Abstract
Bifidobacterium species are difficult to identify and may be underreported or not recovered by many laboratories because of their slow growth. We emphasize the importance of the Gram stain in urine samples and the addition of enriched media and enhanced atmosphere over time for urine cultures with pyuria. This is the first report of a Bifidobacterium scardovii recurrent urinary infection in an elderly woman.
CASE REPORT
An 80-year-old woman presented with dysuria, incontinence, and increased urinary frequency. On admission, the patient was afebrile with normal vital signs. She had a history of breast cancer treated with chemotherapy and radiotherapy (in remission), hypothyroidism, and autoimmune hemolytic anemia. Among other medications, treatment included methylprednisolone (8 mg daily). During this year, the patient developed several episodes of urinary tract infection (UTI).
Because of her symptoms, UTI diagnosis was made, and a new urine sample was taken. The urine analysis showed 12 leukocytes/high-power field (HPF), 1 erythrocyte/HPF, 6 epithelial cells/HPF, a specific gravity of 1.020, a pH of 6, and protein levels within normal limits. Gram stain showed Gram-positive rods, which were reported as indigenous genital bacterial flora. The urine sample was aerobically cultured on CLDE agar (Britania, Buenos Aires, Argentina) and on chromogenic agar CPS ID medium (bioMérieux, Marcy l'Etoile, France) at 35°C. As this sample was not cultured in enriched media, a new urine sample was requested to confirm the above mentioned findings. The new urine sample was received and examined 2 days later. The analysis showed 30 leukocytes/HPF, 2 epithelial cells/HPF, and the same gravity, pH, and protein levels as the previous one. Since Gram stain showed irregularly shaped Gram-positive rods which exhibited bifid forms and often curved forms (Fig. 1), 5% sheep blood agar and chocolate agar were added with incubation at 5% CO2 and anaerobically at 35°C. No growth was observed on CLDE and on the chromogenic agar in both samples. Pinpoint-sized colonies (0.1 to 0.2 mm) were observed after 72 h of incubation on 5% sheep blood agar and on chocolate agar plates, the diameter of the colonies being larger (0.5 to 0.7 mm) under anaerobic conditions. No hemolysis was seen on 5% sheep blood agar. The catalase reaction and test for lipophilia were negative (2). The growth in Mann, Rogosa, and Sharpe (MRS) broth (Difco) was good (slightly turbid) in comparison with Lactobacillus spp., which generally exhibit very good growth. To differentiate from some vancomycin-resistant Lactobacillus species, a 30-μg vancomycin disk was used. The plates were incubated under both aerobic and anaerobic atmospheres. Zones of inhibition of 28 mm and 30 mm with a clear zone around the disk were observed under both conditions. The organism was resistant to SPS disk (Oxoid, Basingstoke, United Kingdom) on a brucella agar plate after 3 days of anaerobic incubation (6).
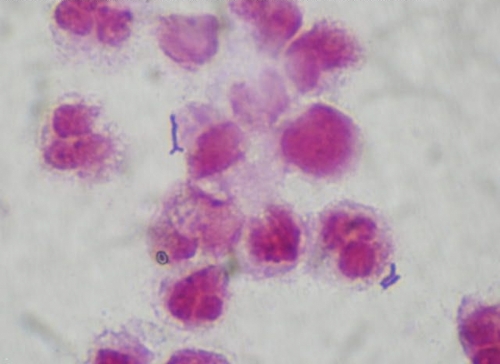
Fig 1.
Gram stain of urine showing irregularly shaped Gram-positive rods of Bifidobacterium scardovii.
Biochemical tests were performed in duplicate using API Rapid ID 32A system and API Coryne system according to the manufacturer's specifications (bioMérieux, Marcy l'Etoile, France) (3, 5). In addition, hippurate hydrolysis was examined at 4 h of incubation using Rosco Diatabs (A/S Rosco, Taastrup, Denmark). The biochemical profiles for the isolates are shown in Table 1.
Table 1.
Biochemical identification of Bifidobacterium scardovii isolates
| Test | Result fora: |
|
|---|---|---|
| Our isolates (cases 1 and 2) | Bifidobacterium scardoviib | |
| Motility | − | − |
| SPS disk | Resistant | NDc |
| Hippurated | − | − |
| API Rapid ID 32A | ||
| Nitrate reduction | − | − |
| Arginine dehydrolase | − | − |
| α-Glucosidase | + | + |
| α-Fucosidase | + | + |
| α-Galactosidase | + | + |
| β-Glucosidase | + | + |
| β-Galactosidase | + | + |
| β-Glucuronidase | − | Variable |
| N-Acetyl-β-glucosaminidase | + | Variable |
| Alkaline phosphatase | − | − |
| Proline arylamidase | − | − |
| Acid production from: | ||
| Raffinose | + | + |
| Mannose | + | + |
| API Coryne | ||
| Nitrate reduction | − | − |
| Urea | − | Variable |
| Catalase | − | − |
| Pyrrolidonyl arylamidase | + | + |
| Esculin | + | + |
| α-Glucosidase | + | − |
| β-Galactosidase | + | + |
| β-Glucuronidase | + | + |
| N-Acetyl-β-glucosaminidase | + | + |
| Alkaline phosphatase | − | − |
| Acid production from: | ||
| Glucose | + | + |
| Maltose | + | + |
| Xylose | − | − |
| Mannitol | − | − |
| Sucrose | + | + |
| Lactose | + | + |
| Glycogen | − | − |
Further identification of the isolates was performed by PCR amplification and sequence analysis of the 16S rRNA gene. The PCR product of the 16S rRNA gene, using the primers described by Weisburg et al. (8), was generated with Taq DNA polymerase based on the manufacturer's specifications (Qiagen). Sequencing of the 1.4-kb PCR product was performed on both DNA strands using an ABI Prism 3100 BioAnalyzer at the Utah State University sequencing facility. The sequences were analyzed with the BLAST V2.0 software available at http://www.ncbi.nlm.nih.gov/BLAST/. Sequence analysis revealed 99% identity with the sequences corresponding to the 16S RNA ribosomal gene of Bifidobacterium scardovii (GenBank accession no. AB437363).
Antibiotic susceptibility was determined by the Etest technique (bioMérieux, Marcy l'Etoile, France) on brucella agar supplemented with hemin (5 μg/ml), vitamin K (1 μg/ml), and 5% sheep blood, and the inoculum size was equivalent to a no. 1 McFarland standard (7). Plates were incubated in anaerobic atmosphere (Anaerocult A system) (Merck, Darmstadt, Germany) at 37°C for 48 h.
The MICs for penicillin and ciprofloxacin were 0.125 μg/ml and 0.75 μg/ml, respectively.
The patient was treated with ampicillin-sulbactam at 500 mg twice daily for 5 days, with a favorable clinical evolution. The urine culture after treatment was negative. However, she returned to the hospital 3 weeks later, complaining of urinary symptoms. A new urine sample was obtained, demonstrating 20 to 30 leukocytes/HPF. The specimen was cultured on 5% sheep blood agar and on chocolate agar and was incubated at 35°C aerobically and anaerobically for 72 h. A pure growth of more than 105 CFU/ml was observed. The Gram stain showed Gram-positive rods, which displayed the same phenotype as the previous isolates.
To determine the relatedness of the two strains, a PCR assay using degenerate oligonucleotide primers (DO-PCR) (4) was carried out. A unique pattern was observed showing that the strains were related.
Most of the species of the genus Bifidobacterium are part of the normal flora of the gastrointestinal tract and oral cavity of humans and animals and have been considered nonpathogenic for humans (1). B. scardovii has been isolated from human blood, urine, and hip specimens (3, 5); however, in some cases the clinical significance of this finding is not clear.
Bifidobacterium species are difficult to identify and may be missed in specimens by many laboratories. This may be caused, in part, by difficulties in distinguishing these organisms by phenotypic methods and because of their fastidious growth requirements. These organisms can also be misidentified as other catalase-negative, Gram-positive rods, such as Actinomyces spp., Lactobacillus spp., Alloscardovia omnicolens, and Actinobaculum schaalii (whose role in UTIs has been described), among others (5). Bifidobacterium can be differentiated from most of Lactobacillus species by simple phenotypic tests, such as its susceptibility to vancomycin and pleomorphic Gram stain morphology; however, it is difficult to differentiate bifidobacteria (Bifidobacterium spp. and A. omnicolens) from Actinomyces species by biochemical tests.
As there were many similarities in the biochemical profile to other related genera, phenotypic tests could not correctly identify the isolates. B. scardovii and A. omnicolens could be differentiated on the basis of α-fucosidase and proline-aminopeptidase tests. The growth in MRS broth was one of the main differences between bifidobacteria and Actinomyces or Actinobaculum spp., since the latter do not grow in MRS broth. However, genotypic methods were necessary to differentiate bifidobacteria from Actinomyces species and to fully identify bifidobacteria.
In a previous report of B. scardovii from human sources, three of five isolates were recovered from urine samples of elderly patients, whose clinical aspects were not mentioned (3). However, in a later report, the clinical significance of two related genera, A. omnicolens and B. scardovii, was suggested (5). In the above mentioned report, four B. scardovii isolates were included, one of them with a level of >105 CFU/ml from a urine sample. Despite the high colony count, a urinary tract infection was not documented (5).
Although the clinical significance of Bifidobacterium spp. in many cases is not clear, we were able to correlate the findings of both B. scardovii isolates with the recurrent urinary infection. These results highlighted the importance of B. scardovii as a potential pathogen in UTIs, especially for patients with predisposing factors.
This episode of recurrent urinary tract infection could be attributed to bacterial persistence or reinfection with the originally infecting strain of B. scardovii.
Bifidobacteria may be underreported or not recovered, as clinical laboratories may consider them normal flora or may not recover these organisms because of their slow growth. These are the reasons why we emphasize the importance of a urine Gram stain and the addition of enriched media, such as blood or chocolate agars incubated under anaerobic or CO2 conditions in cases of urine culture with pyuria but lack of organism recovery on routine culture media.
Nucleotide sequence accession number.
The sequence obtained for the B. scardovii 16S rRNA gene was submitted to GenBank under accession no. JN 180852.
ACKNOWLEDGMENTS
This work was supported by grants from the Secretaría de Ciencia y Técnica de la Universidad de Buenos Aires (UBACyT) to Carlos Vay.
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Felis G, Dellaglio F. 2007. Taxonomy of lactobacilli and bifidobacteria. Curr. Issues Intest. Microbiol. 8:44–61 [PubMed] [Google Scholar]
- 2. Funke G, Bernard KA. 2011. Coryneform gram-positive rods, p 413–442 In Versalovic J, et al. (ed), Manual of clinical microbiology, 10th ed, vol 1 ASM Press, Washington, DC [Google Scholar]
- 3. Hoyles L. 2002. Bifidobacterium scardovii sp. nov., from human sources. Int. J. Syst. Evol. Microbiol. 52:995–999 [DOI] [PubMed] [Google Scholar]
- 4. Limansky A, Viale A. 2002. Can composition and structural features of oligonucleotides contribute to their wide scale applicability as random PCR primers in mapping bacterial genome diversity? J. Microbiol. Methods 50:391–397 [DOI] [PubMed] [Google Scholar]
- 5. Mahlen S, Clarridge J., III 2009. Site and clinical significance of Alloscardovia omnicolens and Bifidobacterium species isolated in the clinical laboratory. J. Clin. Microbiol. 47:3289–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reimer LG, Reller LB. 1985. Use of a sodium polyanetholesulfonate disk for the identification of Gardnerella vaginalis. J. Clin. Microbiol. 21:146–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rosenblatt J, Gustafson E. 1995. Evaluation of the E-test for susceptibility testing of anaerobic bacteria. Diagn. Microbiol. Infect. Dis. 22:279–284, 1995 [DOI] [PubMed] [Google Scholar]
- 8. Weisburg WG, Barns SM, Pelletier DA, Lane LD. 1991. 16S-ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]