Abstract
Two live-attenuated rotavirus group A (RVA) vaccines, Rotarix (G1P[8]) and RotaTeq (G1-G4, P[8]), have been successfully introduced in many countries worldwide, including Belgium. The parental RVA strains used to generate the vaccines were isolated more than 20 years ago in France (G4 parental strain in RotaTeq) and the United States (all other parental strains). At present, little is known about the relationship between currently circulating human RVAs and the vaccine strains. In this study, we determined sequences for the VP7 and VP4 outer capsid proteins of representative G1P[8], G2P[4], G3P[8], G4P[8], G9P[8], and G12P[8] RVAs circulating in Belgium during 2007 to 2009. The analyses showed that multiple amino acid differences existed between the VP7 and VP4 antigenic epitopes of the vaccine viruses and the Belgian isolates, regardless of their G and P genotypes. However, the highest variability was observed among the circulating G1P[8] RVA strains and the G1 and P[8] components of both RVA vaccines. In particular, RVA strains of the P[8] lineage 4 (OP354-like) showed a significant number of amino acid differences with the P[8] VP4 of both vaccines. In addition, the circulating Belgian G3 RVA strains were found to possibly possess an extra N-linked glycosylation site compared to the G3 RVA vaccine strain of RotaTeq. These results indicate that the antigenic epitopes of RVA strains contained in the vaccines differ substantially from those of the currently circulating RVA strains in Belgium. Over time, these differences might result in selection for strains that escape the RVA neutralizing-antibody pressure induced by vaccines.
INTRODUCTION
Group A rotaviruses (RVAs) are a primary cause of gastroenteritis in children under 5 years of age and are associated with over 500,000 deaths annually, of which the majority occur in developing countries (42, 43). RVAs belong to the family Reoviridae, and the infectious RVA virion is a triple-layered icosahedral particle that contains 11 segments of double-stranded RNA (18). The outer capsid layer is composed of the spike protease-sensitive attachment protein VP4 (P) and the glycoprotein VP7 (G). The nucleotide sequences of the VP7- and VP4-encoding segments form the basis of a dual classification system that defines the G and P genotypes of RVAs, respectively. Although 27 G genotypes and 35 P genotypes have been identified to date (31), only a few RVA G and P genotype combinations contribute substantially to the burden of human disease (29, 30, 32, 53). Until the mid 1990s, G1P[8], G2P[4], G3P[8], and G4P[8] RVAs represented the most common and widespread strains infecting humans. Since then, G9P[8] RVA strains have emerged globally and are now also considered a common cause of human disease (33, 35). An increase in prevalence of G12 RVAs, mainly associated with P[8] or P[6] and to a lesser extent with P[4] or P[9] VP4s, was observed around the turn of the century. The G12 RVA strains are now considered the sixth common global genotype (29, 33, 35, 49).
Two live attenuated oral RVA vaccines, Rotarix (GlaxoSmithKline Biologicals, Belgium) and RotaTeq (Merck & Co., Inc., United States), have been licensed for use in many countries around the world. Rotarix is derived from the attenuated human G1P[8] RVA strain 89-12, which was isolated in Cincinnati, OH, in 1988 (65). RotaTeq contains five human-bovine reassortant RVA strains (WI79-9, SC2-9, WI78-9, BrB-9, and WI79-4, referred to as G1, G2, G3, G4, and P1 reassortants, respectively, for simplicity). The G1 to G4 reassortants each express one of the VP7 proteins of the human RVA parental strains WI79 (G1), SC2 (G2), WI78 (G3), and BrB (G4) and the VP4 protein of the bovine RVA strain WC3 (P7[5]), whereas the P1 reassortant expresses the VP4 protein of the human RVA strain WI79 (P1A[8]) and the VP7 protein of the bovine RVA strain WC3 (G6) (9, 34). Human RVA SC-2 was isolated in 1981 at St. Christopher's Hospital of Philadelphia, and the WI79 and WI78 RVAs were isolated in 1983 at the Children's Hospital of Philadelphia, while the BrB (originally Bricout B) RVA strain was isolated in 1984 at L'Hôpital Armand Trousseau (Paris, France) (34). Both RVA vaccines have been proven to be safe and efficacious in large-scale clinical trials (9, 51, 59, 61, 62). The mechanisms by which the vaccines induce immunologic protection in infants have not been clearly elucidated. It likely includes the induction of serum and intestinal serotype-specific neutralizing antibodies directed against VP7 and VP4 and virus-specific cytotoxic T lymphocytes (20, 63, 64). However, proteins other than VP7 and VP4 may be involved in immune protection as well.
As a result of RVA vaccine implementation, substantial reductions in RVA disease burden have been reported in many countries in the world, including Austria, Australia, Belgium, Brazil, El Salvador, Nicaragua, Panama, and the United States (6, 12, 13, 19, 28, 38, 45, 46, 67). Both vaccines have been found to be similarly efficacious against human G1, G3, G4, and G9 RVAs, strains which typically have P[8] VP4 genotypes, the same P genotype of Rotarix and RotaTeq vaccine viruses. Notably, in children vaccinated with Rotarix protection against G2 strains, which usually have P[4] genotypes, is somewhat lower than against other human genotypes. This may be due to the lack of either a G2 VP7 or P[4] VP4 component in the Rotarix vaccine (9, 51, 56, 60, 61). Vaccine efficacy data against human VP7 G12 strains are not yet available.
The serotype-specific efficacy of both RVA vaccines against G1P[8] RVAs has been well established, predominantly because G1P[8] RVA strains were by far the most common circulating strains encountered during phase III clinical trials, which were conducted approximately 2 decades after the vaccine strains were isolated (51, 59–62). However, little is known about the genetic characteristics of the RVA strains that circulated during the clinical trials, and how similar these strains, and their VP7 and VP4 components, were to the strains present in Rotarix and RotaTeq.
For circulating RVA strains identified in clinical trials of RotaTeq, only a small region of the VP7 gene was sequenced, sufficient to allow G genotype assignment (15, 62). For circulating strains identified in clinical trials of Rotarix, reverse transcriptase PCR (RT-PCR), followed by a hybridization assay, was used to determine both viral G and P genotypes (14). Although both molecular assays are suitable to discriminate RVA genotypes, they cannot distinguish subgenotypic lineages or amino acid changes in all relevant antigenic regions. Increasingly, evidence is becoming available that subgenotypic lineages can possess different antigenic properties potentially allowing RVA strains to escape adaptive immunity (2, 4, 21, 24). Such subtle changes can be monitored only by sequencing complete gene segments, an essential approach for comprehending fully the antigenic landscape of circulating RVA strains.
Recently, the complete genomic characterization of the five RVA strains of RotaTeq was reported (34). The VP7 and VP4 sequences of the RVA strain in Rotarix have not been reported yet. Because both RVA vaccines were derived or generated using human RVA strains circulating in the early to mid-1980s for RotaTeq and the late 1980s for Rotarix, it is possible that the currently circulating RVA strains in Belgium cluster in VP7 and VP4 lineages that are different from those of the RVA vaccine strains. As a result of vaccine implementation in national immunization programs such as in Belgium, a varying selective pressure against these different VP7 and VP4 lineages could be induced and over time, this might result in reduced vaccine effectiveness. In the present study, we deduced the VP7 and VP4 amino acid sequences of human RVAs circulating in Belgium, as well as those of Rotarix, as an approach for comparing the VP7 and VP4 lineages of current Belgian strains with those of the two RVA vaccines. In addition, using molecular models, we investigated the potential antigenic disparities between RVA vaccines and the RVA strains currently circulating in Belgium.
MATERIALS AND METHODS
Study samples.
A total of 21 RVA strains (8 G1P[8], 4 G2P[4], 2 G3P[8], 2 G4P[8], 3 G9P[8], and 2 G12P[8] strains) were selected from a large epidemiological study conducted in the 2007 to 2009 RVA seasons at multiple hospitals across Belgium. These strains were chosen based on preliminary phylogenetic analyses of partial sequence information obtained for their VP7 and VP4 genes (∼600 nucleotides each), such that all of the different circulating VP7 and VP4 lineages of every G genotype (G1 to G4, G9, and G12) and P genotype (P[8], P[4]), respectively, were represented (Table 1). For genotypes with a low genetic heterogeneity, two RVA strains were selected. The Belgian G3P[6] strain BE1322, which has previously been deposited in GenBank, was also analyzed in the present study as a representative of recent circulating P[6] RVA strains in Belgium. Subgenotypic lineages were assigned as previously described (33, 34, 44). The nucleotide sequences of the VP7 and VP4 of RotaTeq were published previously (34). Because the VP7 and VP4 sequences of the RVA strain in Rotarix have not been reported, the nucleotide sequences for these gene segments were determined from a commercially obtained dose of the vaccine (lot no. A41CB052A).
Table 1.
Summary of VP7 and VP4 lineage, date of isolation, and available patient information for Belgian rotavirus strains analyzed in this study and rotavirus vaccine strains Rotarix and RotaTeqa
| Strain | VP7 lineage | VP4 lineage | Date of isolation (mo/yr) | Date of birth (mo/yr) | Gender | GenBank identification |
|
|---|---|---|---|---|---|---|---|
| VP7 | VP4 | ||||||
| RVA/Vaccine/USA/Rotarix-A41CB052A/1988/G1P1A[8] | 2 | 1 | JN849114 | JN849113 | |||
| RVA/Vaccine/USA/RotaTeq-WI79-9/1992/G1P7[5] | 3 | NA | GU565057 | GU565055 | |||
| RVA/Human-wt/BEL/BE1520/2009/G1P[8] | 2 | 1 | 7/2009 | 1/2009 | M | JN849152 | JN849151 |
| RVA/Human-wt/BEL/BE1175/2009/G1P[8] | 2 | 3 | 2009 | 8/2005 | F | JN849154 | JN849153 |
| RVA/Human-wt/BEL/BE1280/2009/G1P[8] | 2 | 4 | 4/2009 | 6/2008 | – | JN849150 | JN849149 |
| RVA/Human-wt/BEL/BE1001a/2008/G1P[8] | 2 | 1 | 8/2008 | 8/2007 | M | JN849126 | JN849125 |
| RVA/Human-wt/BEL/BE0253/2008/G1P[8] | 2 | 1 | 3/2008 | 12/2007 | M | JN849120 | JN849119 |
| RVA/Human-wt/BEL/BE1023/2008/G1P[8] | 1 | 3 | 4/2008 | 7/2004 | M | JN849122 | JN849121 |
| RVA/Human-wt/BEL/BE1286/2009/G1P[8] | 1 | 3 | 3/2009 | 2/2008 | M | JN849148 | JN849147 |
| RVA/Human-wt/BEL/BE1128/2009/G1P[8] | 1 | 3 | 2/2009 | 3/2008 | F | JN849136 | JN849135 |
| RVA/Vaccine/USA/RotaTeq-SC2-9/1992/G2P7[5] | 2 | NA | GU565068 | GU565066 | |||
| RVA/Human-wt/BEL/BE1248/2009/G2P[4] | 4 | 3 | 1/2009 | 10/2002 | F | JN849130 | JN849129 |
| RVA/Human-wt/BEL/BE1141/2009/G2P[4] | 4 | 3 | 2009 | JN849156 | JN849155 | ||
| RVA/Human-wt/BEL/BE1058/2008/G2P[4] | 4 | 3 | 5/2008 | 11/2006 | M | JN849124 | JN849123 |
| RVA/Human-wt/BEL/BE1251/2009/G2P[4] | 4 | 3 | 3/2009 | 7/2007 | M | JN849144 | JN849143 |
| RVA/Vaccine/USA/RotaTeq-WI78-8/1992/G3P7[5] | 2 | NA | GU565079 | GU565077 | |||
| RVA/Human-wt/BEL/BE1322/2009/G3P[6] | 3 | NA | 4/2009 | 2/2009 | M | JF460828 | JF460826 |
| RVA/Human-wt/BEL/BE1214/2009/G3P[8] | 1 | 3 | 3/2009 | 1/2008 | M | JN849140 | JN849139 |
| RVA/Human-wt/BEL/BE1259/2009/G3P[8] | 1 | 3 | 3/2009 | 6/2008 | M | JN849146 | JN849145 |
| RVA/Vaccine/USA/RotaTeq-BrB-9/1996/G4P7[5] | 1 | NA | ; | GU565090 | GU565088 | ||
| RVA/Human-wt/BEL/BE1129/2009/G4P[8] | 1 | 3 | 2/2009 | 5/2008 | M | JN849138 | JN849137 |
| RVA/Human-wt/BEL/BE1113/2009/G4P[8] | 1 | 3 | 2/2009 | 4/2006 | M | JN849134 | JN849133 |
| RVA/Vaccine/USA/RotaTeq-WI79-4/1992/G6P1A[8] | NA | 2 | GU565046 | GU565044 | |||
| RVA/Human-wt/BEL/BE1242/2009/G9P[8] | 3 | 3 | 3/2009 | 1/2008 | F | JN849142 | JN849141 |
| RVA/Human-wt/BEL/BE1119/2009/G9P[8] | 3 | 3 | 1/2009 | 10/2008 | F | JN849132 | JN849131 |
| RVA/Human-wt/BEL/BE1032/2008/G9P[8] | 3 | 3 | 12/2008 | 10/2006 | M | JN849128 | JN849127 |
| RVA/Human-wt/BEL/BE0258/2008/G12P[8] | 3 | 3 | 3/2008 | 5/1957 | F | JN849118 | JN849117 |
| RVA/Human-wt/BEL/BE0085/2008/G12P[8] | 3 | 3 | 2/2008 | 2/2006 | F | JN849116 | JN849115 |
NA, not applicable. Shading indicates Rotarix or RotaTeq strains.
RNA extraction and RT-PCR.
Approximately 100 mg of stool sample was diluted in 500 μl of phosphate-buffered saline. Viral RNA was extracted from the diluted stool material using a QIAamp viral RNA minikit (Qiagen, Netherlands) according to the manufacturer's instructions. RNA extracts were diluted 1:1, 10 μl of the dilution was denatured at 95°C for 2 min, and RT-PCR was carried out using a Qiagen One-Step RT-PCR kit (20 μl of H2O, 10 μl of Qiagen OneStep RT-PCR buffer, 10 μl of diluted and denatured RNA, 3 μl of forward primer [8.3 μM], 3 μl of reverse primer [8.3 μM], and 2 μl of enzyme mix). Primers used to amplify VP7 and VP4 genes can be found Table S1 in the supplemental material. RT-PCR was carried out with an initial RT step at 50°C for 30 min; Taq polymerase activation was carried out at 95°C for 15 min, followed by 35 cycles of amplification (30 s at 94°C, 30 s at 45°C, and 1.5 or 3 min at 72°C for VP7 or VP4, respectively), with a final extension of 10 min at 72°C. The 5′ and 3′ terminal sequences of the VP4 open reading frame (ORF) were completed using the single-primer amplification method described previously (36). To recover RNA from a lyophilized dose of Rotarix, the vaccine was reconstituted according to the manufacturer's instructions and then extracted as described above for stool material.
Nucleic acid sequencing and sequence analyses.
PCR amplicons were purified with a MSB Spin PCRapace kit (Invitek, Germany) and sequenced using an ABI Prism BigDye terminator cycle sequencing reaction kit (Perkin-Elmer Applied Biosystems, United States) in an automated sequencer (ABI Prism 3100). Sequencing was performed with forward and reverse primers used for RT-PCR. In addition, primer-walking sequencing was performed to cover the complete sequence of the VP7 and VP4 segments.
Sequencing files were analyzed using Chromas 2.3 (Technelysium, Australia), and consensus sequences were prepared using SeqMan II (DNAstar, USA). Multiple sequence alignments were conducted using the CLUSTAL W plug-in in MEGA 4.0. Phylogenetic analyses, as well as amino acid sequence similarity calculations were also conducted in MEGA 4.0 using the Poisson model and P-distance method, respectively (57). Structural analyses of VP7 (PBD 3FMG) and VP8* (PDB 1KQR) were performed using the UCSF Chimera-Molecular Modeling System (47).
RESULTS
Comparison of VP7 proteins of circulating Belgian RVA strains and vaccine viruses.
The nucleotide sequence of the complete VP7 and VP4 open reading frame (ORF) of the 22 RVA strains (8 G1P[8], 4 G2P[4], 2 G3P[8], 1 G3P[6], 2 G4P[8], 3 G9P[8], and 2 G12P[8]) isolated during the 2007/2008 and 2008/2009 winter seasons (Table 1) were determined. As determined by initial partial sequencing, these strains were representative of all VP7 and VP4 lineages circulating in the Belgian population at these times.
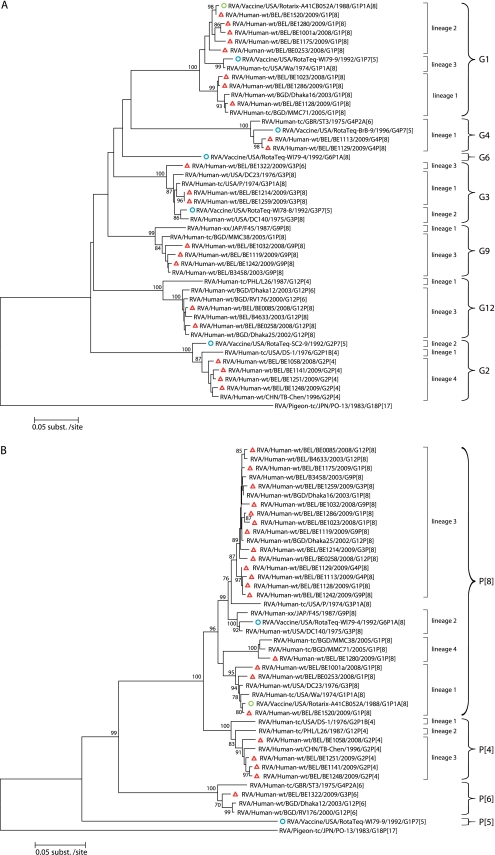
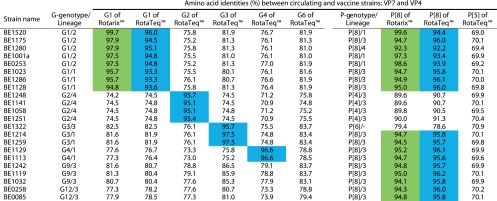
Figure 2A shows the phylogenetic tree of VP7 of the recent circulating Belgian strains as well as those of RotaTeq and Rotarix. Currently circulating G1 RVA strains clustered in lineage 1 and lineage 2. The G1 component of both vaccines clustered in two different lineages: lineage 2 for Rotarix and lineage 3 for RotaTeq. Besides the VP7 of the RVA strain BE1520, which was almost identical (99.7%) to that of Rotarix (Fig. 1), the VP7 of four RVA strains (BE1175, BE1280, BE1001a, and BE0253) clustered in G1 lineage 2 sharing 97.5 to 97.9% amino acid identities with the VP7 of Rotarix. The Belgian RVA strains belonging to the G1 lineage 1 were less related to VP7 of Rotarix (94.8 to 95.7%). VP7 of G1 Belgian RVA strains belonging to either lineage 1 or 2 were more distantly related (93.3 to 93.6% and 94.5 to 96.0%, respectively) to VP7 of the G1 lineage 3 strain (WI79-9) of RotaTeq.
Fig 2.
Phylogenetic analysis of the VP7 (A) and VP4 (B) proteins of circulating Belgian RVA strains and Rotarix and RotaTeq vaccine strains. Neighbor-joining trees were constructed using on the complete amino acid sequences of VP7 and VP4. Pigeon RVA strain PO-13 was used as the outgroup. Belgian strains are indicated with a red triangle. Rotarix and RotaTeq sequences are indicated with green and blue circles, respectively. Bootstrap values (500 replicates) of ≥70% are shown.
Fig 1.
Distance matrix for VP7 and VP4 based on amino acid identities. Intragenotype similarities with Rotarix are colored in green. Intragenotype similarities with RotaTeq are colored in blue.
Belgian G2 RVA strains were all shown to belong to G2 lineage 4 and were rather distantly related to VP7 of the G2 strain (SC2-9) of RotaTeq, which clustered in lineage 2 (95.1 to 95.7%) (Fig. 1 and 2B). The VP7 proteins of the two Belgian G3 RVA strains associated with a P[8] VP4 (BE1214 and BE1259) were identical in sequence and clustered in G3 lineage 1. Although the VP7 proteins of BE1214 and BE1259 were in a lineage different than that of VP7 of the G3 strain (WI78-9) of RotaTeq (lineage 2), they nonetheless were relatively closely related (97.5%). In comparison, VP7 of the G3P[6] strain (BE1322) was less related to the WI78-9 vaccine strain (95.7%). VP7 sequences of the Belgian G4 strains were almost identical (>99%), and both were rather closely related (96.6%) to VP7 of the G4 strain (BrB-9) of RotaTeq. VP7 of the G9 and G12 Belgian RVA strains each belonged to their respective VP7 lineage 3, which is the most common lineage for these emerging G genotypes worldwide (35). The VP7 proteins of the G9 and G12 Belgian strains were 77.6 to 86.5% and 73.3 to 81.0% identical, respectively, to the G1 VP7 component of Rotarix and the G1 to G4, G6 VP7 components or RotaTeq. The G9 and G12 VP7 proteins had the highest amino acid identity with the G3 component of RotaTeq (Fig. 1 and 2B).
Comparison of VP4 proteins of circulating Belgian RVA strains and vaccine viruses.
The phylogenetic tree of the VP4 amino acid sequences of Belgian RVA strains is shown in Fig. 2B. The Belgian VP4 P[8] proteins clustered in three distinct lineages, most belonging to lineage 3. In fact, only P[8] strains with a G1 VP7 were found to cluster in more than one P[8] lineage. VP4 of Rotarix clustered in P[8] lineage 1, while VP4 of the P[8] strain (WI79-4) of RotaTeq clustered in P[8] lineage 2. As observed for VP7 of the Belgian strain BE1520 (Fig. 2A), VP4 of BE1520 was almost identical in sequence to VP4 of Rotarix (99.6%) (Fig. 1). VP4 of Belgian P[8] RVA strains that clustered in lineage 1 were more closely related (97.3 to 98.6%) to VP4 of Rotarix (also in lineage 1) than the lineage 2 P[8] VP4 of RotaTeq (93.4 to 93.9%). VP4 of Belgian P[8] RVA strains clustering in lineage 3 were only 94.1 to 95.0% identical to VP4 protein of Rotarix and 95.7 to 96.2% identical to the VP4 protein of RotaTeq (Fig. 1). The VP4 of RVA strain BE1280 clustered in P[8] lineage 4 and was only distantly related to the P[8] VP4 proteins of Rotarix (92.3%) and RotaTeq (92.2%) (Fig. 1). The VP4 of the P[4] RVA strains circulating in Belgium were more homogeneous in sequence than those of the P[8] RVA strains, as all belonged to a single P[4] lineage (lineage 3). The P[4] VP4 proteins of the Belgian strains showed 89.6 to 90.0% and 90.5 to 91.3% identities to the P[8] VP4 components of Rotarix and RotaTeq, respectively. The VP4 of BE1322, the only P[6] strain analyzed in the present study, clustered together with previously isolated human P[6] RVA strains (49) and was 78.6 to 79.4% identical to the P[8] VP4 proteins of both RVA vaccines. The VP4 proteins of all of the Belgian strains showed low sequence identity (69.0 to 70.9%) with the P[5] VP4 component of RotaTeq (Fig. 1).
VP7 antigenic epitopes of circulating Belgian RVA strains and vaccine viruses.
Amino acid differences in the neutralizing epitopes of the VP7 and VP4 proteins of circulating Belgian RVA strains and the viruses formulating Rotarix and RotaTeq could undermine the effectiveness of these vaccines. To investigate whether such differences might exist, we compared the amino acid composition of the antigenic epitopes on the VP7 trimer and VP4 multimer (VP5* and VP8*) of Belgian and vaccine RVA strains. The location of these epitopes was determined by mapping neutralization escape mutants and identifying surface-exposed amino acids that show intergenotypic variability among prevalent human G and P genotypes (1, 16, 17, 37).
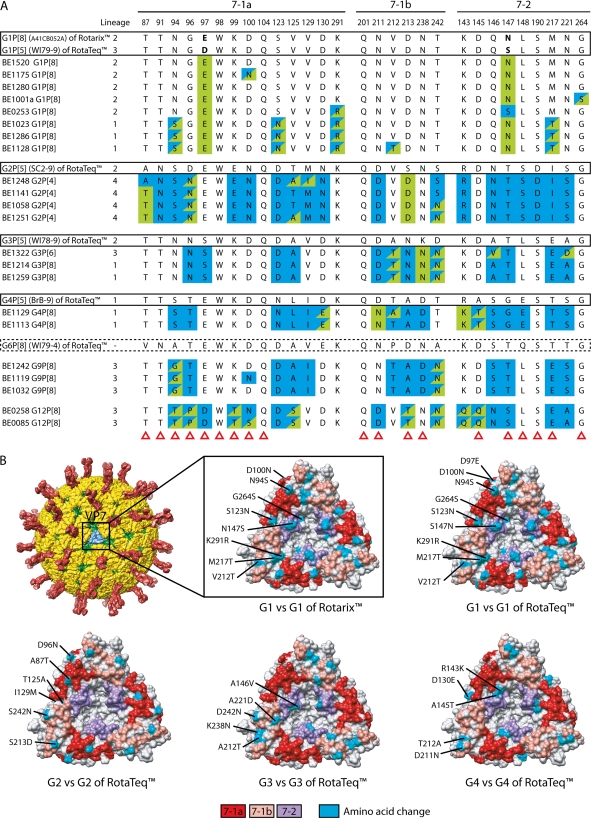
The VP7 trimer contains two structurally defined antigenic epitopes: 7-1 and 7-2. The 7-1 epitope spans the intersubunit boundary and is further subdivided into 7-1a and 7-1b (1). A comparison of residues that constitute the 7-1a, 7-1b, and 7-2 epitopes of the Belgian strains and the vaccine strains of Rotarix and RotaTeq is presented in Fig. 3A. Of the 29 amino acid residues of these epitopes, only three (amino acids 98, 104, and 201) were completely conserved among all Belgian and vaccine RVA strains.
Fig 3.
(A) Alignment of antigenic residues in VP7 between the strains contained in Rotarix and RotaTeq and strains circulating in Belgium. Antigenic residues are divided in three epitopes (7-1a, 7-1b, and 7-2). Amino acids that differ between Rotarix and RotaTeq are indicated in boldface. Blue colored residues are residues that are different from Rotarix, and green colored residues are different from the most similar genotype in RotaTeq. Residues colored in blue/green are different from both Rotarix and RotaTeq, with exception of G9 and G12 strains in which green residues were different from all VP7 genotypes contained in RotaTeq. Amino acid changes that have been shown to escape neutralization with monoclonal antibodies (37) are indicated with a red triangle. (B) Surface representation of the VP7 trimer (PDB 3FMG). Antigenic epitopes are colored in red (7-1a), salmon (7-1b), and purple (7-2). Surface-exposed residues that differ between circulating strains in Belgium and the strains contained in Rotarix or RotaTeq are shown in cyan.
An analysis of the Belgian G1 strains showed that their VP7 antigenic epitopes contained up to five differences (BE1128) with the G1 strain of Rotarix and seven differences (BE1128) with the G1 strain (WI79-9) of RotaTeq. In both situations, most of the differences were located in the 7-1a epitope and, to a lesser extent, in the 7-2 epitope. Amino acids at positions 94, 123, 217, and 291 were the most likely to differ between Belgian G1 strains and the VP7 of Rotarix. In addition to these, the Belgian G1 strains were also likely to differ from the G1 VP7 of RotaTeq at residues 97 and 147. Mapping of the differences in the VP7 epitopes of the Belgian G1 strains and the vaccine strains onto the VP7 trimer revealed that they were distributed fairly heterogeneously across the face of the molecule (Fig. 3B). Of the two lineages (1 and 2) of VP7 proteins analyzed for the Belgian G1P[8] strains, the lineage 1 antigenic epitopes contained markedly more amino acid differences compared to the vaccine strains than those of the lineage 2 epitopes.
The VP7 antigenic epitopes of Belgian G2 strains were compared to the G2 strain (SC2-9) of RotaTeq and the G1 strain of Rotarix (Fig. 3A). The analysis showed that the Belgian G2 strains contained up to 5 amino acid differences (BE1251) with the G2 virus of RotaTeq and up to 19 differences (BE1248) with the G1 virus of Rotarix. Relative to the G2 virus of RotaTeq, the differences were most likely to occur in epitopes 7-1a and 7-1b, while relative to the G1 virus of Rotarix, the differences occurred predominantly in epitopes 7-1a and 7-2. Residues most likely to differ between the Belgian G2 strains and the G2 virus of RotaTeq were located at positions 87, 96, 125, 213, and 242. Residues that differ between the Belgian G2 strains and G2 virus of RotaTeq generally map to the edges of the VP7 trimer (Fig. 3B).
The VP7 epitopes of the Belgian G3P[8] strains (BE1214 and BE1259) contain 3 residues (212,238,242) that differ with those of the G3 strain of RotaTeq (WI78-9), and these are located exclusively within the 7-1b epitope. The Belgian G3P[6] strain (BE1322) has two additional changes (at positions 146 and 221), but these are located in the 7-2 epitope (Fig. 3A). Interestingly, the Belgian strains all include a K238N change, which creates a potential N-linked glycosylation site that is absent in the G3 strain of RotaTeq. Since the VP7-trimer surface N238 is surrounded by other residues of the 7-1a epitope (Fig. 3B), the glycosylation of N238 could have far ranging effects on the antigenicity of this epitope. The VP7 epitopes of the three Belgian G3 strains analyzed here contained 12 residues that differed from those of the G1 strain of Rotarix; the changes were divided equally among the 7-1a, 7-1b, and 7-2 epitopes (Fig. 3A).
The VP7 epitopes of the two Belgian G4 strains contained four (BE1113) or five (BE1129) amino acid differences with the epitopes of the G4 virus (BrB-9) of RotaTeq; these were present in the 7-1a, 7-1b, and 7-2 epitopes. In contrast, the VP7 epitopes of the Belgian G4 strains contained 15 differences with the epitopes of the G1 virus of Rotarix. These differences were mapped to all three VP7 epitopes and involved the majority of 7-1b and 7-2 residues (Fig. 3A).
The VP7 epitopes of the Belgian G9 and G12 virus strains were compared to those of the G1 to G4 and G6 VP7 proteins of RotaTeq and the G1 protein of Rotarix (Fig. 3A). This analysis showed that there were only two amino acids in the Belgian G9 VP7 epitopes that were not present in any of the VP7 epitopes of RotaTeq: residues 94 in 7-1a and 242 in 7-1b. In contrast to the relatively few amino acid differences noted between the VP7 epitopes of the Belgian G9 and strains of RotaTeq, there were as many as 14 differences found between Belgian G9 strains (BE1119) and the virus of Rotarix. These differences involved multiple residues in each of the 7-1a, 7-1b, and 7-2 epitopes.
In comparison to the VP7 proteins of the Belgian G9 strains, those of the Belgian G12 strains were more distantly related to the VP7 proteins of the vaccine strains (Fig. 1 and 2A). Consistent with this, the number of amino acid differences noted between the VP7 epitopes of the Belgian G12 and vaccine strains was high (Fig. 3A). In particular, the Belgian G12P[8] strain BE0085 contained 9 differences with the strains of RotaTeq, most mapping to the 7-1a antigenic epitope. The Belgian G12P[8] strains contained 16 differences with the strain of Rotarix; these involved numerous residues of each of the 7-1a, 7-1b, and 7-2 epitopes.
VP4 antigenic epitopes of circulating Belgian RVA strains and vaccine viruses.
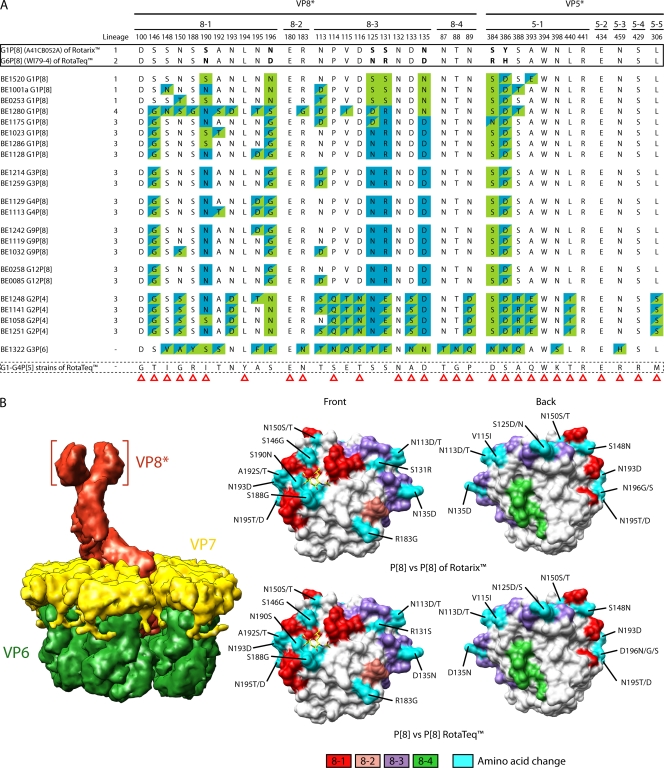
Activation of the VP4 spike protein requires its proteolytic cleavage into VP8* and VP5*. Structural studies have shown that VP8* forms a globular head that sits at the top of a stalk formed by VP5*. The VP8* head and VP5* stalk contain four (8-1 to 8-4) and five (5-1 to 5-5) surface-exposed antigenic epitopes, respectively, which have been predicted to include the 37 amino acids identified in Fig. 4. Among the Belgian RVA strains analyzed in the present study, all contained eight residues that were identical to those in the VP4 P[8] antigenic epitopes of RotaTeq and Rotarix. The VP4 proteins of the Belgian P[8] strains mostly belonged to P[8] lineage 3 but also included a few in lineages 1 and 4. In general, the epitopes of the Belgian P[8] lineage 3 strains showed more differences with the P[8] lineage 1 epitopes of Rotarix (6–9 differences per strain) than with the P[8] lineage 2 epitopes of RotaTeq (4–6 differences per strain). The differences between the Belgian P[8] lineage 3 strains and Rotarix were mostly contained in VP8* epitopes 8-1 and 8-3, while with RotaTeq, the differences were mostly contained in VP8* epitope 8-1 and VP5* epitope 5-1. Not unexpectedly, Belgian P[8] lineage 1 epitopes were much more similar to the P[8] lineage 1 epitopes of Rotarix than to the P[8] lineage 2 epitopes of RotaTeq. The P[8] lineage 4 (OP354-like) epitopes of the Belgian strain BE1280 showed the greatest divergence (16 amino acid changes) from the VP4 P[8] epitopes of the vaccine viruses.
Fig 4.
(A) Alignment of antigenic residues in VP4 between the strains contained in Rotarix and RotaTeq and strains circulating in Belgium. Antigenic residues are divided in three antigenic epitopes in VP8* and five antigenic epitopes in VP5*. Residues are color coded similar as described for Fig. 3. Amino acid changes that have been shown to escape neutralization with monoclonal antibodies (17) are indicated with a red triangle. (B) Surface representation of the VP8* core (PDB 1KQR). The left images show VP8* from the front. The right images are rotated 180° compared to the left images and show VP8* from the back. Antigenic epitopes are colored in red (8-1), pink (8-2), purple (8-3), and green (8-4). Surface-exposed residues that differ between circulating strains in Belgium and the strains contained in Rotarix and RotaTeq are shown in cyan. The N-acetylneuraminic (sialic) acid binding site (17) is indicating by the yellow stick model of a sialoside.
The VP4 antigenic epitopes of the Belgian P[4] strains showed large numbers of differences with vaccine P[8] VP4 epitopes. Specifically, the Belgian P[4] strains contained 16 to 19 residues that differed with the P[8] epitopes of Rotarix and 16 to 18 residues that differed with the P[8] epitopes of RotaTeq (Fig. 4A). Of the amino acid differences, most were located in the VP8* 8-1 and 8-3, and VP5* 5-1 epitopes. The VP4 antigenic epitopes of the Belgian P[6] strain BE1322 showed the most divergence from those of the vaccine strains, with 22 differences noted with P[8] epitopes of Rotarix and 24 differences noted with P[8] epitopes of RotaTeq (Fig. 4A). The differences were concentrated in VP8* 8-1, 8-3, and 8-4, and VP5* 5-1 epitopes (16, 17).
DISCUSSION
In the present study, we contrasted the VP7 and VP4 antigenic epitopes of the two licensed, and available, RVA vaccines in Belgium with those of the currently circulating RVAs in Belgium. Rotarix was introduced in Belgium in June 2006, while RotaTeq became available 1 year later, in June 2007, and quickly thereafter a very high RVA vaccine coverage was reached, which is estimated at 88% (67). Although both vaccines are eligible for administration, Rotarix is used in Belgium most commonly (ca. 80%). By analyzing the ORFs of VP7 and VP4, we identified possible important antigenic disparities between the vaccine strains and Belgian strains.
For VP7 and VP4, significant intragenotypic differences were found with both RVA vaccines. In particular, the G1 and P[8] genotypes displayed a large intragenotypic variety and the G1 and P[8] genotypes of Rotarix and RotaTeq were located in different lineages. Most G1 strains circulating in Belgium belong to lineage 1 (>95%), which is the most distinct G1 lineage compared to the G1 lineage of Rotarix and RotaTeq. Up to seven amino acid differences were found when we compared the VP7 of Belgian G2 strains to the G2 strain of RotaTeq. Since G2 RVA strains are generally associated with P[4] genotypes, protection against G2 RVA strains afforded by RotaTeq would mainly depend on the G2 component of the vaccine. As a consequence, antigenic drift of G2 strains could reduce the vaccine efficacy against G2 strains much faster than against any other of the major human RVA genotypes.
The VP7 of the Belgian G3 strains showed relatively few differences with the VP7 of the G3 strain in RotaTeq but possessed an additional glycosylation site in epitope 7-1a. Analysis of the structural model of VP7 showed that glycosylation of residue 238 could have far-ranging effects on the immunogenicity of the 7-1a epitope. In fact, glycosylation of residue 238 has previously been shown to reduce neutralization of animal RVA strains by hyperimmune sera and monoclonal antibodies (7, 8). Furthermore, glycosylation of viral proteins has been shown to alter immunogenicity dramatically for a number of other viruses, including influenza A virus, human immunodeficiency virus, and human respiratory syncytial virus (41, 54, 58).
Epidemiological data from Australia collected during the “post-vaccine introduction era” suggest a possible rise of G3 strains in Australian states where RotaTeq has been used, although the prevalence of G3 genotypes has declined to 12% in the 2009–2010 season (26, 27). Furthermore, in the United States, the G3 genotype was the dominant genotype in some seasons after RotaTeq was introduced (5, 10, 23). Unfortunately, no sequence information is yet available on these G3 strains to evaluate the presence or absence of the potential glycosylation site at residue 238 of these G3 strains. Although the relative increase of G3 strains in countries where mainly RotaTeq is used could also be the result of the low seroresponse rate to the G3 strain of RotaTeq (62), an extra glycosylation site within an antigenic epitope is another possibility which needs to be further investigated.
For VP4, most variation was observed in P[8] strains. RotaTeq and Rotarix clustered in P[8] lineage 2 and P[8] lineage 1, respectively. The majority of the P[8] strains clustered in P[8] lineage 3, distantly from the P[8] of Rotarix and RotaTeq (66). The Belgian strain BE1280, which clustered in P[8] lineage 4 (also known as “OP354-like”), was distantly related to both RVA vaccine strains. BE1280 clustered closely with previously characterized RVA strains from Bangladesh (39) and showed a remarkable amount of differences in antigenic epitopes with other P[8] lineages. OP354-like P[8] strains are increasingly reported and have been found in combination with various G genotypes throughout Eurasia and Africa (11, 25, 39, 40, 52). Only limited research on the antigenic properties of OP354-like strains has been conducted to date, but neutralization plaque assays performed with antibodies directed against VP5* indicate that the antigenic properties of OP354-like strains appear to be similar as P[8] lineage 3 strains (39). However, most of the genetic variation is found in the VP8* cleavage fragment of VP4 so the overall picture of the antigenicity of OP354-like strains could be a different one.
In the present study, we characterized one G1P[8] strain (BE1520) that clustered very closely to both the VP7 and VP4 of Rotarix G1P[8]. After inquiry with the patient's physician, it was concluded that the patient was vaccinated with Rotarix 69 days before the sample was isolated. Shedding of rotavirus has been documented at least 15 days after Rotarix administration (48) and up to 57 days after hospital admission due to rotavirus gastroenteritis (50) so, possibly, this strain is vaccine derived. Interestingly, it carried some amino acid changes in both VP7 and VP4. The VP7 of strain BE1520 carried one amino acid substitution outside known antigenic epitopes compared to the VP7 of Rotarix: M202→T. However, the VP4 of strain BE1520 had 3 amino acid changes compared to that of Rotarix, located in VP8* (F168→L) and VP5* (Y386→D and A393→E). Residue 168 is not located in an antigenic epitope, whereas both residues 386 and 393 are located in antigenic epitope 5-1 (Fig. 4A). Besides the aforementioned changes, we found another silent nucleotide change (C1524→T) in VP4 and one change in the 5′end noncoding region of VP4: 5A→T. If strain BE1520 is vaccine derived, these amino acid changes could have emerged due to acquired mutations or, alternatively, could be the results of an in vivo selection of minor variants already present in the vaccine. Further studies encompassing all RVA gene segments should be conducted to confirm whether strain BE1520 is vaccine derived. Unfortunately, no further details about the patient could be retrieved, but it is highly unlikely that the possible vaccine strain would be the cause of disease, since it was administered 69 days earlier.
In countries where both RVA vaccines are eligible to be routinely administered, the antigenic landscape is being formed by a complex mixture of selection pressures of both vaccines. For example, P[8] lineage 3 RVA strains are relatively divergent from Rotarix, which clusters within the P[8] lineage 1, but relatively closely related to P[8] lineage 2 of RotaTeq. Genetically, and likely antigenically, P[8] lineage 4 strains are the most divergent from both vaccines. Until now Rotarix is by far the most used vaccine in Belgium and may thus predominantly be shaping the antigenic landscape. An interesting point in this regard is that after the introduction of Rotarix in Belgium, the incidence of G1P[8] strains similar to Rotarix has declined more than the G1P[8] strains belonging to G1 and P[8] lineages that are less related to Rotarix (66).
From the structural analyses conducted in these studies, it seems that some amino acid differences were found at more exposed residues than others, and this could influence their role in antigenicity. However, the precise impact of amino acid changes in the antigenic epitopes cannot be predicted from sequence and structural information alone. A few studies have attempted to correlate intragenotypic nucleotide differences with antigenic differences. For example, Jin et al. showed that antisera raised against a G1 lineage 3 strain neutralized G1 lineage 3 strains more efficiently than G1 lineage 2 strains (24). In another study, Hoshino et al. showed that antisera raised against G9 lineage 1 strains have a broad neutralizing ability against RVAs of all three G9 lineages, while antisera raised against G9 lineage 2 or G9 lineage 3 strains have a lower neutralizing ability to RVAs of other G9 lineages (21). However, more of these studies are needed to obtain a detailed antigenic map of RVA, as has been previously done for influenza A virus and enterovirus 71 (22, 55).
Moreover, since other gene segments are involved in immunity as well (3), complete genome sequencing of RVAs should be applied when possible vaccine escape mutants would emerge. Nonetheless, the present study provides important findings of genetic and possible antigenic differences between circulating RVA strains several years after vaccine introduction in Belgium compared to both RVA vaccines, Rotarix and RotaTeq.
Supplementary Material
ACKNOWLEDGMENTS
M.Z. was supported by the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT Vlaanderen). J.M. was supported by an FWO (Fonds voor Wetenschappelijk Onderzoek) postdoctoral fellowship. J.T.P. was supported by the Intramural Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print 21 December 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Aoki ST, et al. 2009. Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science 324:1444–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arista S, et al. 2006. Heterogeneity and temporal dynamics of evolution of G1 human rotaviruses in a settled population. J. Virol. 80:10724–10733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ball JM, Tian P, Zeng CQ, Morris AP, Estes MK. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272:101–104 [DOI] [PubMed] [Google Scholar]
- 4. Banyai K, et al. 2009. Trends in the epidemiology of human G1P[8] rotaviruses: a Hungarian study. J. Infect. Dis. 200(Suppl 1):S222–S227 [DOI] [PubMed] [Google Scholar]
- 5. Boom JA, et al. 2010. Effectiveness of pentavalent rotavirus vaccine in a large urban population in the United States. Pediatrics 125:e199–e207 [DOI] [PubMed] [Google Scholar]
- 6. Buttery JP, et al. 2011. Reduction in rotavirus-associated acute gastroenteritis following introduction of rotavirus vaccine into Australia's National Childhood vaccine schedule. Pediatr. Infect. Dis. J. 30:S25–S29 [DOI] [PubMed] [Google Scholar]
- 7. Ciarlet M, Hoshino Y, Liprandi F. 1997. Single point mutations may affect the serotype reactivity of serotype G11 porcine rotavirus strains: a widening spectrum? J. Virol. 71:8213–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ciarlet M, Reggeti F, Pina CI, Liprandi F. 1994. Equine rotaviruses with G14 serotype specificity circulate among Venezuelan horses. J. Clin. Microbiol. 32:2609–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ciarlet M, Schodel F. 2009. Development of a rotavirus vaccine: clinical safety, immunogenicity, and efficacy of the pentavalent rotavirus vaccine, RotaTeq. Vaccine 27(Suppl 6):G72–G81 [DOI] [PubMed] [Google Scholar]
- 10. Clark HF, Lawley D, Mallette LA, DiNubile MJ, Hodinka RL. 2009. Decline in cases of rotavirus gastroenteritis presenting to The Children's Hospital of Philadelphia after introduction of a pentavalent rotavirus vaccine. Clin. Vaccine Immunol. 16:382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cunliffe NA, et al. 2001. Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999. J. Clin. Microbiol. 39:836–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Curns AT, et al. 2010. Reduction in acute gastroenteritis hospitalizations among US children after introduction of rotavirus vaccine: analysis of hospital discharge data from 18 US states. J. Infect. Dis. 201:1617–1624 [DOI] [PubMed] [Google Scholar]
- 13. de Palma O, et al. 2010. Effectiveness of rotavirus vaccination against childhood diarrhoea in El Salvador: case-control study. BMJ 340:c2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Vos B, et al. 2009. Live attenuated human rotavirus vaccine, RIX4414, provides clinical protection in infants against rotavirus strains with and without shared G and P genotypes: integrated analysis of randomized controlled trials. Pediatr. Infect. Dis. J. 28:261–266 [DOI] [PubMed] [Google Scholar]
- 15. DiStefano DJ, et al. 2005. Novel rotavirus VP7 typing assay using a one-step reverse transcriptase PCR protocol and product sequencing and utility of the assay for epidemiological studies and strain characterization, including serotype subgroup analysis. J. Clin. Microbiol. 43:5876–5880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dormitzer PR, Nason EB, Prasad BV, Harrison SC. 2004. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 430:1053–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dormitzer PR, Sun ZY, Wagner G, Harrison SC. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Estes M, Kapikian A. 2007. Rotaviruses, p 1917–1974 In Knipe M, Howley P. (ed), Fields virology, 5th ed Lippincott/Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 19. Goveia MG, Ciarlet M, Owen KE, Ranucci CS. 2011. Development, clinical evaluation, and post-licensure impact of RotaTeq®, a pentavalent rotavirus vaccine. Ann. N. Y. Acad. Sci. 1222:14–18 [DOI] [PubMed] [Google Scholar]
- 20. Heaton PM, Ciarlet M. 2007. Vaccines: the pentavalent rotavirus vaccine: discovery to licensure and beyond. Clin. Infect. Dis. 45:1618–1624 [DOI] [PubMed] [Google Scholar]
- 21. Hoshino Y, et al. 2004. Rotavirus serotype G9 strains belonging to VP7 gene phylogenetic sequence lineage 1 may be more suitable for serotype G9 vaccine candidates than those belonging to lineage 2 or 3. J. Virol. 78:7795–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang SW, et al. 2009. Reemergence of enterovirus 71 in 2008 in Taiwan: dynamics of genetic and antigenic evolution from 1998 to 2008. J. Clin. Microbiol. 47:3653–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hull JJ, et al. 2011. United States rotavirus strain surveillance from 2005 to 2008: genotype prevalence before and after vaccine introduction. Pediatr. Infect. Dis. J. 30:S42–S47 [DOI] [PubMed] [Google Scholar]
- 24. Jin Q, et al. 1996. Divergence of VP7 genes of G1 rotaviruses isolated from infants vaccinated with reassortant rhesus rotaviruses. Arch. Virol. 141:2057–2076 [DOI] [PubMed] [Google Scholar]
- 25. Khananurak K, et al. 2010. Prevalence and phylogenetic analysis of rotavirus genotypes in Thailand between 2007 and 2009. Infect. Genet. Evol. 10:537–545 [DOI] [PubMed] [Google Scholar]
- 26. Kirkwood CD, Boniface K, Barnes GL, Bishop RF. 2011. Distribution of rotavirus genotypes after introduction of rotavirus vaccines, Rotarix® and RotaTeq®, into the National Immunization Program of Australia. Pediatr. Infect. Dis. J. 30:S48–S53 [DOI] [PubMed] [Google Scholar]
- 27. Kirkwood CD, Boniface K, Bishop RF, Barnes GL. 2011. Australian Rotavirus Surveillance Program: annual report, 2009/2010. Commun. Dis. Intell. 34:427–434 [DOI] [PubMed] [Google Scholar]
- 28. Lanzieri TM, et al. 2010. Trends in hospitalizations from all-cause gastroenteritis in children younger than 5 years of age in Brazil before and after human rotavirus vaccine introduction, 1998–2007. Pediatr. Infect. Dis. J. 29:673–675 [DOI] [PubMed] [Google Scholar]
- 29. Matthijnssens J, et al. 2009. Rotavirus disease and vaccination: impact on genotype diversity. Future Microbiol. 4:1303–1316 [DOI] [PubMed] [Google Scholar]
- 30. Matthijnssens J, et al. 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 82:3204–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matthijnssens J, et al. 2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 156:1397–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matthijnssens J, et al. 2008. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 153:1621–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matthijnssens J, et al. 2010. Phylodynamic analyses of rotavirus genotypes G9 and G12 underscore their potential for swift global spread. Mol. Biol. Evol. 27:2431–2436 [DOI] [PubMed] [Google Scholar]
- 34. Matthijnssens J, et al. 2010. Molecular and biological characterization of the 5 human-bovine rotavirus (WC3)-based reassortant strains of the pentavalent rotavirus vaccine, RotaTeq. Virology 403:111–127 [DOI] [PubMed] [Google Scholar]
- 35. Matthijnssens J, Rahman M, Ciarlet M, Van Ranst M. 2008. Emerging human rotavirus genotypes, p 171–219 In Palombo KC. (ed), Viruses in the environment. Research Signpost, Trivandrum, India [Google Scholar]
- 36. Matthijnssens J, et al. 2006. Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission. J. Virol. 80:3801–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McDonald SM, et al. 2009. Evolutionary dynamics of human rotaviruses: balancing reassortment with preferred genome constellations. PLoS Pathog. 5:e1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Molto Y, et al. 2011. Reduction of diarrhea-associated hospitalizations among children aged < 5 Years in Panama following the introduction of rotavirus vaccine. Pediatr. Infect. Dis. J. 30:S16–S20 [DOI] [PubMed] [Google Scholar]
- 39. Nagashima S, et al. 2009. Characterization of full-length VP4 genes of OP354-like P[8] human rotavirus strains detected in Bangladesh representing a novel P[8] subtype. Arch. Virol. 154:1223–1231 [DOI] [PubMed] [Google Scholar]
- 40. Nguyen TA, et al. 2008. Use of sequence analysis of the VP4 gene to classify recent Vietnamese rotavirus isolates. Clin. Microbiol. Infect. 14:235–241 [DOI] [PubMed] [Google Scholar]
- 41. Palomo C, Cane PA, Melero JA. 2000. Evaluation of the antibody specificities of human convalescent-phase sera against the attachment (G) protein of human respiratory syncytial virus: influence of strain variation and carbohydrate side chains. J. Med. Virol. 60:468–474 [PubMed] [Google Scholar]
- 42. Parashar UD, et al. 2009. Global mortality associated with rotavirus disease among children in 2004. J. Infect. Dis. 200(Suppl 1):S9–S15 [DOI] [PubMed] [Google Scholar]
- 43. Parashar UD, Gibson CJ, Bresse JS, Glass RI. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12:304–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Parra GI, Bok K, Martinez V, Russomando G, Gomez J. 2005. Molecular characterization and genetic variation of the VP7 gene of human rotaviruses isolated in Paraguay. J. Med. Virol. 77:579–586 [DOI] [PubMed] [Google Scholar]
- 45. Patel M, et al. 2009. Association between pentavalent rotavirus vaccine and severe rotavirus diarrhea among children in Nicaragua. JAMA 301:2243–2251 [DOI] [PubMed] [Google Scholar]
- 46. Paulke-Korinek M, Rendi-Wagner P, Kundi M, Kronik R, Kollaritsch H. 2009. Universal mass vaccination against rotavirus gastroenteritis: impact on hospitalization rates in Austrian children. Pediatr. Infect. Dis. J. 29:319–323 [DOI] [PubMed] [Google Scholar]
- 47. Pettersen EF, et al. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 48. Phua KB, et al. 2005. Evaluation of RIX4414, a live, attenuated rotavirus vaccine, in a randomized, double-blind, placebo-controlled phase 2 trial involving 2464 Singaporean infants. J. Infect. Dis. 192(Suppl 1):S6–S16 [DOI] [PubMed] [Google Scholar]
- 49. Rahman M, et al. 2007. Evolutionary history and global spread of the emerging g12 human rotaviruses. J. Virol. 81:2382–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Richardson S, et al. 1998. Extended excretion of rotavirus after severe diarrhoea in young children. Lancet 351:1844–1848 [DOI] [PubMed] [Google Scholar]
- 51. Ruiz-Palacios GM, et al. 2006. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N. Engl. J. Med. 354:11–22 [DOI] [PubMed] [Google Scholar]
- 52. Samajdar S, et al. 2008. Human group A rotavirus P[8] Hun9-like and rare OP354-like strains are circulating among diarrhoeic children in Eastern India. Arch. Virol. 153:1933–1936 [DOI] [PubMed] [Google Scholar]
- 53. Santos N, Hoshino Y. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 15:29–56 [DOI] [PubMed] [Google Scholar]
- 54. Skehel JJ, et al. 1984. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc. Natl. Acad. Sci. U. S. A. 81:1779–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smith DJ, et al. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376 [DOI] [PubMed] [Google Scholar]
- 56. Soares-Weiser K, et al. 2010. Vaccines for preventing rotavirus diarrhoea: vaccines in use. Cochrane Database Syst. Rev. 2010:CD008521. [DOI] [PubMed] [Google Scholar]
- 57. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software, version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 58. Utachee P, et al. 2010. Two N-linked glycosylation sites in the V2 and C2 regions of human immunodeficiency virus type 1 CRF01_AE envelope glycoprotein gp120 regulate viral neutralization susceptibility to the human monoclonal antibody specific for the CD4 binding domain. J. Virol. 84:4311–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vesikari T, Karvonen A, Ferrante SA, Ciarlet M. 2010. Efficacy of the pentavalent rotavirus vaccine, RotaTeq®, in Finnish infants up to 3 years of age: the Finnish Extension Study. Eur. J. Pediatr. 169:1379–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vesikari T, Karvonen A, Ferrante SA, Kuter BJ, Ciarlet M. 2010. Sustained efficacy of the pentavalent rotavirus vaccine, RV5, up to 3.1 years following the last dose of vaccine. Pediatr. Infect. Dis. J. 29:957–963 [DOI] [PubMed] [Google Scholar]
- 61. Vesikari T, et al. 2007. Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: randomised, double-blind controlled study. Lancet 370:1757–1763 [DOI] [PubMed] [Google Scholar]
- 62. Vesikari T, et al. 2006. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N. Engl. J. Med. 354:23–33 [DOI] [PubMed] [Google Scholar]
- 63. Ward R. 2009. Mechanisms of protection against rotavirus infection and disease. Pediatr. Infect. Dis. J. 28:S57–S59 [DOI] [PubMed] [Google Scholar]
- 64. Ward RL. 2008. Rotavirus vaccines: how they work or don't work. Expert Rev. Mol. Med. 10:e5. [DOI] [PubMed] [Google Scholar]
- 65. Ward RL, Bernstein DI. 2009. Rotarix: a rotavirus vaccine for the world. Clin. Infect. Dis. 48:222–228 [DOI] [PubMed] [Google Scholar]
- 66. Zeller M, Matthijnssens J, Rahman M, Van Ranst M. 2009. Possible immune evasion by G1 lineage I after vaccine introduction in Belgium? Third European Rotavirus Biology Meeting, Loch Lomond, Scotland [Google Scholar]
- 67. Zeller M, et al. 2010. Rotavirus incidence and genotype distribution before and after national rotavirus vaccine introduction in Belgium. Vaccine 28:7507–7513 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.