Abstract
Clonal isolates identified as various nonfermentative Gram-negative bacilli over a 5-year period from sputum cultures of a 30-year-old cystic fibrosis patient were successfully reidentified as Pandoraea sputorum by combining 16S rRNA sequencing and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). Decreased lung function improved after 1 year of azithromycin and inhaled 7%-hypertonic saline treatment.
CASE REPORT
A 30-year-old female cystic fibrosis (CF) patient (genotype F508del/D614G) with pancreatic sufficiency was transferred to our CF unit with a history of chronic bronchopulmonary colonization with Pseudomonas aeruginosa and Candida albicans. When the first microbiological cultures were performed in our laboratory in October 2006, Pandoraea sputorum was detected but was initially reported as nonfermentative Gram-negative bacilli (NFGNB). At this time, her pulmonary function test revealed a forced vital capacity (FVC) of 2.87 liters (85% of the predicted FVC) and a forced expiratory volume in 1 s (FEV1) of 2.22 liters (75% of that predicted). P. sputorum grew in 12 samples from October 2006 until May 2011. During this period she was treated with aerosolized colistin (2 million units twice daily with a Pari Turbo-Boy compressor with a Pari LC-Plus jet nebulizer; Pari Gräfelfing, Germany) and recombinant human DNase (2.5 mg daily). Until January 2008, she did not have any exacerbation and her pulmonary function remained stable. For the next 2 years, her clinical condition worsened and she presented 11 exacerbations requiring 22 weeks of oral antibiotics, including trimethoprim-sulfamethoxazole (160/800 mg twice daily) and 2 weeks of intravenous antibiotics, including piperacillin-tazobactam (4/0.5 g every 8 h). As a result of this, her lung function declined. In January 2010, the FVC and FEV1 were 2.23 liters (67% of the predicted value) and 1.94 liters (67% of that predicted), respectively, and oral azithromycin (500 mg every 48 h) and inhaled 7% hypertonic saline with 0.1% hyaluronic acid (Hyaneb; Praxis Pharmaceutical, Madrid, Spain) twice daily using an electronic vibrating mesh nebulizer (Pari eFlow rapid; Pari) was initiated. Ten months later, her clinical condition improved, with a significant reduction in 24-h sputum volume and in purulence and cough. During this time, the patient had only two pulmonary exacerbations and required 5 weeks of antibiotics (oral ciprofloxacin, 750 mg twice daily, and imipenem given intravenously [i.v.], 500 mg every 8 h). Her FVC and FEV1 increased, to 2.40 liters (71% of that predicted) and 1.88 liters (63% of that predicted), respectively.
Laboratory records indicated that sputum cultures during routine follow-up visits and/or patient's exacerbations until 2009 yielded different NFGNB. Conventional identification of purple colonies recovered on Burkholderia cepacia complex selective agar plates (BCSA plates; BioMériex, Marcy-L′Étoile, France) was performed using two phenotypic tests, including the Api 20NE gallery (bioMérieux) and Wider semiautomatic system (Fco. Soria-Melguizo, Madrid, Spain). The Api 20NE system offered no identification (Api code 0000443), and results from the Wider system were inconsistent; isolates were reported as NFGNB (n = 4), Achromobacter xylosoxidans (n = 3), and P. aeruginosa (n = 1). Afterwards, reassessment of bacterial identification was performed with a matrix-assisted laser desorption ionization—time of flight mass spectrometry (MALDI-TOF MS) method, using the Microflex LT mass spectrometer (Bruker Daltonics GmbH, Leipzig, Germany) with the FlexControl 3.0 and MALDI BioTyper 2.0 and 3.0 software programs.
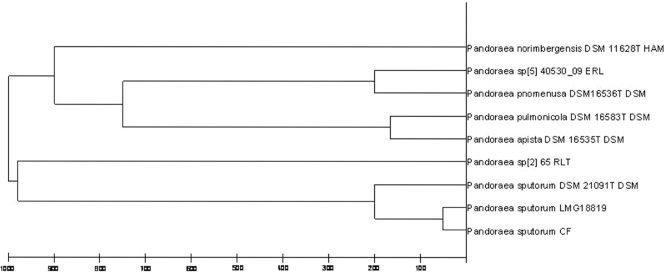
All of these isolates (n = 8) and isolates subsequently obtained from sputum samples (n = 4) (here referred to as CF Pandoraea) recovered until May 2011 were initially identified by MALDI-TOF MS as Pandoraea pnomenusa but with values indicating “secure genus identification, probable species identification” (i.e., a degree of similarity was found between the reference P. pnomenusa LMG 18817 HAM from the database and the CF Pandoraea isolates' MALDI-TOF MS spectra). A subsequently full characterization through PCR and sequencing of 16S rRNA genes was performed using the corresponding specific primers previously published (7, 14). Isolates were identified as Pandoraea sputorum in all of the cases. The length of the amplicon used for sequencing was ca. 1,500 bp, i.e., the complete gene, and the corresponding sequence homology with P. sputorum (LMG 18819 type strain) was 99% (GenBank accession no. AF139176). Consequently, the MALDI Biotyper 2.0 database (containing spectra for 3,995 microorganism) was enlarged in our laboratory to include spectra for the initial P. sputorum isolate and from the P. sputorum LMG 18819 reference type strain (BCCM/LMG bacterial collection, Gent, Belgium). All successive Pandoraea isolates recovered from our patient were identical and were identified as P. sputorum. Hierarchical cluster analysis based on MALDI-TOF MS spectrum comparison using updated MALDI Biotyper 3.0 software showed separate branches for P. pnomenusa and P. sputorum. Moreover, our P. sputorum isolates were clustered in the same branch as reference type strains (Fig. 1).
Fig 1.
Cluster analysis from MALDI-TOF MS spectra of Pandoraea isolates, including seven reference strains (MALDI Biotyper 3.0 database), one P. sputorum LMG 18819 reference type strain, and one clinical isolate (Pandoraea sputorum CF). Distance is displayed in relative units.
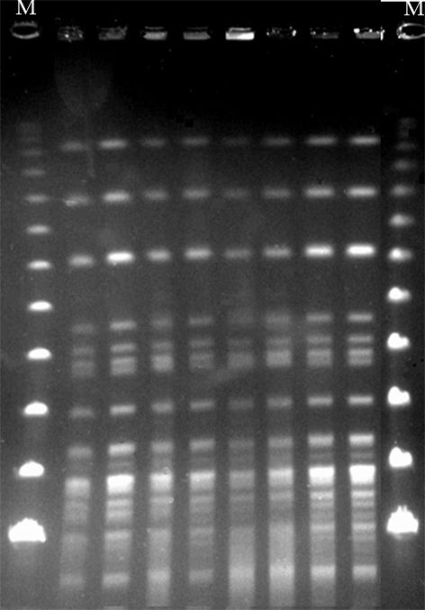
In addition, genetic relatedness was determined by XbaI pulsed-field gel electrophoresis (PFGE) analysis (13), revealing indistinguishable patterns among all isolates (Fig. 2). Susceptibility patterns (standard microdilution, CLSI) were also almost identical for all isolates. In the absence of interpretive susceptibility criteria for strains of Pandoraea, results were interpreted using CLSI criteria for “other non-Enterobacteriaceae” isolates (5). They were resistant to amoxicillin, amoxicillin-clavulanate, cefotaxime, ceftazidime, meropenem, gentamicin, tobramycin, amikacin, ciprofloxacin, colistin, and azithromycin and were susceptible to piperacillin-tazobactam (MIC ≤ 16/4 mg/liter), imipenem (MIC = 4 mg/liter), and trimethoprim-sulfamethoxazole (MIC = 2/38 mg/liter).
Fig 2.
PFGE of XbaI-digested genomic DNA of 8 selected clinical P. sputorum isolates recovered from 2006 to 2011. Lane M, bacteriophage lambda ladder PFGE marker (New England BioLabs).
Pandoraea species are considered emerging pathogens in CF (2, 9). Nevertheless, partially due to the difficulties in their recognition in routine bacteriological cultures, data from the clinical course and outcome in patients colonized/infected with this organism are still scarce. The genus Pandoraea was created in 2000 to accommodate organisms from Pseudomonas rRNA homology group II. Five species were described, including Pandoraea apista, Pandoraea norimbergensis, Pandoraea pulmonicola, P. sputorum and P. pnomenusa. They are motile, aerobic, nonfermentative Gram-negative rods able to grow on BCSA plates. When applying conventional identification phenotypic methods, the microbiology laboratory commonly misidentifies this pathogen as Ralstonia or Burkholderia species, the two genera that are phylogenetically most closely related to Pandoraea spp. (6, 11, 14). In CF, correct identification of the different pathogens is extremely important for the clinical management of the patient colonized by these organisms, which might exhibit distinct degrees of pathogenicity (4, 8).
Pandoraea has been isolated from environmental samples and occasionally recovered, but at an increasing rate, from the respiratory tract of CF patients and is considered an emerging opportunistic pathogen. Also, it has been recovered from blood cultures from non-CF patients (8, 9, 13). Within this genus, P. apista has been isolated from the lungs and blood cultures of CF patients, supporting an invasive role for this pathogen in this disease (12), unlike the case with P. aeruginosa. In addition, chronic bronchopulmonary infection with a single clone of P. apista has been associated with decreased lung function (13), and different P. apista strains have been implicated in the deterioration of lung function, although it is unclear whether this deterioration could be directly attributed to this organism (1). Interestingly, P. sputorum was persistently recovered from respiratory secretions of a CF patient under consideration for lung transplantation (14). Furthermore, cross-infection with Pandoraea spp. has been reported in winter camps and/or hospitals (13). More recently, among the Irish CF population, P. pulmonicola was the species most predominantly identified, but since all Pandoraea-colonized patients were cocolonized with other CF pathogens, it was difficult to establish clinical correlations between microorganisms and the severity of pathogenic colonization (8).
Here we report chronic bronchopulmonary colonization by a P. sputorum isolate that was successfully identified by combining a molecular technique based on 16S rRNA PCR and sequencing and a mass spectrometry method. This organism, previously identified in respiratory secretions of CF patients (6, 14), has demonstrated pathogenicity in an in vitro model with lung cells (4). P. sputorum isolates triggered a pronounced proinflammatory response, with elevation of interleukin 8 (IL-8) similar to that seen with other Pandoraea species and well-established CF pathogens, such as P. aeruginosa and Burkholderia cepacia complex (4). A lung function decline with frequent exacerbations was observed in our patient. Azithromycin and inhaled 7%-hypertonic saline were used, and patient lung function improved with no further exacerbations. As in patients chronically colonized by P. aeruginosa, the potential role of azithromycin, which might interfere with biofilm formation and exert an immunomodulatory effect, requires further investigation.
Since it is known that different Pandoraea species are capable of causing infections in CF subjects, the correct identification of these organisms is important. Pandoraea species, including P. sputorum, have never been identified by conventional methods in our laboratory, even when using a widely accepted phenotypic method, such as the Api 20NE gallery (3, 10). This fact was reinforced in a recent quality assurance trial of CF microbiology in which a P. pnomenusa strain was not even detected or was misclassified by many laboratories (11). In addition, not all the 16S rRNA PCR and sequencing schemes are able to correctly identify all the Pandoraea species (14). One factor limiting the use of MALDI-TOF MS is the scant reference data sets for microorganisms that are infrequently isolated from clinical specimens. Nevertheless, the possibility of updating the database by enlarging it with new bacterial species is valuable (2). In our CF patient, all isolates were accurately identified as P. sputorum, combining 16S rRNA PCR and sequencing and the MALDI-TOF MS proteomic platform after the extension of its reference database.
Finally, to determine persistence over time of the same P. sputorum isolate, as generally occurs with P. aeruginosa in CF patients, a molecular epidemiological analysis of P. sputorum isolates was performed. PFGE showed indistinguishable patterns among all isolates, indicating chronic colonization with a single clone rather than repeated infections (Fig. 2).
In conclusion, the presence of Pandoraea spp. can be underestimated in CF patients when using conventional identification methods. MALDI-TOF MS appears to be a promising means of accurate identification of organisms of this genus, which may have a potential role in pulmonary function deterioration when chronically colonizing CF patients.
ACKNOWLEDGMENTS
A.F.-O. is a recipient of a Rio Hortega contract (CM08/166) from the Instituto Carlos III, Ministry of Health, Spain. Part of this study was funded by the Microbial Sciences Foundation (Madrid, Spain).
Footnotes
Published ahead of print 14 December 2011
REFERENCES
- 1. Atkinson RM, LiPuma JJ, Rosenbluth DB, Dunne WM. 2006. Chronic colonization with Pandoraea apista in cystic fibrosis patients determined by repetitive-element-sequence PCR. J. Clin. Microbiol. 44:833–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bittar F, Rolain JM. 2010. Detection and accurate identification of new or emerging bacteria in cystic fibrosis patients. Clin. Microbiol. Infect. 16:809–820 [DOI] [PubMed] [Google Scholar]
- 3. Brisse S, et al. 2002. Comparative evaluation of the BD Phoenix and VITEK 2 automated instruments for identification of isolates of the Burkholderia cepacia complex. J. Clin. Microbiol. 40:1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caraher E, et al. 2008. Evaluation of in vitro virulence characteristics of the genus Pandoraea in lung epithelial cells. J. Med. Microbiol. 57:15–20 [DOI] [PubMed] [Google Scholar]
- 5. Clinical Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI document M100-S21. CLSI, Wayne, PA [Google Scholar]
- 6. Coenye T, et al. 2000. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int. J. Syst. Evol. Microbiol. 50:887–899 [DOI] [PubMed] [Google Scholar]
- 7. Coenye T, Liu LX, Vandamme P, LiPuma JJ. 2001. Identification of Pandoraea species by 16S ribosomal DNA-based PCR assays. J. Clin. Microbiol. 39:4452–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costello A, et al. 2011. Virulence of an emerging respiratory pathogen, genus Pandoraea, in vivo and its interactions with lung epithelial cells. J. Med. Microbiol. 60:289–299 [DOI] [PubMed] [Google Scholar]
- 9. Davies JC, Rubin BK. 2007. Emerging and unusual gram-negative infections in cystic fibrosis. Semin. Respir. Crit. Care Med. 28:312–321 [DOI] [PubMed] [Google Scholar]
- 10. Fernandez-Olmos A, et al. 2012. MALDI-TOF MS improves routine identification of non-fermenting Gram negative isolates from cystic fibrosis patients. J. Cyst. Fibros. 11:59–62 [DOI] [PubMed] [Google Scholar]
- 11. Hogardt M, Ulrich J, Riehn-Kopp H, Tummler B. 2009. EuroCareCF quality assessment of diagnostic microbiology of cystic fibrosis isolates. J. Clin. Microbiol. 47:3435–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson LN, et al. 2004. Pandoraea bacteremia in a cystic fibrosis patient with associated systemic illness. Pediatr. Infect. Dis. J. 23:881–882 [DOI] [PubMed] [Google Scholar]
- 13. Jorgensen IM, et al. 2003. Epidemic spread of Pandoraea apista, a new pathogen causing severe lung disease in cystic fibrosis patients. Pediatr. Pulmonol. 36:439–446 [DOI] [PubMed] [Google Scholar]
- 14. Pimentel JD, MacLeod C. 2008. Misidentification of Pandoraea sputorum isolated from sputum of a patient with cystic fibrosis and review of Pandoraea species infections in transplant patients. J. Clin. Microbiol. 46:3165–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]