Abstract
Clostridium difficile is an important nosocomial pathogen associated particularly with diarrheal disease in elderly individuals in hospitals and long-term care facilities. We examined the carriage rate of Clostridium difficile by culture as a function of fecal microbiota composition in elderly subjects recruited from the community, including outpatient, short-term respite, and long-term hospital stay subjects. The carriage rate ranged from 1.6% (n = 123) for subjects in the community, to 9.5% (n = 43) in outpatient settings, and increasing to 21% (n = 151) for patients in short- or long-term care in hospital. The dominant 072 ribotype was carried by 43% (12/28) of subjects, while the hypervirulent strain R027 (B1/NAP1/027) was isolated from 3 subjects (11%), 2 of whom displayed C. difficile associated diarrhea (CDAD) symptoms at the time of sampling. Emerging ribotypes with enhanced virulence (078 and 018) were also isolated from two asymptomatic subjects. Pyrosequencing of rRNA gene amplicons was used to determine the composition of the fecal microbiota as a surrogate for the microbial population structure of the distal intestine. Asymptomatic subjects (n = 20) from whom C. difficile was isolated showed no dramatic difference at the phylum or family taxonomic level compared to those that were culture negative (n = 252). However, in contrast, a marked reduction in microbial diversity at genus level was observed in patients who had been diagnosed with CDAD at the time of sampling and from whom C. difficile R027 was isolated.
INTRODUCTION
Changes in gut physiology and function occur during aging, and the decline in the immune system (so-called “inflammaging”) may be the result of dysbiosis of the gut microbiota (12). A number of studies have reported changes in gut microbiota composition of the elderly at both phylum and species levels (16, 39, 40). Initial studies by the ELDERMET consortium (http://eldermet.ucc.ie) compared the fecal microbiota compositions of 161 elderly subjects and 9 healthy young controls through pyrosequencing, which demonstrated that while each individual showed a distinct profile, at the phylum level the microbiota of the elderly subjects is dominated by Bacteroidetes, whereas in the younger subjects analyzed the Firmicutes phylum is dominant (7).
Antibiotic treatment has been shown to alter the gut microbiota causing severe dysbiosis, which can have a long-term health impact (20). The classic example of the protective role played by a normal gut microbiota is demonstrated by the dysbiosis of the gut microbiota that occurs after antibiotic treatment leading to C. difficile infection. After antibiotic therapy, virulent strains of C. difficile can colonize the gut, producing toxins and shedding spores, leading to a wide spectrum of illness from mild diarrhea to fulminant relapsing diarrhea and pseudomembranous colitis (PMC) (29). Current antibiotic therapy for C. difficile-associated diarrhea (CDAD) almost exclusively involves the use of the broad spectrum antibiotics vancomycin and metronidazole. Although the development of resistance to these antibiotics is infrequent, there is a trend toward decreased susceptibility, which is often manifested as increased recurrence of symptoms (5, 13, 18). In a study by Baines et al. (3), 24% of C. difficile 001 ribotypes demonstrated reduced susceptibility to metronidazole, which may raise questions regarding the use of metronidazole to treat very refractory cases (3). The increased rate of disease recurrence has driven the search for alternative treatments, and antibiotics such as rifaximin, nitazoxanide, tigecycline, and fidaxomicin (OPT-80) are currently being proposed as different therapies with various degrees of success (31). In addition, bacteriocins have been suggested as substitute or adjunct therapies to conventional antibiotics (4, 25–27).
The risk of developing CDAD varies between individuals and is thought to be dependent on host factors and the type, dose, and duration of the antibiotic treatment (37). C. difficile infection is primarily regarded as a nosocomial or health care-related disease that is most prominent among the elderly population (29). This is most likely due to a reduced immune status, an increased likelihood of antibiotic use, and more frequent hospitalization. C. difficile is the major cause of nosocomial diarrhea in adults in the developed world, being responsible for virtually all cases of PMC and up to 25% of all antibiotic-associated diarrhea (11, 22). The worldwide emergence of the hypervirulent strain R027 (B1/NAP1/027) has resulted in an increased number and severity of outbreaks of CDAD (15). Numerous studies have reported C. difficile carriage from both healthy and diseased adults, ranging from 2% for asymptomatic healthy adults in the community to 10 to 51% of patients in hospitals or long-term care facilities (10, 28, 30, 38).
The ELDERMET consortium (http://eldermet.ucc.ie) is using culture-independent methods to determine the composition of the fecal microbiota from 500 subjects aged 65 years and older, and a baseline microbiota composition survey for several hundred subjects has been published recently (7–9). In this phase of the project, we analyzed fecal samples for culturable C. difficile to determine the carriage rate among healthy elderly subjects and those attending outpatient clinics in hospitals, short-term inpatient respite care, or long-stay patients. While the C. difficile isolates were characterized in terms of ribotype and potential to produce toxin, the main focus of the study was to investigate whether carriage of C. difficile was related to the composition of the fecal microbiota of elderly subjects. In addition, the fecal microbiota of two subjects who had active CDAD at the time of sampling were also investigated.
MATERIALS AND METHODS
Subject recruitment and sample collection.
The present study was approved by the Clinical Research Ethics Committee of the Cork Teaching Hospitals. Subjects aged 65 years and older were recruited and clinically investigated in two local hospitals. Exclusion criteria included a history of alcohol abuse, participation in an investigational drug evaluation within the previous 30 days, or advanced organic disease (.i.e., patients who have end-stage renal failure, end-stage pulmonary disease, life-threatening tumors, or anything deemed terminal). Informed written consent was obtained from all subjects and, in cases of cognitive impairment, next of kin in accordance with the local research ethics committee guidelines. The data collected included clinical history and status and medication history, including antibiotic usage. Some C. difficile culture-positive subjects were on a variety of antibiotics in the 4 weeks prior to sampling; these antibiotics are listed in Table 1.
Table 1.
Summary data for subjects from whom C. difficile was isolated and classification of isolates regarding ribotype and the presence of a pathogenicity locus
| Subject | Sex | Age (yr) | Ribotype | Stratificationa | Antibiotic usage within 4 weeks prior to sampling | tcdA/tcdB |
|---|---|---|---|---|---|---|
| EM011 | F | 86 | 027 | D | No antibiotic usage recorded | +/+ |
| EM012 | F | 85 | 002 | D | Unknown | +/+ |
| EM045 | M | 67 | 072 | D | No antibiotic usage recorded | +/+ |
| EM069 | M | 83 | 072 | R | No antibiotic usage recorded | +/+ |
| EM112 | F | 84 | 027 | LS | Amoxicillin, tazocin, flagyl, ciprofloxacin | +/+ |
| EM113 | M | 75 | 072 | R | Penicillin, moxifloxacin | +/+ |
| EM124 | M | 79 | 072 | LS | Ciproxin, augmentin | +/+ |
| EM126 | M | 75 | 072 | LS | No antibiotic usage recorded | +/+ |
| EM136 | M | 73 | 014 | LS | No antibiotic usage recorded | +/+ |
| EM139 | F | 69 | 072 | LS | No antibiotic usage recorded | +/+ |
| EM140 | F | 78 | 072 | LS | No antibiotic usage recorded | +/+ |
| EM146 | F | 88 | 308 | LS | No antibiotic usage recorded | +/+ |
| EM148 | M | 71 | 072 | R | No antibiotic usage recorded | +/+ |
| EM149 | F | 82 | 010 | C | Flucloxacillin | –/– |
| EM152 | F | 87 | 050 | R | No antibiotic usage recorded | +/+ |
| EM156 | M | 74 | 220 | LS | No antibiotic usage recorded | +/+ |
| EM186 | F | 87 | 308 | LS | Trimethoprim, augmentin | +/+ |
| EM188 | F | 96 | 018 | LS | Augmentin | +/+ |
| EM218 | F | 90 | 072 | LS | Cephradine, co-amoxiclav | +/+ |
| EM243 | F | 81 | 072 | LS | No antibiotic usage recorded | +/+ |
| EM255 | F | 91 | 072 | LS | Augmentin | +/+ |
| EM304 | F | 77 | 027 | R | Klacid, clindamycin, flucoxacillin, penicillin, metronidazole, trimethoprim, ciprofloxacin, vancomycin, co-amoxiclav, tazocin | +/+ |
| EM306 | F | 87 | 027 | R | Vancomycin, flagyl | +/+ |
| EM308 | M | 82 | 078 | R | No antibiotics | +/+ |
| EM321 | F | 84 | 026 | R | No antibiotics | –/– |
| EM324 | F | 81 | 216 | R | Co-amoxiclav | +/+ |
| EM334 | M | 83 | 216 | R | No antibiotics | +/+ |
| EM336 | F | 70 | 216 | C | Clarithromycin, co-amoxiclav | +/+ |
D, subjects in hospital as day patients; R, subjects in rehabilitation, i.e., less than 6 weeks as an in-patient; LS, long stay,i.e., subjects more than 6 weeks as an in-patient; C, community-dwelling subjects.
Isolation of C. difficile.
Samples were collected from subjects and stored at refrigeration temperatures prior to delivery on ice to the laboratory within 24 h of sampling; at the laboratory they were frozen immediately at −20°C before ethanol shocking and plating on cycloserine cefoxitin egg yolk agar (Lab M, Bury, United Kingdom) as described previously (10). Presumptive C. difficile isolates, i.e., Gram-positive anaerobic spore-forming rod-shaped bacteria with typical “horse-stable” odor, nonhemolytic, lecithinase negative, and l-proline aminopeptidase positive, which fluoresced under long wave UV light, were subsequently confirmed as C. difficile by 16S ribosomal DNA sequencing using the primer pairs and PCR cycling conditions as described by Simpson et al. (31a). Strains were maintained on Microbank beads (Pro-Lab Diagnostics, Ontario, Canada) at −80°C. For routine use, strains were subcultured onto Fastidious Anaerobic Agar (Lab M) containing 7.5% defibrinated horse blood and grown anaerobically in a Don Whitley anaerobic chamber at 37°C.
Bacterial strains used.
C. difficile VPI 10463 (ATCC 43255; A+/B+), C. difficile CUG 20309 (A−/B+), and C. difficile ATCC 43593 (A−/B−) were used as positive and negative controls for toxin production in vitro and for the presence of tdcA and tcdB genes.
PCR ribotyping.
PCR ribotyping for the present study was carried out by the Microbiology Department, Leeds Teaching Hospital NHS Trust, Old Medical School, Leeds General Infirmary, Leeds, West Yorkshire, United Kingdom.
Toxin testing.
Toxin production, in vitro, was assessed from culture supernatants of strains grown in reinforced Clostridium medium (Merck) for up to 48 h using the commercial kits “Toxin A+/B+: ProSpecT II” (Remel, Lenexa, KS) and “Toxin A+: ImmunoCard Stat! Clostridium difficile Toxin A” (Meridian Bioscience, Inc., Cincinnati, OH). Assays were carried out according to the manufacturers' instructions.
Detection of tcdA and tcdB by PCR.
C. difficile toxin genes tcdA and tcdB were amplified from the isolates as described by Terhes et al. (35) with minor modifications. The DNA was extracted using 5% Chelex 100 as follows. Five to six colonies from an overnight culture of C. difficile grown on fastidious anaerobic agar containing 7% defibrinated horse blood were resuspended in 200 μl of 5% Chelex 100 (Sigma), heated for 30 min at 56°C, and subsequently boiled for 8 min before centrifugation at 13,000 rpm for 3 min.
Sequencing and bioinformatics.
A total of 274 fecal samples were collected, 22 from C. difficile culture-positive subjects and 252 from C. difficile culture-negative subjects. Pyrosequencing yielded, on average, 28,000 reads per sample. DNA was extracted from fecal samples according to standard protocol (Qiagen, West Sussex, United Kingdom). Universal 16S primers, designed to amplify from highly conserved regions corresponding to those flanking the V4 region, i.e., the forward primer F1 (5′-AYTGGGYDTAAAGNG) and a combination of four reverse primers—R1 (5′-TACCRGGGTHTCTAATCC), R2 (5′-TACCAGAGTATCTAATTC), R3 (5′-CTACDSRGGTMTCTAATC), and R4 (5′-TACNVGGGTATCTAATC) (Ribosomal Database Project [RDP] Pyrosequencing Pipeline [http://pyro.cme.msu.edu/pyro/help.jsp])—were used for Taq-based PCR amplification. Subsequently, the V4 region of the 16S rRNA gene was amplified and sequenced on a 454 Genome Sequencer FLX platform (Roche Diagnostics) according to 454 protocols as previously described (8). Raw sequencing reads were quality trimmed using a locally installed version of the RDP Pyrosequencing Pipeline, applying the following criteria: (i) exact matches to primer sequences and barcode tags, no ambiguous bases (N′s) and (ii) read lengths no shorter than 150 bp. Trimmed FASTA sequences were then BLASTed (1) against a previously published 16S rRNA-specific database (36) using default parameters. The resulting BLAST output was parsed using MEGAN (19). MEGAN is tool for taxonomic classification based on BLAST output. Although not a true phylogenetic representation (branch lengths are not illustrative of evolutionary time), the MEGAN graphic exploits a tree-like representation to illustrate the taxonomical spread of samples in a hierarchical manner (i.e., phylum > class > order > family > genus > species). MEGAN assigns reads to National Center for Biotechnology Information taxonomies by using the lowest-common-ancestor algorithm. Bit scores were used from within MEGAN for filtering the results prior to tree construction and summarization. A bit-score of 86 was selected as previously used for 16S ribosomal sequence data (36). Phylum and family counts for each subject were extracted from MEGAN. Each subject's DNA was individually sequenced and analyzed; the resulting data were subsequently pooled for further examination.
Statistical methods.
Statistical analyses were carried out using Minitab Release 15.1.1.0 (Minitab, Inc.). P values were calculated using the nonparametric Kruskal-Wallis test, and statistical significance was accepted at P < 0.05.
RESULTS
Characterization of C. difficile isolates.
In the present study, fecal samples of patients, who were recruited primarily as part of the ELDERMET Consortium, were analyzed to determine whether carriage of C. difficile impacted on the gut microbiota. Carriage of C. difficile was determined through standard culturing techniques, and subsequently the gut microbiota of culture-positive (n = 22) and culture-negative subjects were sequenced using 454 sequencing to determine the impact, if any, of C. difficile carriage on the gut microbiota.
Subjects recruited from the community were classified as healthy (n = 123), subjects attending outpatient clinics/day wards in hospitals as outpatients (n = 43), those with 6 weeks hospitalization or less were classified as in rehabilitation (n = 48), and those who had spent greater than 6 weeks hospitalized in geriatric units were termed long-stay subjects (n = 103). C. difficile was isolated from 28 subjects (9%). Table 1 summarizes the data obtained here on the subjects who were culture positive for C. difficile carriage with regard to (i) location of the subject, sex, age, and previous antibiotic usage and (ii) the isolates, i.e., ribotype and the presence of pathogenicity locus. Two subjects (EM304 and EM306) were clinically diagnosed with CDAD at the time of sampling and two subjects. (EM124 and EM186) had been clinically diagnosed as suffering from CDAD for ca. 9 months previously all other culture-positive subjects had no history of CDAD. The carriage rates for community, outpatients, rehabilitation, and long-stay patients were 1.6, 9.5, 8, and 13%, respectively. Although the most dominant ribotype isolated from asymptomatic patients was 072, R027 was isolated from three subjects, two of whom were diagnosed with CDAD at the time of sampling. Both symptomatic subjects had a history of metronidazole and vancomycin usage within the 4 weeks prior to sampling (Table 1).
Sequence-based fecal microbiota composition from C. difficile culture-positive and culture-negative subjects.
The V4 region of the 16S rRNA gene was amplified and sequenced from DNA isolated from the stool samples of 22 C. difficile culture-positive subjects, and the microbiota composition established bioinformatically was compared to that of 252 subjects from whom C. difficile was not detected by culture. The phylogenetic profile of each subject was elucidated using a combination of BLAST and MEGAN algorithms, and resulting taxa were compared at the phylum, family, and genus levels. At the phylum level, the only statistically significant difference was a reduction in the proportion of Spirochaetes in the C. difficile-positive population (P = 0.017). All other phyla remained at approximately the same abundance across the populations, with the exception of the Proteobacteria, which was elevated from 2.3% in the C. difficile-negative population to 4.3% in the C. difficile-positive population.
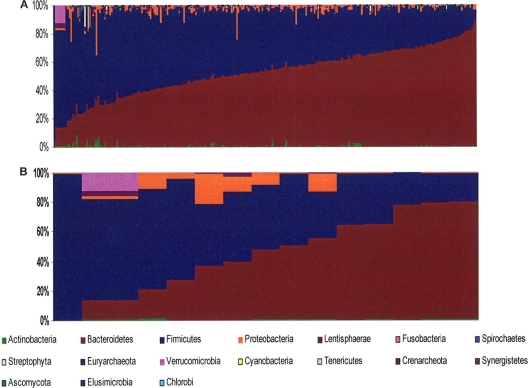
There was large variability among the subjects in the individual phylogenetic profile, in both the C. difficile-negative and the C. difficile-positive groups (Fig. 1). At the phylum level, Bacteroidetes ranged from 4 to 92% of the total microbiota in the culture-negative group and from 0.1 to 79% in the positive group. The Firmicutes ranged from 8 to 96% in the negative group and from 19 to 99% in the positive group. The Actinobacteria ranged from 0 to 8% in the negative group and from 0 to 1.2% in the positive group.
Fig 1.
Bar chart of interindividual variability of C. difficile-negative (A) and C. difficile-positive (B) subjects. The color-coding is defined at the bottom of the figure.
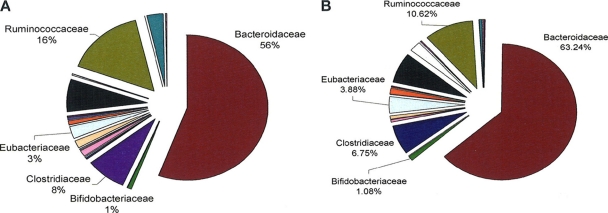
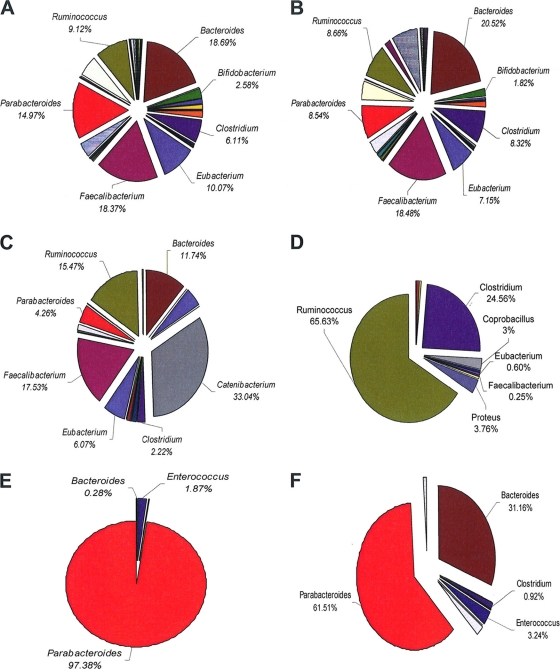
At the family level, some significant differences in the compositional data were observed (Fig. 2). The families Erysipelotrichaceae (P = 0.011), Aerococaccae (P = 0.000) and Flavobacteriaceae (P = 0.032) were elevated in the C. difficile-positive population compared to the culture-negative group. However, Flavobacteriaceae and Aerococaccae were only present in one subject each in the culture-positive group and in none of the culture-negative group. Conversely, the levels of Enterococcaceae (P = 0.009), Leuconostocaceae (P = 0.026), Prevotellaceae (P = 0.018), and Spirochaetaceae (P = 0.045) were reduced in the culture-positive population compared to the C. difficile culture-negative population. At the genus level, significant differences were observed only in genera that were present in low numbers of subjects; these differences are listed in Table 2. In all, the fecal microbiota of 22 subjects from whom C. difficile was isolated was determined by amplicon sequencing. Eighteen of these were asymptomatic carriers, two subjects had a previous episode of CDAD recorded but were asymptomatic at the time of sampling, and two subjects had active CDAD at the time of sampling. We analyzed the microbiota compositional differences at the genus level between the C. difficile culture-negative subjects (Fig. 3A), asymptomatic carriers of C. difficile (Fig. 3B), the two subjects with a previous episode of CDAD recorded (Fig. 3C and D), and the two subjects with active CDAD at time of sampling (Fig. 3E and F). The most obvious difference between the microbiota of these subjects was the dramatic alterations identified at the genus level in the subjects with active CDAD. The dominant genus in the subjects with active CDAD is Parabacteroides (97.4 and 61.5% compared to 14.97 and 8.5% for the C. difficile culture-negative group and asymptomatic carriers, respectively). However, in subjects with a history of CDAD, the dominant species were either Catenibacterium (33.0%) or Ruminococcus (65.6%). These genera were present at <2% (Catenibacterium) and <10% (Ruminococcus) in the C. difficile culture-negative or asymptomatic carrier groups. In addition, no Bifidobacteria spp. were detected in subjects with current or a history of CDAD, but they were present in the asymptomatic carriers (1.8%) and the C. difficile culture-negative group (2.6%). Where C. difficile was isolated from the same subject at 3 and 6 months, there appeared to be little alteration in the microbiota composition over time (see Fig. S1 in the supplemental material), which indicates the stability of the gut microbiota of these individuals. This observation was apparent at the phylum (see Fig. S1 in the supplemental material), family, and genus levels (data not shown).
Fig 2.
Family level assignments of gut communities of C. difficile culture-positive subjects (A [n = 22]) compared to C. difficile culture-negative controls (B [n = 252]) expressed as a percentage of assignable tags.
Table 2.
Significant differences in the abundance of genera between C. difficile culture-positive and culture-negative populations
| Genus | Significance (P) | Change in abundance in C. difficile-positive population |
|---|---|---|
| Aerococcus | 0.001 | Increase |
| Anaerococcus | 0.028 | Increase |
| Enterococcus | 0.002 | Decrease |
| Helcococcus | 0.029 | Increase |
| Leuconostoc | 0.009 | Decrease |
| Weeksella | 0.001 | Increase |
Fig 3.
Genus level assignments of gut communities in 252 C. difficile-negative subjects (A), 18 asymptomatic carriers of C. difficile (B), individual subjects with previous documented incidence of C. difficile infection (C and D), and individual subjects with active C. difficile infection at the time of sampling (E and F).
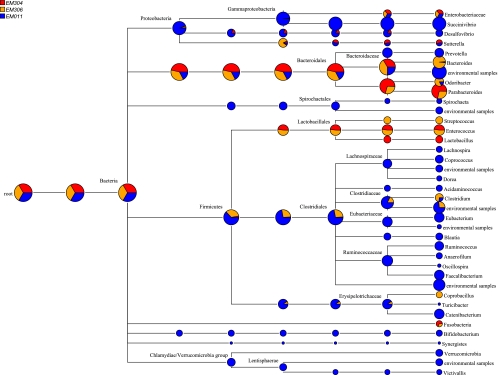
C. difficile R027 was detected by culture in just three subjects, two of whom had active CDAD at the time of sampling and one of whom was an asymptomatic carrier of C. difficile. A comparative MEGAN illustration of the overall gut microbiota of these three subjects was developed in which the asymptomatic carrier of ribotype 027 is depicted in blue and subjects with active C. difficile subjects are depicted in red and orange (Fig. 4). There are obvious differences, in that the subjects with active CDAD have a reduced overall number of assigned taxa compared to the asymptomatic carrier of R027. In the Bacteroides phyla, the active CDAD subjects harbor Parabacteroides as the dominant genus, whereas lactobacilli are only present in the active C. difficile subjects, and these subjects also have increased Enterobacteriaceae populations. Members of the Lachnospiraceae, Succinivibrionaceae, Ruminococcaceae, Synergistaceae, Bifidobacteriaceae, and Victivallaceae families are only present in the asymptomatic carrier group.
Fig 4.
MEGAN comparison of three (EM011, EM304, and EM306) subjects harboring C. difficile strain R027. This MEGAN illustration provides a comparison view in which each node shows the number of reads assigned to it for each of the data sets. Pie charts at each node display the relative abundance for each taxonomic level. Reads are assigned according to BLAST assignments. EM304 and EM306 were diagnosed with CDAD at the time of sampling. EM3011 was an asymptomatic carrier with no history of CDAD.
DISCUSSION
The study outlined here examined C. difficile carriage in the elderly population in the Cork region of Ireland, based upon mixed urban-suburban-country town catchment areas. Although the main focus of the present study was to determine the effect of carriage of C. difficile among the elderly Irish population, preliminary characterization of the isolates was carried out to determine ribotype and toxin production capability. Although five ribotypes (027, 078, 018, 014, and 026) associated with CDAD outbreaks (17) were isolated from the largely asymptomatic population of elderly people, of particular interest here was the isolation of the hypervirulent ribotype 027 from three subjects, one of whom was a day hospital patient and asymptomatic at the time of sampling. Ribotype 027 has come to prominence since the beginning of this millennium and has been isolated in at least 16 European countries (21), its increased virulence being associated with increased toxin and spore production (6, 13, 23).
The human gut microbiota is a complex ecosystem that encompasses an estimated 15,000 to 36,000 different species living in a mutualistic relationship with each other and with the human host. Although traditional culturing techniques identify only a small proportion of the total intestinal microbiota (<25%) (33), recent advances in sophisticated culture independent techniques allow us to make a more global assessment of the composition of the intestinal microbiota. The emerging method of choice to study the complexity of the human microbiome is high-throughput amplicon sequencing. This high-throughput method is not, however, without its limitations; a previous study by this group (24) demonstrated both the strengths and the limitations of both culture-independent methods, such as the one implemented here, and culture-dependent methods in-depth. These limitations include the inability of amplicon sequencing to identify populations present in an environment below levels of 106 and the inability to assign data to the species level.
A study by Andersson et al. (2) of healthy adults showed that in six fecal samples the phylum Firmicutes was predominant (81%), followed by Actinobacteria (14.6%), Bacteroidetes (2.5%), and Proteobacteria 1.7%. The dominant class among the Firmicutes was Clostridium (92%), and the genera Ruminococcus, Clostridium, and Eubacterium were also well represented (2). Similarly, a study by Tap et al. of 17 healthy adults showed a similar phylum distribution (34) More recently, a study by Claesson et al. (7) showed that the dominant phylum of nine healthy subjects between 28 and 46 years old was also Firmicutes (51%), while in contrast Bacteroidetes (57%) were predominant in 161 elderly subjects and the Firmicutes phylum was reduced to 40% of the total number of reads assigned.
Although there were no dramatic differences between the C. difficile culture-positive and culture-negative groups at the phylum level, there was a dramatic interindividual variation in the gut microbiota of these elderly subjects that has been reported previously for ELDERMET subjects (7). While some significant differences were observed at family level between the culture-positive and -negative subjects, these groups comprised a very small percentage of the total families identified (0.03 to 0.05% of the total population). Therefore, the isolation of ribotypes from asymptomatic subjects that can produce TcdA and TcdB suggests that the commensal flora in such subjects can protect the host by preventing potentially pathogenic C. difficile from overcoming colonization resistance, proliferating within the colon, and producing TcdA and TcdB. Antibiotic therapy is generally considered a risk factor for the development of CDAD; however, 50% of the culture-positive but asymptomatic subjects had a history of broad-spectrum antibiotic usage within the previous 4 weeks (Table 1). Nevertheless, no usage of clindamycin, which is frequently associated with development of CDAD, was recorded by these subjects. Broad-spectrum antibiotic therapy not only targets the pathogenic bacteria but also impacts on the wider microbiota of the gut, leading to perturbations of the commensal flora and therefore decreasing their colonization resistance, potentially leading to the outgrowth of pathogens such as C. difficile. In the present study, ribotype 027 was isolated from three subjects two of whom had CDAD at the time of sampling. A MEGAN comparison of the bacterial populations in these three subjects shows a dramatic decrease in taxa assigned in the patients with active CDAD compared to the asymptomatic carrier of 027, their microbiota being dominated by members of the Bacteroidetes phylum and, to a lesser extent, members of the Lactobacillaceae family (Fig. 4). However, it must be stated that these two subjects were on antibiotics prior to sampling (Table 1), which poses the question of whether the reduced number of taxa is due to the prior antibiotic therapy leading to the proliferation of C. difficile or whether the reduction is due to the antibiotic treatment for CDAD However, the more diverse microflora in the asymptomatic subject carrying ribotype 027 who was not receiving antibiotics supports the theory that the gut microbiota may have protected this subject from developing CDAD.
A number of studies using a variety of culture-dependent and culture-independent methods have examined the microbiota composition of patients with CDAD in an effort to elucidate the impact of gut flora on CDAD. A study by Hopkins and MacFarlane (16) used cellular fatty acid profiles to identify bacteria from fecal samples to the species level. CDAD patients were characterized by decreased numbers of Bacteroides, Prevotella, and Bifidobacteria, together with increased numbers of facultative species, i.e., Clostridium and Lactobacillus spp. These results are largely in agreement with our study. We also observed decreased numbers of Bacteroides, Prevotella, and Bifidobacteria in C. difficile-positive subjects, and while our study cannot identify species at the genus level, we did observe, in agreement with the Hopkins and McFarlane study, a decrease in Lactobacillus sp. However, in contrast to the Hopkins and McFarlane study, there was an increase in Clostridium sp. in C. difficile-positive subjects. Chang et al. (6a), using culture-independent phylogenetic analysis of the 16S rRNA encoding gene sequences, compared the fecal communities of control subjects to those with initial or recurrent CDAD and demonstrated that the fecal microbiota of patients with recurrent CDAD consistently displayed a decreased phylotype richness compared to the controls. Interestingly, this reduction in species richness was not observed in patients with antibiotic-associated diarrhea not linked to C. difficile. However, in contrast to our study, 16S phylotypes related to C. difficile were predominant in the fecal microflora. The absence of reads assigned to C. difficile in our study may be explained by the antibiotic usage (metronidazole and vancomycin) by these subjects prior to sampling and also to the inability of pyrosequencing to assign reads to the species level (24). In contrast, a study by Manges et al. (21a) using rRNA microarrays compared the differences in the microbiota of control subjects to the microbiota of subjects prior to and after development of CDAD, taking a number of risk factors into account. Probe intensities for the phyla Firmicutes, Proteobacteria, and Actinobacteria were increased in CDAD patients, whereas the phylum Bacteroides decreased. However, when an adjustment was made for the use of nonsteroidal anti-inflammatory drugs and fluoroquinolones, only a small subset of organisms within the Bacteroidetes and Firmicutes phyla remained significantly and independently associated with CDAD. These researchers, however, did make an association between increased probe intensities in Lactobacillaceae and Enterococcaceae but not with Enterobacteriaceae. Our study demonstrates a decrease in Enterococcaceae but an increase in Lactobacillaceae and Enterobacteriaceae in C. difficile-positive subjects. The differences observed in our study compared to the findings of Manges et al. could be accounted for by the fact that the dominant phylum changes from Firmicutes in a younger population to Bacteroidetes in the elderly population. We also acknowledge that in the study reported here the patients with active or a history of CDAD were sampled after the disease was established and in the case of the two patients with active CDAD after the administration of the antibiotics vancomycin and metronidazole, which may contribute to the differences observed at the phylum and genus levels and to the species richness for the symptomatic versus the asymptomatic group.
The four subjects who had active or a history of C. difficile infection had no reads assigned to the genus Bifidobacterium. These data are in broad agreement with a culture-based study for patients with CDAD, wherein Bifidobacterium spp. were not detected in significant numbers. In addition, in the present study there was a trend in subjects with active or a history of CDAD to harbor lower numbers of Faecalibacterium spp. (17.53 and 0.25% in subjects with a history of CDAD and 0% in subjects with a current CDAD versus 18.48% for the asymptomatic group). The levels of Faecalibacterium prausnitzii and Bifidobacterium are reduced in patients with chronic gut inflammation, and F. prausnitzii, in particular, has been shown to exert anti-inflammatory effects on inflammatory bowel disease models of colitis (14, 32).
Studies such as ELDERMET are investigating, in depth, the gut microbiota of healthy and diseased elderly subjects, including the changes in the gut microbiota of patients with bowel disease, including CDAD. By elucidating the changes that occur in the microbiota of patients suffering from intestinal disorders, these studies may facilitate the development of prophylactic therapies for elderly patients prior to antibiotic therapy, through supplementation of their diet with pre/probiotics to encourage the growth of beneficial or protective bacteria such as Bifidobacterium spp. and F. prausnitzii. As a result, this may reduce the major dysbiosis of the gut microbiota caused by antibiotic treatment and therefore reduce the chances of susceptible patients developing CDAD.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the assistance of Mark Wilcox and Peter Parnell of Microbiology Department, Leeds Teaching Hospital NHS Trust, for performing the ribotyping of the C. difficile isolates for this study. We thank all the people who kindly donated their time, effort, and samples to this study. We are also grateful to Edel Flannery, Caitriona Guinane, Rachel Greene, Nessa Gallwey, Karen O'Donovan, and Patricia Egan for technical and clinical help and to Siobhan Cusack for Project Management.
This study was performed as part of the ELDERMET project (http://eldermet.ucc.ie) and was funded by the Government of Ireland's Department of Agriculture Fisheries and Food and the Health Research Board through the Food and Health Initiative 2007–2011. The ELDERMET consortium (http://eldermet.ucc.ie) also include principal investigators Ted Dinan, Gerald Fitzgerald, Tony Fitzgerald, Albert Flynn, Denis O'Mahony, Cillian Twomey, Douwe van Sinderen, Julian Marchesi, and physicians Michael O'Connor, Norma Harnedy, Colm Henry, and Kieran O'Connor.
Footnotes
Published ahead of print 7 December 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andersson AF, et al. 2008. Comparative analysis of human gut microbiota by bar-coded pyrosequencing. PLoS One 3:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baines SD, et al. 2008. Emergence of reduced susceptibility to metronidazole in Clostridium difficile. J. Antimicrob. Chemother. 62:1046–1052 [DOI] [PubMed] [Google Scholar]
- 4. Bartoloni A, et al. 2004. In vitro activity of nisin against clinical isolates of Clostridium difficile. J. Chemother. 16:119–121 [DOI] [PubMed] [Google Scholar]
- 5. Brazier JS, et al. 2008. Distribution and antimicrobial susceptibility patterns of Clostridium difficile PCR ribotypes in English hospitals, 2007–08. Euro Surveill. 13:1–4 [DOI] [PubMed] [Google Scholar]
- 6. Cartman ST, Heap JT, Kuehne SA, Cockayne A, Minton NP. 2010. The emergence of ‘hypervirulence’ in Clostridium difficile. Int. J. Med. Microbiol. 300:387–395 [DOI] [PubMed] [Google Scholar]
- 6a. Chang J, et al. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197:435–438 [DOI] [PubMed] [Google Scholar]
- 7. Claesson MJ, et al. 2010. Microbes and Health Sackler Colloquium: composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4586–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Claesson MJ, et al. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Claesson MJ, et al. 2010. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clayton EM, et al. 2009. The vexed relationship between Clostridium difficile and inflammatory bowel disease: an assessment of carriage in an outpatient setting among patients in remission. Am. J. Gastroenterol. 104:1162–1169 [DOI] [PubMed] [Google Scholar]
- 11. Cramer JP, Burchard GD, Lohse AW. 2008. Old dogmas and new perspectives in antibiotic-associated diarrhea. Medizinische Klinik Munich Germany 103:325–340 (In German.) [DOI] [PubMed] [Google Scholar]
- 12. Franceschi C, et al. 2000. Inflammaging: an evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 908:244–254 [DOI] [PubMed] [Google Scholar]
- 13. Freeman J, et al. 2010. The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 23:529–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friswell M, Campbell B, Rhodes J. 2010. The role of bacteria in the pathogenesis of inflammatory bowel disease. Gut Liver 4:295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gerding DN. 2010. Global epidemiology of Clostridium difficile infection in 2010. Infect. Control Hosp. Epidemiol. 31(Suppl. 1):S32–S34 [DOI] [PubMed] [Google Scholar]
- 16. Hopkins MJ, Macfarlane GT. 2002. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J. Med. Microbiol. 51:448–454 [DOI] [PubMed] [Google Scholar]
- 17. HPA 2011. Clostridium difficile ribotyping network (CDRN) for England and Northern Ireland. Health Protection Agency, London, England: http://www.hpa.org.uk/Publications/InfectiousDiseases/GastrointestinalOutbreaksAndIllnessReports/1102cdiffcdrnCDRNannualreportEW/ [Google Scholar]
- 18. Huang H, Weintraub A, Fang H, Nord CE. 2009. Antimicrobial resistance in Clostridium difficile. Int. J. Antimicrob. Agents 34:516–522 [DOI] [PubMed] [Google Scholar]
- 19. Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res. 17:377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jernberg C, Lofmark S, Edlund C, Jansson JK. 2010. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology 156:3216–3223 [DOI] [PubMed] [Google Scholar]
- 21. Kuijper EJ, et al. 2008. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill. 13:1–7 [PubMed] [Google Scholar]
- 21a. Manges AR, et al. 2010. Comparative metagenomic study of alterations due to the intestinal microbiota and risk of nosocomial Clostridium difficile-associated disease. J. Infect. Dis. 202:1877–1884 [DOI] [PubMed] [Google Scholar]
- 22. McFarland LV, Mulligan ME, Kwok RY, Stamm WE. 1989. Nosocomial acquisition of Clostridium difficile infection. N. Engl. J. Med. 320:204–210 [DOI] [PubMed] [Google Scholar]
- 23. Merrigan M, et al. 2010. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J. Bacteriol. 192:4904–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Sullivan O, et al. Correlation of rRNA gene amplicon pyrosequencing and bacterial culture for microbial compositional analysis of faecal samples from elderly Irish subjects. J. Appl. Microbiol. 111:467–473 [DOI] [PubMed] [Google Scholar]
- 25. Rea MC, et al. 2007. Antimicrobial activity of lacticin 3,147 against clinical Clostridium difficile strains. J. Med. Microbiol. 56:940–946 [DOI] [PubMed] [Google Scholar]
- 26. Rea MC, et al. 2010. Microbes and Health Sackler Colloquium: effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4639–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rea MC, et al. 2010. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. U. S. A. 107:9352–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Riggs MM, et al. 2007. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin. Infect. Dis. 45:992–998 [DOI] [PubMed] [Google Scholar]
- 29. Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 30. Ryan J, et al. 2010. Asymptomatic carriage of Clostridium difficile in an Irish continuing care institution for the elderly: prevalence and characteristics. Irish J. Med. Sci. 179:245–250 [DOI] [PubMed] [Google Scholar]
- 31. Shah D, et al. Clostridium difficile infection: update on emerging antibiotic treatment options and antibiotic resistance. Expert Rev. Anti-Infective Ther. 8:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a. Simpson PJ, Stanton C, Fitzgerald GF, Ross RP. 2003. Genomic diversity and relatedness of bifidobacteria isolated from a porcine cecum. J. Bacteriol. 185:2571–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sokol H, et al. 2006. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm. Bowel Dis. 12:106–111 [DOI] [PubMed] [Google Scholar]
- 33. Suau A, et al. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Applied Environ. Microbiol. 65:4799–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tap J, et al. 2009. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11:2574–2584 [DOI] [PubMed] [Google Scholar]
- 35. Terhes G, Urban E, Soki J, Hamid KA, Nagy E. 2004. Community-acquired Clostridium difficile diarrhea caused by binary toxin, toxin A, and toxin B gene-positive isolates in Hungary. J. Clin. Microbiol. 42:4316–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Urich T, et al. 2008. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walk ST, Young VB. 2008. Emerging insights into antibiotic-associated diarrhea and Clostridium difficile infection through the lens of microbial ecology. Interdiscip. Perspect. Infect. Dis. 2008:125081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wistrom J, et al. 2001. Frequency of antibiotic-associated diarrhoea in 2462 antibiotic-treated hospitalized patients: a prospective study. J. Antimicrob. Chemother. 47:43–50 [DOI] [PubMed] [Google Scholar]
- 39. Woodmansey EJ. 2007. Intestinal bacteria and ageing. J. Appl. Microbiol. 102:1178–1186 [DOI] [PubMed] [Google Scholar]
- 40. Woodmansey EJ, McMurdo ME, Macfarlane GT, Macfarlane S. 2004. Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Appl. Environ. Microbiol. 70:6113–6122 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.