Abstract
Babesiosis is an emerging zoonosis with important public health implications, as the incidence of the disease has risen dramatically over the past decade. Because the current gold standard for detection of Babesia is microscopic examination of blood smears, accurate identification requires trained personnel. Species in the genus cannot be distinguished microscopically, and Babesia can also be confused with the early trophozoite stage (ring forms) of Plasmodium parasites. To allow more accurate diagnosis in a format that is accessible to a wider variety of laboratories, we developed a real-time PCR assay targeting the 18S rRNA gene of Babesia microti, the dominant babesiosis pathogen in the United States. The real-time PCR is performed on DNA extracted from whole-blood specimens and detects Babesia microti with a limit of detection of ∼100 gene copies in 5 μl of blood. The real-time PCR assay was shown to be 100% specific when tested against a panel of 24 organisms consisting of Babesia microti, other Babesia species, Plasmodium species, tick-borne and other pathogenic bacteria, and other blood-borne parasites. The results using clinical specimens show that the assay can detect infections of lower parasitemia than can be detected by microscopic examination. This method is therefore a rapid, sensitive, and accurate method for detection of Babesia microti in patient specimens.
INTRODUCTION
Babesiosis is a disease caused by infection with intraerythrocytic parasites of the genus Babesia. Typical presentation includes fever, chills, and body aches, although symptoms can be variable. Babesiosis may be asymptomatic in healthy, immunocompetent persons. However, severe disease, including hemolytic anemia, thrombocytopenia, organ failure, and even death, may occur in immunodeficient individuals. Particularly at risk are the elderly, asplenic individuals, and those with coincident Lyme disease (7, 11). Although babesiosis usually resolves in 2 to 3 weeks in immunocompetent individuals, there are reports of individuals carrying a low-grade asymptomatic infection for more than 2 years (11a).
Babesia parasites are transmitted to humans and animals by ixodid ticks. In the United States the primary tick vector for Babesia microti is Ixodes scapularis. Ixodid ticks, which also transmit Borrelia burgdorferi and Anaplasma phagocytophilum, take blood meals from vertebrate reservoir hosts such as white-footed mice and white-tailed deer. Humans are inadvertent hosts and can become infected when ixodid ticks feed on them. Since the first report of human infection in the United States in 1966 (13), babesiosis has been considered an emerging infectious disease, with increasing numbers of cases occurring annually; in many areas, incidence has increased 4- to 20-fold in the last decade (15). In addition to tick-associated infections, rarer cases of transmission during pregnancy or via blood transfusion have also been reported. Transmission by transfusion is of particular concern, since recipients are likely to be in suboptimal health and less able to mount an efficient immune response: this could lead to rapid development of high parasitemia and difficulty clearing the infection. Because babesiosis may be asymptomatic, blood donors may not realize that they are infected, which poses a risk to the blood supply in areas of high endemicity. Between 1979 and 2009 over 159 transfusion-related cases of Babesia infection, including nine deaths, have been documented (4, 5, 8). The Centers for Disease Control and Prevention (CDC) responded to the increased public health threat by making babesiosis a nationally notifiable disease as of January 2011.
Most U.S. cases of babesiosis occur in the Northeast, where the range has extended from coastal regions to areas such as inland counties of the lower Hudson River Valley of New York State (6, 10). Babesia microti infection has also been reported in the upper Midwest, where the Minnesota Department of Health reported 35 cases in 2009. The incidence of the disease in states outside this region is much lower than in the Northeast and is usually caused by infection with B. duncani (2, 13), although another strain (MO-1) has also been reported (7). Human babesiosis is rarer in Europe, where it is most often caused by infection with Babesia divergens (16). Although the incidence is lower, infection with B. divergens is often more severe and can be fatal if not treated properly.
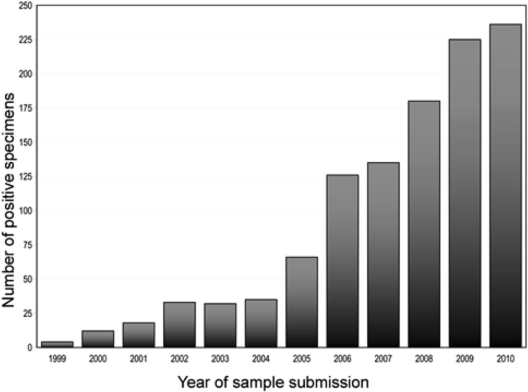
Both babesiosis and Lyme disease are transmitted by ixodid ticks, and New York reports the highest numbers of these diseases in the United States (1). The incidence of reported cases of Lyme disease increased markedly in New York State in the late 20th century, and the yearly incidence of Babesia infection has increased in a similar fashion over the last decade. Twelve cases were reported to the Parasitology Laboratory at the Wadsworth Center, New York State Department of Health (NYSDOH), in 2000, but the number had grown to 126 by 2006. Babesiosis has been a reportable disease in New York State since 1986, so the increases are unlikely to be a consequence of reporting practices. The yearly caseload is now more than 200 and continues to increase (Fig. 1).
Fig 1.
Patient specimens identified as positive for Babesia sp. in New York State from 1999 to 2010.
The long-standing gold standard assay for diagnosis of active babesiosis has been microscopic examination of Giemsa-stained thick and thin blood smears, an assay that requires specially trained personnel for analysis. There are limitations to this method, since Babesia can only be identified to the genus level based on morphological criteria. Furthermore, Babesia parasites can be difficult to distinguish from the early trophozoite (ring form) of Plasmodium parasites, particularly P. falciparum, in which ring forms are often the only stage that is observed. There are no U.S. Food and Drug Administration-approved tests for Babesia. Although the use of real-time PCR has been described in a report on transfusion-associated babesioses (14), we describe here for the first time the development, validation, and clinical use of a real-time PCR assay for detection of Babesia microti in human patients.
Our laboratory previously developed a conventional PCR assay for babesiosis derived from the amplification and hybridization method of Persing et al. (12). This modified assay consisted of two independent PCRs targeting the 18S rRNA gene, one using primers to produce a product of 238 bp and the other using a different primer set to produce a product of 154 bp. However, this assay requires handling of amplified target DNA, which is a risk factor for contamination of the workspace and is relatively time-intensive. To overcome these limitations, we designed a real-time PCR assay to detect Babesia microti, the primary Babesia pathogen in the northeastern United States, where this pathogen is highly endemic. The region of the gene targeted in this assay shows interspecies variation and is therefore appropriate for extension of the assay to multiplex detection of other pathogenic Babesia species.
MATERIALS AND METHODS
Primers, probes, and control plasmids.
The target for this assay is a variable region of the 18S rRNA gene that contains sequence that is species specific for B. microti. As a positive control, a plasmid was constructed that contains an insert encoding 514 bp of the 5′ end of the B. microti 18S rRNA gene (corresponding to positions 1 to 514 of GenBank accession number M93660). The sequences of the B. microti specific primers and a minor groove binding probe are given in Table 1. In addition, a Babesia-bicoid inhibition control (BBIC) plasmid was generated that contains the binding sites for the B. microti primers, flanking an insert encoding a portion of the bicoid gene of D. melanogaster (positions 2935 to 3115 of GenBank accession number X07870). This fragment was produced by amplifying bicoid DNA with long primers containing Babesia-specific sequence on the 5′ ends, and bicoid-specific sequence on the 3′ ends (see Table 1 for details). Control plasmids were constructed by ligation of the appropriate PCR amplification products into pCR4-TOPO vector, followed by transformation into TOP10 Escherichia coli (Invitrogen).
Table 1.
Primers and probesa
| Primer or probe | Sequence (5′-3′) | Target in assay | Position in reference sequence |
|---|---|---|---|
| Primers | |||
| Babesia 8 forward | CAGGGAGGTAGTGACAAGAAATAACA | B. microti DNA and both control plasmids | 417–442 |
| Babesia 8 reverse | GGTTTAGATTCCCATCATTCCAAT | B. microti DNA and both control plasmids | 465–488 |
| Probes | |||
| Babesia microti 8 | 6FAM-TACAGGGCTTAAAGTCT-MGBNFQ | B. microti DNA, genomic and plasmid | 444–461 |
| BBIC | VIC-CTACAACTTCAACAGCTCGTA-MGBNFQ | Bicoid plasmid | 2971–2992 |
The full sequence of the inhibition control insert was as follows: CAGGGAGGTAGTGACAAGAAATAACACTGTCGCGTCCTGGTCAAGGACGAACCGGAGGCCGACTACAACTTCAACAGCTCGTACTACATGCGATCGGGAATGTCTGGCGCCACTGCATCGGCATCCGCTGTGGCCCGAGGCGCTGCCTCGCCGGGCTCCGAGGTCTACGAGCCATTAACACCCAAGAATGACGAAAGTCCGAGTCTGATTGGAATGATGGGAATCTAAACC. This insert was constructed by amplifying bicoid DNA with chimeric forward and reverse primers. Underlining indicates the sequence corresponding to the Babesia primers used in this assay, and boldfacing indicates the 5′ and 3′ termini of the bicoid sequence. The italics indicate the position of the bicoid probe.
Samples.
Aliquots of EDTA-preserved whole blood were submitted to the Wadsworth Center by laboratories that test samples from patients in New York State. Samples were submitted for testing during the periods from 2008 to 2009 (validation samples) and from 2009 to 2011 (clinical samples). Clinical samples were submitted for identification or confirmation of infection with Babesia. Samples used for validation purposes may have been submitted for this reason or may have been submitted for diagnosis of other blood-borne pathogens. Samples submitted for blood-borne parasite identification are evaluated by Giemsa stained thin blood smears, as well as nucleic acid amplification. At least 300 oil-immersion fields were examined before calling a specimen negative.
Extraction of DNA from blood samples.
Blood samples were shipped at room temperature and stored at 4°C. Unless otherwise indicated, patient specimens were extracted and tested within, at most, 4 days of receipt and typically within 24 h. For the blinded validation study, patient blood was extracted from older samples, but the results were highly comparable to those from recent samples. DNA was extracted from 200 μl of EDTA-preserved whole blood using the QIAamp DNA minikit blood and body fluids protocol (Qiagen) according to the manufacturer's instructions and eluted in an equal volume of elution buffer. DNA was extracted from two additional samples as controls: a negative control consisting of previously tested blood that did not contain B. microti and a positive control consisting of the same negative blood spiked with ∼100 copies (2 × 10−5 ng) of BBIC plasmid DNA. To test the clinical sensitivity of the assay, we also constructed a dilution series consisting of known quantities of the B. microti control plasmid spiked into negative blood.
Real-time PCR.
Extracted DNA (5 μl) or plasmid containing B. microti 18S rRNA gene was used as a template for real-time PCR using the FastStart DNA Master mix without ROX (Roche Applied Science, Indianapolis, IN) in a 25-μl reaction mixture with 5.5 mM MgCl2, 500 nM concentrations of the primers, and 250 nM concentrations of the probes. A two-step PCR cycling protocol was used for the amplification as follows: 95°C 10 min and 45 cycles of 95°C for 15 s and 60°C for 1 min. Ramp rates for all steps were set at maximum, and fluorescence data were acquired at the end of each annealing step. Experiments were performed on ABI 7500 and ABI 7500 Fast real-time PCR instruments.
For comparison, conventional PCR was also performed on the samples used in the validation assay, using AmpliTaq Gold (Roche, Branchburg, NJ). Primer sequences were identical to those in Persing et al. (12), but the two primer pairs were used in independent PCRs, with correct amplification of both products being required for a positive result. The cycling conditions were 95°C for 10 min and 35 cycles of 94°C 1 min, 58°C 1 min, and 72°C 1 min, with a final extension at 72°C for 4 min. The products were resolved on a 3% agarose gel and visualized with ethidium bromide.
Analytical specificity of primer and probe sets.
Real-time PCR with the 18S gene primer and probe set was performed with template DNA from other pathogens (see Table 3) to demonstrate specificity for Babesia microti. Specificity within the genus was tested by using a sample set comprising cervine blood infected with B. odocoilei, supplied by the New York State Department of Environmental Conservation, and DNA from five species of Babesia provided by the College of Veterinary Medicine, Texas A&M University. In addition, 18 other parasitic and bacterial blood-borne pathogens provided by the Bacteriology and Parasitology Laboratories of the Wadsworth Center, NYSDOH, were tested. The source material for these samples consisted of primary patient specimens (Escherichia coli, Anaplasma phagocytophila, and Ehrlichia chaffeensis), commercial preparations (Streptococcus pneumophila, ATCC 49619; Bartonella henselae, ATCC 49882; Staphylococcus aureus MRDA, ATCC 33591; Borrelia B31, ATCC 35210; Rickettsia rickettsiae, DNA extracted from CDC RA2296 Yolk Sac Antigen for IFA), and other cultured specimens (Treponema denticola, T. vincentii, and T. pallidum) Approximately 106 genome copies of each organism were used for specificity testing.
Table 3.
Organisms tested for the analytical specificity panel
| Type | Organismsa |
|---|---|
| Babesia species | Babesia bigemina, Babesia caballi, Babesia divergens*, Babesia gibsoni, Babesia microtib*, Babesia odocoilei |
| Other parasites | Leishmania amazonensis, Leishmania guyanensis, Plasmodium falciparum, Plasmodium malariae, Plasmodium ovale (two sequence variants), Plasmodium vivax, Trypanosoma brucei |
| Bacteria | Anaplasma phagocytophilum, Bartonella henselae, Borrelia burgdorferi B31, Ehrlichia chaffeensis, Escherichia coli, Rickettsia rickettsii, Staphylococcus aureus, Streptococcus pneumoniae, Treponema pallidum, Treponema denticola |
Of the 24 pathogens listed, only B. microti produced a positive result. *, Species of Babesia that are known human pathogens.
Babesia microti yielded the only DNA sample that gave a positive result by real-time PCR.
RESULTS
Analytical sensitivity and specificity.
The sensitivity of the real-time PCR assay was determined using nine samples of human blood spiked with a dilution series of the positive control plasmid, which contains a portion of the 18S rRNA gene from B. microti. Total DNA was extracted from each sample as described, and the cycle threshold was determined for each sample in triplicate. Approximately 100 copies of 18S rRNA gene in a real-time PCR can be reliably detected, and the standard deviation between replicates is approximately one cycle. At higher concentrations (103 copies/reaction and above), the standard deviation is negligible (Table 2).
Table 2.
Assay sensitivity and reproducibilitya
| Babesia microti copies/reaction (extrapolated) |
CT |
Mean | SD | %CVb | ||
|---|---|---|---|---|---|---|
| Value 1 | Value 2 | Value 3 | ||||
| 1.01 × 107 | 17.14 | 17.20 | 17.12 | 17.15 | 0.043 | 0.251 |
| 1.01 × 106 | 20.50 | 20.44 | 20.49 | 20.48 | 0.031 | 0.151 |
| 1.01 × 105 | 24.00 | 23.77 | 24.03 | 23.93 | 0.139 | 0.581 |
| 11,000 | 27.03 | 26.99 | 27.38 | 27.13 | 0.213 | 0.785 |
| 1100 | 31.29 | 31.34 | 31.15 | 31.26 | 0.098 | 0.314 |
| 100 | 34.02 | 35.94 | 34.23 | 34.73 | 1.051 | 3.026 |
| 10 | ND | ND | ND | NA | NA | NA |
| 1 | ND | ND | ND | NA | NA | NA |
| 0.1 | ND | ND | 38.58 | NA | NA | NA |
ND, not determined, i.e., a sample that did not yield a CT value after 45 cycles of amplification; NA, the calculations were not applicable for this dilution.
%CV, percent coefficient of variation.
The linearity of the assay was determined by plotting cycle threshold versus copy number for the dilution series of the control plasmid used in the previous experiment. The slope was found to be −3.53, with an R2 of 0.995, which is very close to the theoretical optimum of 1.0. The calculated efficiency was 93.5%. Therefore, this assay could be expanded to estimate gene copy number and, by extension, percent parasitemia in clinical samples. For use as a quantitative assay, the positive control plasmid would be included as a set of calibration samples.
The taxonomic specificity of the assay was determined by testing DNA from 24 species of pathogens, comprising six members of the genus Babesia as well as several other blood-borne parasites and bacteria (Table 3). Approximately one million genome copies of each organism were used to test for cross-reactivity in this real-time PCR assay. Of the Babesia species tested B. caballi has the fewest nucleotide changes in the amplified region, with a single deletion and a single mismatch in the probe sequence and four nucleotide changes in the downstream primer. However, neither this species nor any other organism tested showed cross-reactivity. In addition, we compared the sequences of primers and probe sequences from known human-infective Babesia in GenBank (B. duncani = Babesia sp. strain WA-1, B. divergens, Babesia sp. strain MO-1, and Babesia sp. strain EU-1). The primers have at least one mismatch to these sequences, and the probe has several mismatches (Fig. 2). Because differences of this magnitude were sufficient to prevent nonspecific amplification of the non-target Babesia species we tested, we conclude that the assay is unlikely to erroneously detect other species in the genus.
Fig 2.
Diagnostic region targeted by this assay in B. microti and other members of the genus. Bases which differ from the sequence of B. microti (boxed) are highlighted. GenBank accession numbers, in order: AY027815, AY048113, GU647159, U16370, M93660, X59604, Z15104, AF175300, U16369, GQ411405, AJ871610, Z15105, DQ200887, AB049999, AF244912, AY260176, DQ312442, AY726009, DQ111765, AY533146, L19079, DQ111764, AY596279, EU376017, EF551335, AY260180, HQ150006, AF419313.
Comparison of real-time PCR with conventional PCR and Giemsa staining using clinical samples.
The clinical sensitivity and specificity of the real-time PCR assay were compared to those of the assays previously in use in our laboratory, namely, microscopic examination of Giemsa-stained thin blood smears and conventional PCR. For initial assay validation, 40 peripheral blood samples which had been submitted to the Wadsworth Center for confirmation of Babesia infection were blinded and randomized. These specimens consisted of 30 that had been confirmed as positive for Babesia sp. by conventional PCR and Giemsa staining and 10 that had been found to be negative. DNA was extracted from each sample as described above and tested in duplicate in the real-time PCR assay.
Of the 40 samples, 10 were negative by all three assays and 30 were positive by all assays. The specificity of the conventional and real-time PCR assays, which amplify 238- and 72-bp regions of the 18S rRNA gene, respectively, was also confirmed by sequencing. The products of the 30 successful reactions were found to be 100% identical to GenBank accession no. M93660 (Babesia microti 18S rRNA gene). Cycle threshold (CT) values for the positive samples were low (mean, 21.6; median, 20.7; range, 18.2 to 29.3), a finding consistent with the high parasitemia often exhibited during Babesia infection.
Performance of the assay in routine clinical testing.
The assay we describe was validated for diagnostic use in mid-2009. Since that time, the assay has been used to evaluate 671 peripheral blood samples submitted to the laboratory for identification or confirmation of Babesia infection. Samples in this group were not evaluated by conventional PCR, since the real-time method was intended to replace this assay. Of the 671 samples, 638 were of sufficient quality to permit examination by Giemsa staining, which is routinely used in our laboratory as an independent confirmation of diagnosis. The remaining 33 comprised specimens that were hemolyzed and therefore could not be properly analyzed by staining methods. No sample exhibited PCR inhibition, which we have previously observed to be very rare in whole blood extracts.
Of the 638 samples tested by both methods, 542 were positive by both real-time PCR and Giemsa staining, and 93 were negative by both methods (Table 4). The 33 samples that were not able to be analyzed by Giemsa staining were all positive by real-time PCR. Three samples were positive by real-time PCR but negative by Giemsa staining. The distribution of CT values for the samples tested is shown in Fig. 3. To be reported as positive, a sample must be tested in duplicate, with both replicates exhibiting a CT of ≤38 for the B. microti probe and with the CT of the inhibition control being no more than 3 CT higher than that of the positive extraction control. As shown in the figure, CT values in the 39 to 45 range are only rarely observed. Two of the 638 specimens we tested yielded CT values of greater than 38. In each case, one replicate gave a positive but very high CT result, whereas the duplicate reaction was negative. Upon retesting, both replicates were negative.
Table 4.
Summary of clinical resultsa
| Real-time PCR result | Giemsa result (no. of samples) |
||
|---|---|---|---|
| Positive | Negative | NA | |
| Positive | 542 | 3 | 33 |
| Negative | 0 | 93 | 0 |
The CT values of the three samples that were negative by Giemsa but positive by PCR were 24.4, 27.7, and 30.8. NA, not applicable.
Fig 3.
CT values of patient specimens tested from 2009 to 2011. Samples lying to the left of the dotted line were reported as positive. Those to the right were retested. In each case, one replicate gave a high positive CT result, whereas the duplicate reaction was negative. Upon retesting both replicates were negative.
The assay developed here was also able to improve parasite detection in a patient who was slowly clearing an infection. It is not known whether the patient was immunocompromised or had been splenectomized, which would affect the patient's ability to clear the infection. Six blood samples were taken over the course of a month in late summer 2010 and were submitted for testing. The first five samples were positive by both microscopic examination and real-time PCR. The sixth was negative by Giemsa staining, but parasite DNA was still detectable, with a CT of 24.4.
DISCUSSION
We have demonstrated that this real-time PCR assay is highly effective as a diagnostic tool for the detection of Babesia microti in whole blood samples. This assay is 100% specific against all of the pathogens or infectious agents tested, including several other members of the genus Babesia. We conclude that this assay permits speciation of the infecting parasite, unlike Giemsa staining, which permits identification only to the genus level. Although B. microti is by far the dominant Babesia pathogen in the northeastern United States, the region of the 18S rRNA gene targeted by the assay shows species-specific sequence variation and the assay can easily be modified to permit detection of other species in the genus, such as B. duncani and the MO-1 and EU-1 strains. Unfortunately, we were unable to obtain samples of these pathogens for testing, which is a limitation of the present study.
We also provide evidence that the assay can detect gene copy numbers far below those which would be expected in a clinical infection. The clinical sensitivity has been determined to be ∼100 gene copies. Although the number of copies of the 18S rRNA gene in the Babesia microti genome is not known, other apicomplexan parasites contain between two and five copies of this gene, which would suggest a limit of detection corresponding to a parasitemia of ca. 5 to 10 parasites/μl of blood or ∼0.0001% parasitemia (3). This sensitivity exceeds that of Giemsa stain, which is estimated to be 10 to 50 parasites/μl (0.0002 to 0.001% parasitemia) under ideal conditions but ∼100 parasites/μl (0.002%) for routine diagnostic screening (9). Thus, the assay described here is up to 20-fold more sensitive than examination of stained blood smears. In addition, since babesiosis typically presents with a parasitemia of >0.1%, the assay is orders of magnitude more sensitive than required for the majority of clinical specimens (see also Fig. 3). With the inclusion of a set of samples to generate a calibration curve, the excellent linearity of response also permits calculation of the percent parasitemia. This calculation is potentially important for treatment since the disease can reach high levels of parasitemia quickly.
The assay is also helpful in cases where parasites may be difficult to detect by microscopic methods. This can occur when there is a delay between collection of the sample and receipt by the laboratory, and the whole blood sample is received hemolyzed. Parasites in samples from patients that have been drug-treated can also be difficult to detect, and we speculate that this is the case when treated patients still have a relatively high CT but no morphologically detectable parasites.
This method may also be effective for screening donated blood, an important consideration in the face of the reports of transmission-associated infections described above. Finally, the use of this assay in conjunction with Giemsa stain may be able to provide early warning of the appearance of new Babesia species in North American patients, since these would produce a positive result by Giemsa stain but would be expected to give a negative result by PCR.
New York State has experienced a 17-fold increase in human Babesia infections since 2000, and other states, such as Minnesota, have seen equally dramatic increases (Annual Summary of Communicable Diseases Reported to the Minnesota Department of Health, 2009 [http://www.health.state.mn.us/divs/idepc/newsletters/dcn/sum09/sum09.pdf]).
The real-time PCR assay described here and validated as a clinical assay is a rapid and sensitive method for detection of B. microti and a valuable new tool for diagnostic laboratories, especially since the incidence of this parasite has risen dramatically in the last decade.
ACKNOWLEDGMENTS
We are indebted to Ivana Pysova (supported in part by the New York State International Training and Research Program Grant 2D43TW000233, NIH Fogarty International Center) for assistance with real-time PCR; to the Wadsworth Center Applied Genomic Technologies Core for sequencing; to Patricia Holman at CVM, Texas A&M, for Babesia DNA; and to Ward Stone, NYS DEC, for a sample of cervine Babesia.
Footnotes
Published ahead of print 14 December 2011
REFERENCES
- 1. Centers for Disease Control 2002. Lyme disease–United States, 2000. MMWR Morb. Mortal. Wkly. Rep. 51:29–31 [PubMed] [Google Scholar]
- 2. Conrad PA, et al. 2006. Description of Babesia duncani n.sp. (Apicomplexa: Babesiidae) from humans and its differentiation from other piroplasms. Int. J. Parasitol. 36:779–789 [DOI] [PubMed] [Google Scholar]
- 3. Garcia LS. 2007. Diagnostic medical parasitology, 5th ed, p 896 ASM Press, Washington, DC [Google Scholar]
- 4. Gubernot DM, et al. 2009. Babesia infection through blood transfusions: reports received by the US Food and Drug Administration, 1997–2007 Clin. Infect. Dis. 48:25–30 [DOI] [PubMed] [Google Scholar]
- 5. Gubernot DM, et al. 2009. Transfusion-transmitted babesiosis in the United States: summary of a workshop. Transfusion 49:2759–2771 [DOI] [PubMed] [Google Scholar]
- 6. Hatcher JC, Greenberg PD, Antique J, Jimenez-Lucho VE. 2001. Severe babesiosis in Long Island: review of 34 cases and their complications. Clin. Infect. Dis. 32:1117–1125 [DOI] [PubMed] [Google Scholar]
- 7. Herwaldt B, et al. 1996. A fatal case of babesiosis in Missouri: identification of another piroplasm that infects humans. Ann. Intern. Med. 124:643–650 [DOI] [PubMed] [Google Scholar]
- 8. Herwaldt B, et al. 2011. Transfusion-associated babesiosis in the United States: a description of cases. Ann. Intern. Med. 155:509–519 [DOI] [PubMed] [Google Scholar]
- 9. Kamau E, et al. 8 June 2011. Development of a highly sensitive genus-specific qRT-PCR assay for detection and quantitation of Plasmodium by amplifying RNA and DNA of the 18S rRNA genes. J. Clin. Microbiol. doi:10.1128/JCM.00276-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kogut SJ, et al. 2005. Babesia microti, Upstate New York. Emerg. Infect. Dis. 11:476–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krause PJ, et al. 1996. Concurrent Lyme disease and babesiosis: evidence for increased severity and duration of illness. JAMA 275:1657–1660 [PubMed] [Google Scholar]
- 11a. Krause PJ, et al. 1998. Persistent parasitemia after acute babesiosis. N. Engl. J. Med. 339:160–165 [DOI] [PubMed] [Google Scholar]
- 12. Persing DH, et al. 1992. Detection of Babesia microti by polymerase chain reaction. J. Clin. Microbiol. 30:2097–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scholtens RG, Braff EH, Healy GR, Gleason N. 1968. A case of babesiosis in man in the United States. Am. J. Trop. Med. Hyg. 17:810–813 [DOI] [PubMed] [Google Scholar]
- 14. Tonnetti L, et al. 2009. Transfusion complications: transfusion-transmitted Babesia microti identified through hemovigilance. Transfusion 49:2557–2563 [DOI] [PubMed] [Google Scholar]
- 15. Vannier E, Gewurz BE, Krause PJ. 2008. Human Babesiosis. Infect. Dis. Clin. N. Am. 22:469–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zintl A, Mulcahy G, Skerrett HE, Taylor SM, Gray JS. 2003. Babesia divergens, a bovine blood parasite of veterinary and zoonotic importance. Clin. Microbiol. Rev. 16:622–636 [DOI] [PMC free article] [PubMed] [Google Scholar]