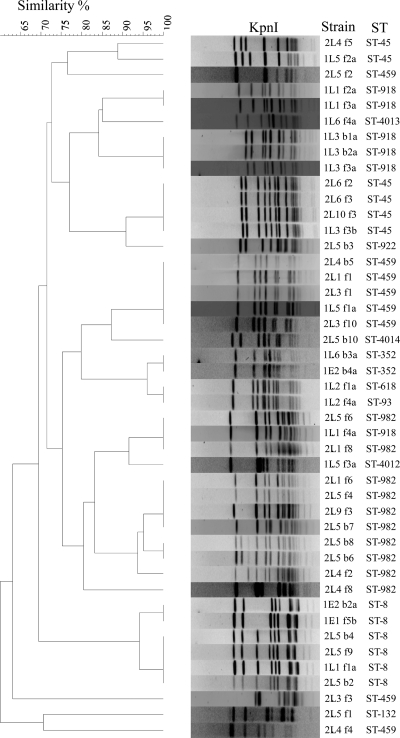
Fig 2.
PFGE-based dendrogram depicting the relatedness of 48 C. jejuni isolates from a lamb slaughterhouse. Strain names and STs are given next to the restriction profiles. The components of the strain prefixes, from left to right, are as follows: number 1 or 2, sampling occasion; E or L, ewe or lamb, respectively; lot number; B or F, bile or fecal sample, respectively; sample number; a or b, identifier of the C. jejuni colony when multiple colonies from the same sample were saved.
Distribution of clone SA in healthy sheep.
To help understand the distribution of C. jejuni clone SA in animal reservoirs, we conducted a survey to determine if the clone was present in healthy sheep. A local abattoir slaughtering lambs from multiple states (Iowa, Idaho, Kansas, Minnesota, Missouri, Nebraska, and South Dakota) was visited twice during the fall of 2008 to collect samples. On each sampling day, small-intestine contents and bile were obtained from randomly chosen carcasses soon after evisceration. Depending on the sizes of the lots, 3 to 10 specimens for each sample type were collected from different animals within the same lot (defined as a cohort of animals from a particular farm). In total, 103 intestinal contents and 124 bile samples (from 22 different lots) were obtained. Direct plating was used for isolation of Campylobacter from bile samples, while both direct plating and enrichment were employed with the intestinal contents (47, 56). Campylobacter-like colonies were selected, and a single isolate from each sample was subjected to species identification by PCR (36, 47) and molecular typing by PFGE and MLST, as described above.
Antimicrobial susceptibility testing.
A standard broth microdilution test, as recommended by the CLSI and the National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) (8, 12), was used for the antimicrobial susceptibility test on the C. jejuni isolates from the sheep slaughterhouse and was conducted using commercially available Sensititre Campylobacter plates (Trek Diagnostics, Cleveland, OH), which include nine antimicrobial agents.
RESULTS
Association of clone SA with human infections.
Based on the PFGE patterns, clone SA has a single SmaI type and two closely related KpnI subtypes, I (lanes 2,3,4 and 5) and II (lanes 6 and 7), as shown in Fig. 1 and in our previous study (48). According to the PulseNet designations for PFGE profiles, the SmaI pattern of clone SA is DBRS16.0008, while the KpnI patterns for subtypes I and II are DBRK02.0148 and DBRK02.0028, respectively. The combined pattern designation of DBRS16.0008 and DBRK02.0028 has been seen 76 times among human isolates in the PulseNet National database at the CDC, among which 42 were from sporadic infections and 34 were associated with outbreaks (no. 2, 3, 4, 5, 7, and 9) (Table 1). These cases occurred in multiple states (Arkansas, Colorado, Kansas, Michigan, Pennsylvania, Rhode Island, South Carolina, South Dakota, Utah, Vermont, Wisconsin, and Wyoming) from 2004 to 2010 (Table 1). Additionally, the combined pattern designation of DBRS16.0008 and DBRK02.0148 had exact matches to 47 human C. jejuni isolates, 14 of which were from sporadic infections (occurring in Massachusetts, South Dakota, Utah, and Wisconsin between 2004 and 2010) and 33 were associated with outbreaks (no. 1, 5, 6, 9, and 10) in the PulseNet national database at the CDC (Table 1). Two outbreaks (no. 5 and 9) had both pattern combinations associated with them. Most of the outbreak cases were related to raw milk (Table 1). Altogether, the combined SmaI and KpnI PFGE pattern designations of clone SA were indistinguishable from those of 123 human C. jejuni isolates (representing 9.03% of 1,361 human C. jejuni isolates in the CDC database), 56 of which were from sporadic infections while 67 were associated with outbreaks known in the PulseNet database through 2010. Although limited epidemiological information is available on the matches, most implicated cases were gastroenteritis, and only two had evidence of systemic (blood) infection (Table 1). In addition to these human isolates, two C. jejuni isolates with pattern designations DBRS16.0008 and DBRK02.0028 were from raw-milk samples purchased from an implicated dairy during the 2008 outbreak that occurred in Pennsylvania (Table 1). Twenty-seven of the 56 sporadic human C. jejuni isolates were available for further characterization by MLST; all 27 isolates were ST-8.
Table 1.
Human C. jejuni isolates indistinguishable by PFGE from clone SA identified in the PulseNet National Campylobacter database at the CDC through 2010
| Case | No. of isolates | State | Yr | Isolation source | Exposure |
|---|---|---|---|---|---|
| Outbreak 1 | 1 | VT | 2003 | Unknown | Raw milk |
| Outbreak 2 | 4 | SC | 2007 | Stool | Raw milk |
| Outbreak 3 | 16a | PA | 2008 | Unknown | Raw milk |
| Outbreak 4 | 4 | RI | 2008 | Unknown | Chicken |
| Outbreak 5 | 32 | WI | 2009 | Stool | Raw milk |
| Outbreak 6 | 2 | MA | 2010 | Stool/blood | Raw milk |
| Outbreak 7 | 7 | MI | 2010 | Stool | Raw milk |
| Outbreak 8 | 1 | MT | 2010 | Unknown | Well water |
| Outbreak 9 | 2 | VT | 2010 | Stool | Raw milk |
| Sporadicb | 56 | Multiple | 2004–2010 | Stool | Unknown |
Includes two raw milk isolates from the dairy implicated in the outbreak.
Sporadic cases were from multiple states (Arkansas, Colorado, Kansas, Massachusetts, Utah, Sout Dakota, Vermont, Wisconsin, and Wyoming); the isolates were mainly from stool samples (except a few from blood or unknown origin), and sources of exposure were unknown for most cases (raw milk was implicated in one case).
In total, nine C. jejuni isolates from the FDA database (which comprised over 3,000 Campylobacter isolates at the time of the database search) showed matches with clone SA, among which seven, from retail chicken meat (all from Minnesota in 2004), had a combined SmaI-KpnI restriction pattern indistinguishable from that of clone SA subtype I. The remaining two isolates were of human origin (both from Iowa) and had PFGE profiles identical or closely related to that of clone SA subtype I. To further confirm their relationship to clone SA, four of the FDA isolates, two from chickens and two from humans, were subjected to PFGE and MLST. All 4 isolates had the same SmaI restriction profile as the SA clone (Fig. 1A). With respect to the KpnI restriction pattern (Fig. 1B), both chicken isolates (lanes 3 and 4) and one of the human isolates (lane 2) were indistinguishable from clone SA subtype I (lane 5), whereas the other human isolate (lane 1) had a single-band difference from subtype I. All of the isolates had the same MLST type, ST-8.
Isolates of clone SA belong to serotype HS:1,8.
Four representative C. jejuni clone SA isolates were examined using the Penner HS scheme, which identified all of them as HS:1,8. The four isolates were IA5908 and IA3902 (both from sheep abortions), FDA 17848 (from human gastroenteritis), and 1E2B2a (from the bile of a healthy sheep). These isolates are either indistinguishable from or very closely related (a single-band difference) to the PFGE subtype of clone SA (Fig. 1 and 2), and all belong to ST-8 by MLST.
Presence of clone SA in nonsheep ruminants.
Of the 13 C. jejuni isolates associated with cattle abortions, 5 (38%) were identified as clone SA by MLST (ST-8) and PFGE, 3 of which occurred in California between 2003 and 2004, one in Iowa in 2009, and one in North Dakota in 2005 (results not shown). Out of the 14 total goat abortion isolates, 6 (43%; all from Iowa between 2004 and 2010) were identified as clone SA by PFGE (partly shown in Fig. 1) and MLST (ST-8; results not shown). In addition, there were 18 C. jejuni isolates with PFGE matches identical to those of clone SA from nonhuman sources (15 bovine and 3 ovine) in the PulseNet National Campylobacter database at the CDC from 2003 to 2010. Of these, 14 were from bovine stool samples investigated during raw-milk outbreaks that occurred in Wisconsin, Massachusetts, and Vermont in 2009 and 2010. These findings indicate that C. jejuni clone SA is broadly distributed in the ruminant reservoirs.
Presence of clone SA in healthy sheep.
Of the 103 small-intestine contents and 124 bile samples collected from a lamb slaughterhouse, Campylobacter was isolated from 45 (44%) and 14 (11%) of the samples, respectively. Most of the isolates were C. jejuni (n = 48), and the remaining ones were C. coli (n = 7); 4 Campylobacter isolates were not recovered from frozen stocks in subsequent passages. A total of 14 different STs were identified among the 48 C. jejuni isolates, indicating a substantial level of genetic diversity and revealing the presence of ST-8 (n = 6; 12.5%) in healthy sheep (Fig. 2). The ST-8 isolates were from both bile (n = 3) and intestinal contents (n = 3) collected from four different lots.
PFGE typing with KpnI on the 48 C. jejuni isolates generated 17 major PFGE types at a 90% similarity cutoff level, representing 23 unique macrorestriction profiles (Fig. 2). Notably, the six ST-8 isolates had SmaI and KpnI PFGE patterns indistinguishable from or highly similar (ca. 95% genetic similarity) to those of clone SA (Fig. 2). This finding and the MLST result indicated that C. jejuni clone SA is present in clinically healthy animals in both intestinal contents and bile.
The IAs (reflecting the degree of clonality in a population) for the entire group of C. jejuni isolates from the slaughterhouse was 0.4358 (P < 0.01), indicating the presence of significant linkage disequilibrium for the alleles from different loci. This finding, plus the relatively high level of mean genetic diversity (H = 0.7055 ± 0.0306), indicates that the C. jejuni population from the lamb slaughterhouse showed a moderate level of clonality with a low level of recombination between different strains. These values are in overall agreement with the population structure of Campylobacter from different sources (16, 29).
The vast majority of C. jejuni isolates derived from the sheep slaughterhouse (ca. 84%; n = 40) were resistant to tetracycline, with a MIC90 of >64 μg/ml (Table 2). All of the ST-8 slaughterhouse isolates (n = 6) were resistant to tetracycline (MIC > 64 μg/ml), consistent with other clone SA isolates reported previously (48). The sheep slaughterhouse isolates were generally susceptible to the rest of the antibiotics tested in this study (Table 2).
Table 2.
Antimicrobial susceptibilities of 48 C. jejuni isolates from a sheep slaughterhouse as determined by a broth microdilution test
| Antibiotic | Range tested (μg/ml) | MIC (μg/ml) |
Resistance breakpoint (μg/ml)a | No. (%) of resistant isolates | ||
|---|---|---|---|---|---|---|
| Range | 50% | 90% | ||||
| Azithromycin | 0.015–64 | 0.03–0.12 | 0.06 | 0.12 | ≥8 | 0 (0) |
| Ciprofloxacin | 0.015–64 | 0.06–16 | 0.12 | 0.12 | ≥4 | 2 (4.1) |
| Clindamycin | 0.03–16 | 0.06–1 | 0.12 | 0.25 | ≥8 | 0 (0) |
| Erythromycin | 0.03–64 | 0.25–2 | 0.5 | 1 | ≥32 | 0 (0) |
| Florfenicol | 0.03–64 | 0.5–4 | 1 | 1 | ≥16 | 0 (0) |
| Gentamicin | 0.12–32 | 0.25–2 | 0.5 | 1 | ≥8 | 0 (0) |
| Nalidixic acid | 4–64 | <4–>64 | <4 | 8 | ≥32 | 3 (6.2) |
| Telithromycin | 0.015–8 | 0.25–2 | 1 | 1 | ≥16 | 0 (0) |
| Tetracycline | 0.06–64 | 0.25–>64 | >64 | >64 | ≥16 | 40 (83.3) |
Evolution of the ST-8 genotype.
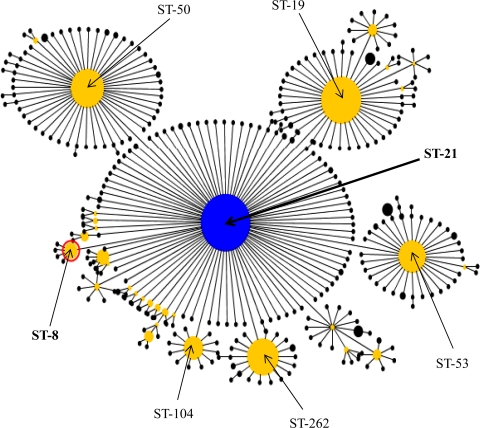
The ST-21 complex comprises C. jejuni isolates from diverse sources, such as humans, farm animals (cattle, sheep, and poultry), wild birds, and the environment, including ST-8 strains (http://pubmlst.org/campylobacter/). eBURST analysis of the entire ST-21 complex isolates present in the Campylobacter MLST database as of February 2010 was performed to provide clues to the evolutionary descent and the emergence of the ST-8 genotype, to which isolates of clone SA belong. The analysis assigned 393 STs to two eBURST-based clonal groups (STs sharing identical alleles at six or more of the seven MLST loci) and 34 STs as singletons (STs that do not share at least six out of seven loci with any other STs in the data set). The largest clonal group comprised 357 STs representing 1,144 isolates, of which the founding genotype was ST-21, and included the isolates within ST-8 (Fig. 3). The founder (ST-21) of this eBURST group differs from ST-8 at only one locus, and the only difference is a single-nucleotide polymorphism in the uncA gene (data not shown). These analyses suggest that ST-8 originated from ST-21. Also, ST-8 itself is a predicted founder of a much smaller subgroup comprising four STs that each differed from ST-8 at a single locus (Fig. 3).
Fig 3.
eBURST diagram depicting the evolutionary descent of the entire ST-21 complex in the Campylobacter MLST database. The database was accessed on 10 February 2010. The largest eBURST group with the default group definition (six of seven shared alleles), in which all STs are single locus variants of least one other ST in the group, is displayed. Each circle denotes an ST (only the major ones are shown for clarity), and the size of the circle is proportional to the number of isolates present in that genotype. Group and subgroup founders are shown as blue and yellow circles, respectively. The predicted group founder (ST-21; 99% bootstrap support) and clone SA (ST-8) are labeled.
DISCUSSION
Many pathogenic bacteria comprise a population structure in which several highly successful genetic lineages predominate for a certain time and are associated with the majority of a particular disease syndrome (6, 34). Although the Campylobacter population exhibits certain structured clonal diversity and some serotypes are associated with a particular disease outcome (e.g., Guillain-Barré syndrome), no particular genetic clones have been linked to a specific disease or severity of illness (10). However, our recent work demonstrated the emergence of the highly virulent C. jejuni clone SA associated with sheep abortion in the United States (7, 48). In the present study, we further demonstrated the association of clone SA with human disease, including multiple outbreak cases of human gastroenteritis linked mainly to raw-milk consumption (Table 1). This finding provides compelling evidence for zoonotic transmission of this pathogenic clone from animals to humans. Epidemiologically, contaminated chicken meat is recognized as a main source of human Campylobacter infections; however, the significance of the ruminant reservoir in Campylobacter transmission is increasingly recognized (13, 14, 28, 51, 59). The results from this study further highlight the importance of ruminant Campylobacter in public health and underscore the need for enhanced efforts in the surveillance and investigation of ruminant sources for better control of the zoonotic transmission of Campylobacter.
Based on MLST analysis, certain clonal complexes and STs of C. jejuni are overrepresented within a particular host species (10, 35, 38, 39, 45, 50). For example, ST-21 is frequently observed in human, ruminant, and poultry isolates (29, 35, 45), but ST-61 and ST-42 are most common among cattle and sheep isolates (22, 40, 50). A recent study of C. jejuni strains isolated from chickens, cattle, and sheep showed that ST-8 was found only among sheep and cattle populations of Campylobacter (39). Clone SA belongs to ST-8, but not all ST-8 isolates are necessarily clone SA, since a single ST type may harbor divergent genotypes, as determined by more discriminatory typing methods, such as PFGE. Considering the facts that clone SA has recently become the predominant cause of sheep abortion in the United States (48, 53) and that multiple human cases associated with clone SA have been identified in recent years (the increasing number of matches to clone SA are shown in Table 1), it is tempting to speculate that the cross-species transmission (from ruminants to humans) of C. jejuni clone SA in the United States might be a recent event. This possibility cannot be ascertained from the PulseNet database, because limited information is associated with the matched isolates. Thus, additional studies are required to assess if C. jejuni clone SA is an emerging pathogen in humans. Regardless, the results from this study clearly indicate that clone SA is a significant threat to public health, at least in the United States. At this stage, it is unknown if C. jejuni clone SA is also a significant problem for other countries. However, published data on Campylobacter-associated sheep abortions from other regions of the world (e.g., New Zealand and the United Kingdom) showed high genetic diversity and a lack of predominant clones among the isolates (37, 53), suggesting that the dominance of C. jejuni clone SA in sheep abortion might be a situation unique to the United States.
How clone SA is transmitted to humans is not entirely clear, but raw milk appeared to be the main source of infection for the outbreak cases (Table 1). Besides raw milk, ruminants can also transmit pathogenic Campylobacter to humans via other means, including direct contact and environmental contamination of water and produce (11, 26, 32, 59), which may explain the fact that clone SA was also associated with a number of sporadic cases of campylobacteriosis (Table 1). Given that red meat (beef and lamb) is rarely contaminated by Campylobacter (33), it is unlikely that it plays a significant role in the transmission of clone SA. The raw-milk-associated outbreaks of human campylobacteriosis due to clone SA (Table 1) clearly implicate cattle and milk as important sources and/or transmission vehicles for C. jejuni clone SA. Importantly, detection of clone SA in the human patients and the raw milk implicated in the outbreak (Table 1) that occurred in Pennsylvania in 2008 provides both epidemiological and genetic evidence for food-borne transmission of clone SA via milk. Additionally, identification of clone SA in bovine stool samples during several raw-milk-related outbreaks (Table 1) further highlights the significance of ruminants in the epidemiology of human illness caused by this particular C. jejuni clone. The findings in this study are consistent with previous reports that implicate Campylobacter as the most common cause of milk-borne disease outbreaks in the United States (43) and unpasteurized milk and associated dairy products as major sources for Campylobacter infections in humans (19, 32, 44).
The observation that clone SA has become a persistently predominant cause of Campylobacter-associated ovine abortions in the United States (48, 53) is quite intriguing and suggests that the clone is ecologically well adapted and pathologically hypervirulent in ruminants. Also, clone SA was detected in healthy sheep and cattle and abortion cases in cows and goats, indicating that the clone is present in the ruminants as both a commensal and a pathogen. The genetic diversity and the lack of a predominant genotype among the C. jejuni isolates from healthy sheep (Fig. 2) clearly contrast with the dominance of clone SA among the isolates obtained from clinical abortion cases, further indicating that clone SA is much more virulent than other C. jejuni strains to pregnant sheep. The identification of C. jejuni clone SA in the bile of healthy sheep suggests that it may colonize the gall bladder, which would facilitate the persistence of clone SA in sheep. At present, we do not know how prevalent clone SA is in bovine feces or how frequently it contaminates milk. These questions are important for understanding the transmission of C. jejuni clone SA and will be addressed in future studies.
To gain insights into the evolution of clone SA, eBURST analysis was performed using the MLST data on the entire set of isolates within the ST-21 clonal complex. The analysis implied that the ST-8 genotype, to which clone SA belongs, emerged from the founding genotype, ST-21, of the clonal complex (Fig. 3). Evolution of ST-8 from ST-21 was likely accompanied by acquisition of new genetic traits that may have enhanced its virulence and fitness in ruminant animals and its transmissibility to humans. It should be noted that evolutionary inferences made from MLST could be inaccurate and that further studies using high-resolution molecular analysis (e.g., genome-wide single-nucleotide polymorphism or whole-genome sequencing) should be performed to confirm the results. What has driven the selection of clone SA remains to be elucidated, but the fact that all clone SA isolates (regardless of their sources of isolation) are resistant to tetracyclines, which are commonly used for prevention and treatment of sheep abortion in the United States (25), suggests that the use of this class of antibiotics might have facilitated the emergence of the clone in sheep. Other factors, such as the use of vaccines nonhomologous to clone SA, might also have facilitated the emergence of this pathogenic clone in the sheep production systems. These possibilities remain to be examined in future studies.
Currently there is no evidence implicating clone SA as a cause of abortion in humans, but the possibility cannot be excluded, considering the high virulence of clone SA in pregnant animals and the fact that C. jejuni has been implicated in human abortions, stillbirths, and neonatal deaths (4, 5, 55). Laboratory challenge studies using pregnant guinea pigs confirmed that clone SA was highly abortifacient compared to other C. jejuni strains (7). Previously, it was reported that animal caretakers acquired C. jejuni from aborting ewes and developed enteritis (18). In addition, recent epidemiological studies have revealed a significant overlap between human and sheep Campylobacter genotypes and identified contact with sheep as an important risk factor for human infections (13, 51). In ewes that abort due to Campylobacter, a large number of organisms are present in maternal and fetal tissues/fluids, and environmental contamination occurs readily (54), which may facilitate the zoonotic transmission of the abortifacient strains to susceptible persons. Campylobacter-induced abortions in humans and sheep share similarities, including the route of infection (via the intestinal tract), pathogenesis (bacteremia), and clinical outcomes (abortion, stillbirths, and premature births), as well as the usually asymptomatic nature of infected mothers (52). Considering the highly pathogenic and zoonotic nature of clone SA, further studies are warranted to assess if this C. jejuni clone is involved in abortion in humans, in addition to causing gastroenteritis.
ACKNOWLEDGMENTS
This work was supported by the National Research Initiative Competitive Grants Program of the National Institute of Food and Agriculture at the USDA (grant no. 2010-65110-20419) and the Iowa Livestock Health Advisory Council (grant no. 109-05-14).
We thank Joyce Knutsen, Carissa Pursell, Chris Carlson, and Wanda Manley for providing PFGE data and human C. jejuni isolates for MLST characterization. We also thank all PulseNet participants who contributed PFGE data to the PulseNet National Campylobacter database and Wayne Muraoka for his assistance in analyzing the PFGE data from the sheep isolates.
Footnotes
Published ahead of print 21 December 2011
REFERENCES
- 1. Adak GK, Meakins SM, Yip H, Lopman BA, O'Brien SJ. 2005. Disease risks from foods, England and Wales, 1996–2000. Emerg. Infect. Dis. 11:365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allos BM, Moore MR, Griffin PM, Tauxe RV. 2004. Surveillance for sporadic foodborne disease in the 21st century: the FoodNet perspective. Clin. Infect. Dis. 38(Suppl.):S115–S120 [DOI] [PubMed] [Google Scholar]
- 3. Blaser MJ. 1997. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176(Suppl.):S103–S105 [DOI] [PubMed] [Google Scholar]
- 4. Blaser MJ, Engberg J. 2008. Clinical aspects of Campylobacter jejuni and Campylobacter coli Infections, p 99–121 In Nachamkin I, Szymanski CM, Blaser MJ. (ed), Campylobacter, 3rd ed ASM Press, Washington, DC [Google Scholar]
- 5. Blaser MJ, et al. 1986. Extraintestinal Campylobacter jejuni and Campylobacter coli infections: host factors and strain characteristics. J. Infect. Dis. 153:552–559 [DOI] [PubMed] [Google Scholar]
- 6. Buckee CO, et al. 2008. Role of selection in the emergence of lineages and the evolution of virulence in Neisseria meningitidis. Proc. Natl. Acad. Sci. U. S. A. 105:15082–15087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burrough ER, Sahin O, Plummer PJ, Zhang QJ, Yaeger MJ. 2009. Pathogenicity of an emergent, ovine abortifacient Campylobacter jejuni clone orally inoculated into pregnant guinea pigs. Am. J. Vet. Res. 70:1269–1276 [DOI] [PubMed] [Google Scholar]
- 8. CDC 2009. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2007. CDC, Atlanta, GA [Google Scholar]
- 9. CDC 2009. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2008. MMWR Morb. Mortal. Wkly. Rep. 58:333–337 [PubMed] [Google Scholar]
- 10. Champion OL, et al. 2005. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc. Natl. Acad. Sci. U. S. A. 102:16043–16048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clark CG, et al. 2003. Characterization of waterborne outbreak-associated Campylobacter jejuni, Walkerton, Ontario. Emerg. Infect. Dis. 9:1232–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clinical and Laboratory Standards Institute 2006. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria: approved guideline. CLSI document M45-A. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 13. Danis K, et al. 2009. Risk factors for sporadic Campylobacter infection: an all-Ireland case-control study. Euro Surveill. 14:pii: 19123 [PubMed] [Google Scholar]
- 14. de Haan CPA, Kivisto RI, Hakkinen M, Corander J, Hanninen ML. 2010. Multilocus sequence types of Finnish bovine Campylobacter jejuni isolates and their attribution to human infections. BMC Microbiol. 10:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delong WJ, Jaworski MD, Ward AC. 1996. Antigenic and restriction enzyme analysis of Campylobacter spp. associated with abortion in sheep. Am. J. Vet. Res. 57:163–167 [PubMed] [Google Scholar]
- 16. Dingle KE, et al. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doorduyn Y, et al. 2010. Risk factors for indigenous Campylobacter jejuni and Campylobacter coli infections in The Netherlands: a case-control study. Epidemiol. Infect. 138:1391–1404 [DOI] [PubMed] [Google Scholar]
- 18. Duffell SJ, Skirrow MB. 1978. Shepherd's scours and ovine Campylobacter abortion—a “new” zoonosis? Vet. Rec. 103:144. [DOI] [PubMed] [Google Scholar]
- 19. DuPont HL. 2007. The growing threat of foodborne bacterial enteropathogens of animal origin. Clin. Infect. Dis. 45:1353–1361 [DOI] [PubMed] [Google Scholar]
- 20. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fitzgerald C, et al. 2001. Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39:2386–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. French N, et al. 2005. Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7:1116–1126 [DOI] [PubMed] [Google Scholar]
- 23. Friedman CR, et al. 2004. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 38(Suppl.): S285–S296 [DOI] [PubMed] [Google Scholar]
- 24. Friedman CR, Neimann J, Wegener HC, Tauxe RV. 2000. Epidemiology of C. jejuni infections in the United States and other industrialized nations., p 121–138 In Nachamkin I, Blaser MJ. (ed), Campylobacter, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 25. Giguere S, Prescott JF, Baggot JD, Walker RD, Dowling PM. 2006. Antimicrobial therapy in veterinary medicine, 4th ed Blackwell Publishing, Ames, IA [Google Scholar]
- 26. Gilpin BJ, Scholes P, Robson B, Savill MG. 2008. The transmission of thermotolerant Campylobacter spp. to people living or working on dairy farms in New Zealand. Zoonoses Public Health 55:352–360 [DOI] [PubMed] [Google Scholar]
- 27. Gormley FJ, et al. 2011. A 17-year review of foodborne outbreaks: describing the continuing decline in England and Wales (1992–2008). Epidemiol. Infect. 139:688–699 [DOI] [PubMed] [Google Scholar]
- 28. Greig JD, Ravel A. 2009. Analysis of foodborne outbreak data reported internationally for source attribution. Int. J. Food Microbiol. 130:77–87 [DOI] [PubMed] [Google Scholar]
- 29. Habib I, Miller WG, Uyttendaele M, Houf K, De Zutter L. 2009. Clonal population structure and antimicrobial resistance of Campylobacter jejuni in chicken meat from Belgium. Appl. Environ. Microbiol. 75:4264–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hannon SJ, et al. 2009. Genomics-based molecular epidemiology of Campylobacter jejuni isolates from feedlot cattle and from people in Alberta, Canada. J. Clin. Microbiol. 47:410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haubold B, Hudson RR. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847–849 [DOI] [PubMed] [Google Scholar]
- 32. Hunt DC. 2009. Campylobacter jejuni infection associated with unpasteurized milk and cheese—Kansas, 2007. MMWR Morb. Mortal. Wkly. Rep. 57:1377–1379 [PubMed] [Google Scholar]
- 33. Jacobs-Reitsma W, Lyhs U, Wagenaar J. 2008. Campylobacter in the food supply, p 627–644 In Nachamkin I, Szymanski CM, Blaser MJ. (ed), Campylobacter, 3rd ed ASM Press, Washington, DC [Google Scholar]
- 34. Keim PS, Wagner DM. 2009. Humans and evolutionary and ecological forces shaped the phylogeography of recently emerged diseases. Nat. Rev. Microbiol. 7:813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levesque S, Frost E, Arbeit RD, Michaud S. 2008. Multilocus sequence typing of Campylobacter jejuni isolates from humans, chickens, raw milk, and environmental water in Quebec, Canada. J. Clin. Microbiol. 46:3404–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Linton D, Lawson AJ, Owen RJ, Stanley J. 1997. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35:2568–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mannering SA, West DM, Fenwick SG, Marchant RM, O'Connell K. 2006. Pulsed-field gel electrophoresis of Campylobacter jejuni sheep abortion isolates. Vet. Microbiol. 115:237–243 [DOI] [PubMed] [Google Scholar]
- 38. Manning G, et al. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370–6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McCarthy ND, et al. 2007. Host-associated genetic import in Campylobacter jejuni. Emerg. Infect. Dis. 13:267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McTavish SM, et al. 2009. Multilocus sequence typing of Campylobacter jejuni, and the correlation between clonal complex and pulsed-field gel electrophoresis macrorestriction profile. FEMS Microbiol. Lett. 298:149–156 [DOI] [PubMed] [Google Scholar]
- 41. Mullner P, et al. 2010. Molecular and spatial epidemiology of human Campylobacteriosis: source association and genotype-related risk factors. Epidemiol. Infect. 138:1372–1383 [DOI] [PubMed] [Google Scholar]
- 42. Nachamkin I, Allos BM, Ho T. 1998. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11:555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Newkirk R, Hedberg C, Bender J. 2011. Establishing a milkborne disease outbreak profile: potential food defense implications. Foodborne Pathog. Dis. 8:433–437 [DOI] [PubMed] [Google Scholar]
- 44. Olson CK, Ethelberg S, van Pelt W, Tauxe RV. 2008. Epidemiology of Campylobacter jejuni infections in industrialized nations, p 163–189 In Nachamkin I., Szymanski C. M., Blaser M. (ed), Campylobacter, 3rd ed ASM Press, Washington, DC [Google Scholar]
- 45. Ragimbeau C, Schneider F, Losch S, Even J, Mossong J. 2008. Multilocus sequence typing, pulsed-field gel electrophoresis, and fla short variable region typing of clonal complexes of Campylobacter jejuni strains of human, bovine, and poultry origins in Luxembourg. Appl. Environ. Microbiol. 74:7715–7722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ribot EM, Fitzgerald C, Kubota K, Swaminathan B, Barrett TJ. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sahin O, Kobalka P, Zhang Q. 2003. Detection and survival of Campylobacter in chicken eggs. J. Appl. Microbiol. 95:1070–1079 [DOI] [PubMed] [Google Scholar]
- 48. Sahin O, et al. 2008. Emergence of a tetracycline-resistant Campylobacter jejuni clone associated with outbreaks of ovine abortion in the United States. J. Clin. Microbiol. 46:1663–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scallan E, et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sheppard SK, et al. 2009. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int. J. Food Microbiol. 134:96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sheppard SK, et al. 2009. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 48:1072–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Simor AE, Karmali MA, Jadavji T, Roscoe M. 1986. Abortion and perinatal sepsis associated with Campylobacter infection. Rev. Infect. Dis. 8:397–402 [DOI] [PubMed] [Google Scholar]
- 53. Sippy R, et al. 2011. Genetic diversity of Campylobacter isolates associated with sheep abortion in the United States and the United Kingdom, abstr P-Z1490. Abstr. 111th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC [Google Scholar]
- 54. Skirrow MB. 1994. Diseases due to Campylobacter, Helicobacter and related bacteria. J. Comp. Pathol. 111:113–149 [DOI] [PubMed] [Google Scholar]
- 55. Smith JL. 2002. Campylobacter jejuni infection during pregnancy: long-term consequences of associated bacteremia, Guillain-Barré syndrome, and reactive arthritis. J. Food. Prot. 65:696–708 [DOI] [PubMed] [Google Scholar]
- 56. Stanley KN, Wallace JS, Currie JE, Diggle PJ, Jones K. 1998. Seasonal variation of thermophilic campylobacters in lambs at slaughter. J. Appl. Microbiol. 84:1111–1116 [DOI] [PubMed] [Google Scholar]
- 57. Tauxe RV. 1997. Emerging foodborne diseases: an evolving public health challenge. Emerg. Infect. Dis. 3:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Williams PD. 2010. Darwinian interventions: taming pathogens through evolutionary ecology. Trends Parasitol. 26:83–92 [DOI] [PubMed] [Google Scholar]
- 59. Wilson DJ, et al. 2008. Tracing the source of campylobacteriosis. PLoS Genet. 4:e1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]