Abstract
Drug resistance surveillance and strain typing of Mycobacterium leprae are necessary to investigate ongoing transmission of leprosy in regions of endemicity. To enable wider implementation of these molecular analyses, novel real-time PCR–high-resolution melt (RT-PCR-HRM) assays without allele-specific primers or probes and post-PCR sample handling were developed. For the detection of mutations within drug resistance-determining regions (DRDRs) of folP1, rpoB, and gyrA, targets for dapsone, rifampin, and fluoroquinolones, real-time PCR-HRM assays were developed. Wild-type and drug-resistant mouse footpad-derived strains that included three folP1, two rpoB, and one gyrA mutation types in a reference panel were tested. RT-PCR-HRM correctly distinguished the wild type from the mutant strains. In addition, RT-PCR-HRM analyses aided in recognizing samples with mixed or minor alleles and also a mislabeled sample. When tested in 121 sequence-characterized clinical strains, HRM identified all the folP1 mutants representing two mutation types, including one not within the reference panel. The false positives (<5%) could be attributed to low DNA concentration or PCR inhibition. A second set of RT-PCR-HRM assays for identification of three previously reported single nucleotide polymorphisms (SNPs) that have been used for strain typing were developed and validated in 22 reference and 25 clinical strains. Real-time PCR-HRM is a sensitive, simple, rapid, and high-throughput tool for routine screening known DRDR mutants in new and relapsed cases, SNP typing, and detection of minor mutant alleles in the wild-type background at lower costs than current methods and with the potential for quality control in leprosy investigations.
INTRODUCTION
Leprosy is an infectious disease of skin and nerves caused by Mycobacterium leprae. The disease remains endemic in many parts of the world and is now listed as a neglected tropical disease (27) by the World Health Organization. The drug dapsone was introduced in 1950 and was administered in the form of long-term monotherapy for treatment of leprosy; unfortunately, drug resistance emerged during the 1960s and 1970s (4). For this reason, in 1982, the World Health Organization (WHO) formally recommended multidrug therapy (MDT) which includes dapsone, rifampin, and clofazimine for the treatment and control of multibacillary (MB) leprosy (43). Sporadic reports of clinical resistance to dapsone and rifampin started appearing in several countries such as Vietnam, Mexico, India, and Philippines (1, 8, 13, 19, 24, 25, 26). Noncompliance and inadequate therapy may be the causes of the clinical resistance, particularly for MB leprosy. The drug targets and the mutations in the coding genes folP1, rpoB, and gyrA that lead to clinical resistance to dapsone, rifampin, and the fluoroquinolones (used in an alternative leprosy drug regimen), respectively, have been identified and characterized (5, 10, 14, 39). The in vivo drug susceptibility or resistance phenotypes of various mutations seen in clinical strains in patient skin biopsy specimens have been determined empirically by the traditional mouse footpad (MFP) inoculation assays (3, 20, 34). Mice are given drugs (in diet or by gavage) at different concentrations, and bacterial growth in the footpads is measured at different time points. These assays have corroborated the hypothesis that clinical resistance to rifampin, dapsone, and oxfloxacin highly correlates (almost always) with detection of specific mutations within the M. leprae rpoB, folP1, and gyrA genes (21). For M. leprae, mutations have been reported in one or several codons located within short DNA regions in each of the target genes, thus defining the drug resistance-determining regions (DRDRs). Although it is possible that there are mutations outside the DRDRs of rpoB, folP1, and gyrA or in other genes or that alternative mechanisms of resistance exist, these have yet to be identified. The MFP assays are labor, time, and cost intensive; moreover, the results are not available in time to influence treatment options. Therefore, the MFP methods have given way to molecular methods for screening proven resistance-related mutations (42).
Furthermore, despite global MDT programs, the new case detection rates have not declined as expected in many of the countries of high endemicity (41), which indicates continued transmission of the pathogen. In 2001, the first reference M. leprae genome of the TN strain from a Tamil Nadu, India, leprosy patient was sequenced, offering new insight and opportunities for development of tools in investigating bacteriology, pathogenesis, and epidemiology. Mapping polymorphic loci, such as variable-number tandem repeats (VNTRs) (7, 15) and single nucleotide polymorphisms (SNPs), has applications in strain typing for tracing transmission of leprosy. Four M. leprae lineages (SNP types 1, 2, 3, and 4) have been described on the basis of unique haplotypes derived from three SNPs that were identified by comparative genome sequencing following the availability of the TN strain genome sequence (28, 29).
Although it has become possible to perform amplification of the target loci by PCR followed by DNA sequencing of the amplicons for detecting genetic variants (10, 11, 12, 32, 38, 40), the labor and costs involved in PCR-DNA sequencing are still limiting factors for routine drug resistance surveillance and SNP strain typing. Several surrogate methods, using techniques that involve single-strand conformation polymorphism (SSCP), reverse hybridization on membranes, or microarrays, have been developed for allele-specific detection of DRDR mutations from clinical specimens (23, 33), which require dedicated reagents or kits. PCR-restriction fragment length polymorphism (PCR-RFLP) assays developed by us expedited SNP typing by eliminating the sequencing steps and replacing them with conventional DNA electrophoresis for distinguishing the variants (31). DRDRs are not suitable for PCR-RFLP, as there are different mutations at one or more positions. Numerous PCR assays exist for allele discrimination, but such assays often require multiple PCRs, allele target-specific primers, or additional expensive probes (35).
In this context, we exploited the emerging real-time PCR technologies that can eliminate post-PCR procedures for genotyping any M. leprae genomic target of interest, particularly those suitable for leprosy epidemiology applications. Real-time PCR–high-resolution melt (PCR-HRM) analysis is a novel simple post-PCR step that exploits thermal characteristics of the amplicons for detection of sequence variants. This report describes the method of development and validation of real-time PCR-HRM assays for two applications: global drug resistance surveillance and SNP-based strain typing of M. leprae. These are referred to as DRDR and SNP typing assays in this report. The technical and practical considerations related to the two methods and the advantages and current limitations are discussed in the context of leprosy.
MATERIALS AND METHODS
M. leprae reference and clinical specimens.
M. leprae clinical strains maintained in mouse footpad (MFP) systems and in armadillo animal systems were utilized as reference strains. Eighteen MFP strains were obtained from the Leprosy Research Centre (LRC), National Institute of Infection Diseases, Tokyo, Japan (22); those strains are therefore designated MFP-LRC in this report. Suspensions of bacilli (108) were preserved in 1 ml of 70% ethanol. The armadillo-derived M. leprae (ADML) clinical strains NHDP63, Br4923, Thai-53, and 3039/210 obtained from infected tissues have been described previously (15).
The clinical skin biopsy samples used in this study were obtained from patients presenting at the Cebu Skin Clinic, Leonard Wood Memorial Leprosy Research Centre, Philippines, as previously described (n = 121) (18, 31) and at the Anandaban Hospital, Kathmandu, Nepal (n = 25; collected during 2000 to 2010). The Philippine samples were stored in 70% ethanol at the time of collection. The Nepal samples were obtained from a repository of homogenized skin biopsy specimens stored frozen in phosphate saline buffer containing 0.1% bovine serum albumin (BSA). An aliquot of the homogenate was transferred to a fresh vial and suspended in 70% ethanol. All of the procedures involving biological sample collections and testing were performed following approval from the governing human research ethical committees and informed consent procedures as necessary.
DNA extraction from reference M. leprae cells and clinical tissue specimens.
As template DNA quality affects the amplification and the analysis by HRM, all the DNAs were prepared under uniform conditions using a DNeasy tissue kit (Qiagen, Valencia, CA) as described previously (15). The method involves proteinase K digestion and spin column chromatography. DNA was eluted in 100 to 200 μl of the AE buffer provided in the kit. For use as DNA concentration standards and as reference genotypes for the DRDR and SNP typing assays, approximately 1 mg of purified cells of NHDP63 M. leprae purified from infected armadillo tissues as described previously were processed using the DNeasy tissue kit. These cells were prepared and obtained via the Leprosy Research Support-NIH-NIAID contract at Colorado State University. The DNA was eluted in AE elution buffer. The DNA concentration was estimated by measuring UV light absorbance at a wavelength of 260 nm. An aliquot of the stock DNA was adjusted with AE buffer to a final working stock of 10 μg/ml, from which the DNA standards (1 ng/μl, 100 pg/μl, 10 pg/μl, 1 pg/μl, and 0.1pg/μl) were prepared by 10-fold serial dilutions in AE buffer. The DNA derived from MFP-LRC M. leprae (∼108 cells) was diluted 1:100 in AE buffer; 1 μl of DNA was typically sufficient for one PCR.
Primers for real-time PCR-HRM DRDR and SNP typing assays.
Primer sets used for amplification of the DRDRs in the rpoB, folP1, and gyrA genes for real-time PCR-HRM are shown in Fig. 1. The primers for the SNP typing assays are shown in Table 1. The primer sequences and nucleotide numbering system refer to those of the genome sequence of the TN strain (http://genolist.pasteur.fr/Leproma/). Primers for target loci were designed per the recommended amplicon size (less than 200 bp) for optimal real-time PCR-HRM genotyping using the freely available software Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast) (20, 37). The target sequences were analyzed by MFOLD software to select the products with the least secondary structure (http://mfold.rna.albany.edu/?q = mfold) (45).
Fig 1.
The M. leprae targets of dapsone (ML0224, folP1), rifampin (ML1891c, rpoB), and fluoroquinolones (ML0006, gyrA). The partial nucleotide (upper) and corresponding amino acid (lower) sequences containing the drug resistance-determining regions (DRDR) of the target genes are presented. The nucleotides and the amino acid numbers are shown with reference to the open reading frames of the genes for the M. leprae TN strain as found in the Leproma website (http://genolist.pasteur.fr/Leproma/). The codons/amino acids implicated in drug resistance are shown within boxes. The primer sequences selected for real-time PCR-HRM are underlined.
Table 1.
Primer sequences for SNP typing by real-time PCR-HRM analysis
| SNP target and locationa,b | Primer name | Primer locationb | Primer sequence (5′–3′) |
|---|---|---|---|
| Locus 1: 14,676 | HRM14F | 14601–14621 | TGAACAGTCTCGTAACCGTG |
| HRMM14Ra | 14721–14701 | CAATGCATGCTAGCCTTAATG | |
| Locus 2: 1,642,875 | HRM16F | 1642813–1642836 | CTCGTCACAAATCCGAGTTTGAAT |
| HRM16R | 1642925–1642902 | GTAGTAGTCTTCCAAGTTGTGGTG | |
| Locus 3: 2,935,685 | HRM29F | 2935599–2935616 | TGGTGTCGGTCTCCATCC |
| HRM29Ra | 2935716–2935699 | ACCGGTGAGCGCACTAAG |
Data are from the study by Monot et al. (28).
Data correspond to the M. leprae TN genome sequence (http://genolist.pasteur.fr/Leproma/).
Real-time PCR conditions and DNA template quantitation.
The real-time PCR (20 μl) was composed of 10 μl of 2× Precision Melt Supermix (Bio-Rad Laboratories, Hercules, CA), forward and reverse primers (0.5 μl each of 10 μM working stocks was used for five of the PCR amplicons; for rpoB, 0.4 μl of each primer was used), nuclease-free water (8 or 8.2 μl), and DNA extracted from clinical and reference materials (1 μl). The reactions were set up in triplicate in a 96-well PCR plate and processed on a CFX96 real-time PCR system (Bio-Rad). CFX Manager software (Bio-Rad) was utilized to set up the sample arrangement on the PCR plate, to define PCR conditions, to monitor the amplification in real time, to view melting curves, and to calculate DNA concentrations and other PCR parameters.
The cycling parameters of PCR were as follows: 95°C for 2 min followed by 45 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s and then a hetero-duplex formation step, including 95°C for 10 s and 60°C for 1 min. After the PCR amplification steps, melt curves for the products were generated by heating in 0.2°C increments at a rate of 10 s/step for the temperature range 65 to 95°C.
DNA quantitation of templates was based on a standard curve developed with the five NHDP63 standards, each tested in triplicate for each target. The results were presented as cycle threshold (CT) values and starting quantity (SQ).
Real-time PCR-HRM analysis for cluster detection.
Post-PCR-HRM analyses of the melt curves were performed using Precision Melt Analysis software (Bio-Rad), which analyzes the temperature and shapes of the melting curves. The software classifies the data into different clusters. Data that are similar are “clustered” by the software and assigned a cluster number. The melt curves corresponding to each cluster are color coded for easy visualization. For each of the three targets, in DRDR (folp1, rpoB, and gyrA) and SNP typing (locus 1, 2, and 3) assays, NHDP63 DNA was used to select the reference cluster. Clustering results are influenced by the melt region selected and cluster detection settings. The melt region can be autodetected or manually defined by selecting the pre- and postmelt temperature ranges. The cluster detection settings include melt curve shape sensitivity (default value of 50% clustering) and melting temperature (Tm) difference threshold (default of 0.15 degrees). These settings can be adjusted to determine the stringency of the clusters. The instrument and software manual indicates that for most applications, default settings produce acceptable results; these were used as the starting points for analyzing melt curves obtained for each of the six amplicons and mutation types within the reference and clinical samples.
RESULTS
Development of DRDR assays based on real-time PCR-HRM analysis and assay validation with reference specimens.
First, the performance characteristics of the real-time PCR assays using the primers, reagents, and reaction conditions were investigated using NHDP63 DNA. As shown in Table 2, it can be seen that all three DRDRs were reliably amplified using the primer sequences shown in Fig. 1, with high-quality performance values for PCR efficiency and correlation of determination (see Table 2 footnote). A standard DNA curve (0.1 to 1,000 pg) determined using NHDP63 DNA was established to enable the estimation of the M. leprae DNA levels in test samples and also used for assay evaluation. The corresponding CT values ranged from 39 to 22. The cycle quantification CT values obtained for the three DRDRs are comparable.
Table 2.
Comparison of RT-PCR CT values and estimates of starting quantities
| Sample typea | Strain (quantity) | DRDRb |
|||||
|---|---|---|---|---|---|---|---|
|
folP1 |
rpoB |
gyrA |
|||||
| CT | SQ (pg) | CT | SQ (pg) | CT | SQ (pg) | ||
| ADML | NHDP63 (1 ng) | 22.02 | 1,000.00 | 23.61 | 1,000.00 | 22.87 | 1,000.00 |
| NHDP63 (100 pg) | 25.36 | 100.00 | 27.41 | 100.00 | 26.41 | 100.00 | |
| NHDP63 (10 pg) | 28.83 | 10.00 | 31.37 | 10.00 | 30.28 | 10.00 | |
| NHDP63 (1 pg) | 32.29 | 1.00 | 35.25 | 1.00 | 33.95 | 1.00 | |
| NHDP63 (0.1 pg) | 35.11 | 0.10 | 39.1 | 0.10 | 37.3 | 0.10 | |
| MFP | Airaku-2 | 26.17 | 59.97 | 28.66 | 51.13 | 27.22 | 64.48 |
| Airaku-3 | 26.63 | 43.98 | 28.99 | 41.29 | 28.12 | 36.43 | |
| Amami | 25.54 | 92.43 | 28.25 | 66.69 | 26.81 | 83.70 | |
| Hoshizuka-4 | 29.20 | 7.60 | 31.58 | 7.84 | 30.55 | 7.81 | |
| Kusatsu-3 | 28.61 | 11.36 | 31.26 | 9.60 | 29.96 | 11.42 | |
| Kusatsu-6 | 26.64 | 43.60 | 29.13 | 37.81 | 28.07 | 37.61 | |
| Ryukyu-6 | 27.44 | 25.24 | 29.56 | 28.59 | 28.53 | 28.11 | |
| Zensho-2 | 26.49 | 48.24 | 28.85 | 45.26 | 27.56 | 51.82 | |
| Zensho-4 | 26.84 | 38.04 | 29.44 | 30.91 | 28.12 | 36.44 | |
| Zensho-5 | 27.40 | 25.95 | 29.97 | 22.06 | 29.05 | 20.28 | |
| Zensho-9 | 26.72 | 41.14 | 29.39 | 31.85 | 28.25 | 33.60 | |
| Zensho-15 | 28.03 | 16.92 | 30.69 | 13.80 | 29.35 | 16.75 | |
| Gushiken | 25.64 | 86.33 | 27.75 | 91.88 | 26.47 | 103.63 | |
| Hoshizuka-5 | 27.74 | 20.50 | 29.92 | 22.67 | 28.72 | 24.99 | |
| Indonesia-1 | 26.89 | 37.00 | 29.47 | 30.27 | 27.96 | 40.34 | |
| Korea-3-2 | 27.48 | 24.63 | 29.44 | 30.96 | 28.51 | 28.56 | |
| Thai-53 | 27.29 | 27.89 | 29.84 | 23.92 | 28.22 | 34.26 | |
| Thai-311 | 25.80 | 77.56 | 28.18 | 69.74 | 26.77 | 85.79 | |
ADML, armadillo-derived M. leprae; MFP, mouse footpad-derived M. leprae.
Percent efficiency, correlation of coefficient of determination (R2), and slope are 95.4, 0.997, and 3.373 for folP1, 83, 0.998, and 3.811 for rpoB, and 91.3, 0.997, and 3.549 for gyrA, respectively. SQ, starting quantity (SQ). All DNA templates were tested in triplicate for each target and quantitated according to the NHDP63 DNA standard curve.
Next, the melt curves were analyzed by HRM. For the five concentrations of NHDP63 tested in triplicate, the melt curves clustered together as a tight group; the standard with the smallest amount of input DNA (0.1 pg) occasionally separated into a different cluster. The normalized relative fluorescence unit (RFU) values and differential RFU curves produced by the HRM Precision Melt Analysis software in the melt region are shown in Fig. 2. These results indicated that the PCR assay conditions were suitable for testing other DNA samples and that cluster classification for amplicons produced at CT values of >35 may not be reliable; the data should be individually analyzed, repeated, and assessed by other tests.
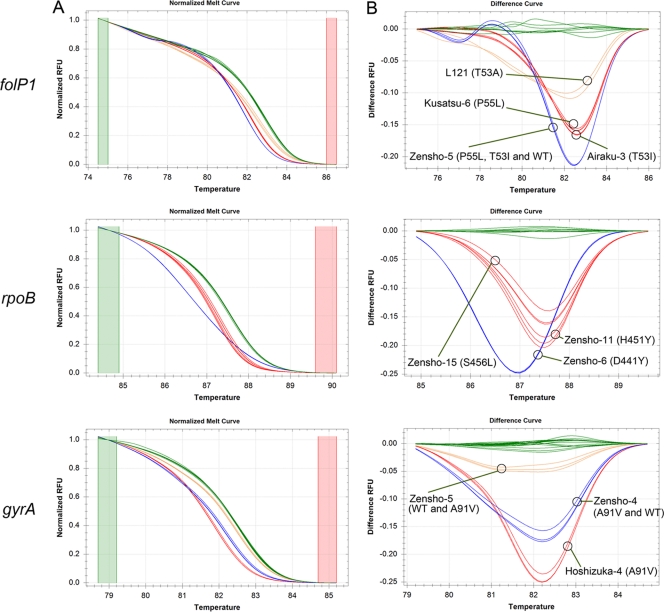
Fig 2.
Real-time PCR-HRM analysis for detection of mutations in M. leprae drug resistance-determining regions (DRDR assays). Representative real-time-PCR normalized melt curves (A) and differential curves (B) for M. leprae strains analyzed at the DRDRs of folP1, rpoB and gyrA are shown. The green color was assigned to the NHDP63 strain serving as the wild type for each DRDR. The mutant or mixed-genotype strains are shown in red, orange, and blue. The genotypes of the mutants are indicated within parentheses next to the sample name.
The objective of the real-time PCR-HRM assays was to determine if the samples that have DRDR mutations of interest could be identified. By using NHDP63 DNA for the “wild-type” (WT) sequence for the DRDR analyses performed with folP1, rpoB, and gyrA, melt curves from the test samples that did not cluster with it corresponded to putative sequence variations occurring anywhere in the amplicon (and hence were designated “cluster V”), while those that clustered with it corresponded to the wild-type sequence (and hence were designated “cluster WT”). Prior to testing HRM assays on samples derived from leprosy patients, a panel of 18 mouse footpad (MFP) and one or more armadillo-derived M. leprae (ADML) strains with known DRDR mutations were utilized for validating the assays (Table 3).
Table 3.
Real-time PCR-HRM assay for M. leprae DRDR mutation detection (DRDR assays)
| Sample typea | Template | Assay resultb |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
folP1 |
rpoB |
gyrA |
||||||||
| Reported DRDR genotype | HRM cluster | HRM cluster verification by sequencing | Reported DRDR genotype | HRM cluster | HRM cluster verification by sequencing | Reported DRDR genotype | HRM cluster | HRM cluster verification by sequencing | ||
| ADML | NHDP63 | No mutation | WT | No mutation | WT | No mutation | WT | No mutation | ||
| MFP | Airaku-2c | P(CCC)55L(CTC) | WT | No mutation | S(TCG)456L(TTG) | WT | No mutation | No mutation | WT | No mutation |
| Airaku-3 | T(ACC)53I(ATC) | V | No mutation | WT | No mutation | WT | ||||
| Amami | P(CCC)55L(CTC) | V | No mutation | WT | No mutation | WT | ||||
| Hoshizuka-4 | P(CCC)55S(CTC) | V | S(TCG)456L(TTG) | V | A(GCA)91V(GTA) | V | ||||
| Kusatsu-3 | T(ACC)53I(ATC) | V | No mutation | WT | No mutation | WT | ||||
| Kusatsu-6 | P(CCC)55L(CTC) | V | D(GAT)441Y(TAT) | V | No mutation | WT | ||||
| Ryukyu-6 | No mutation | WT | No mutation | WT | A(GCA)91V(GTA) | V | ||||
| Zensho-2 | P(CCC)55L(CTC) | V | No mutation | WT | No mutation | WT | ||||
| Zensho-4d | T(ACC)53I(ATC) | V | S(TCG)456L(TTG) | V | A(GCA)91V(GTA) | V | A(GCA)91V(GTA) and no mutation | |||
| Zensho-5e | P(CCC)55L(CTC) | V | P(CCC)55L(CTC), T(ACC)53I(ATC), and no mutation | S(TCG)456L(TTG) | V | S(TCG)456L(TTG) | No mutation | V | No mutation; A(GCA)91V(GTA) | |
| Zensho-9 | No mutation | V | P(CCC)55L(CTC) | H(CAC)451Y(TAC) | V | H(CAC)451Y(TAC) | No mutation | WT | ||
| Zensho-15 | Unknown | V | P(CCC)55L(CTC) | Unknown | V | S(TCG)456L(TTG) | Unknown | V | A(GCA)91V(GTA) | |
| Gushiken | No mutation | WT | No mutation | WT | No mutation | WT | ||||
| Hoshizuka-5 | No mutation | WT | No mutation | WT | No mutation | WT | ||||
| Indonesia-1 | No mutation | WT | No mutation | No mutation | WT | WT | No mutation | WT | No mutation | |
| Korea-3-2 | No mutation | WT | No mutation | WT | No mutation | WT | ||||
| Thai-53 | No mutation | WT | No mutation | WT | No mutation | WT | ||||
| Thai-311 | No mutation | WT | No mutation | WT | No mutation | WT | ||||
ADML, armadillo-derived M. leprae; MFP, mouse footpad-derived M. leprae.
Reported DRDR genotype sequence data were verified by the method of Matsuoka (22). HRM cluster data are designated WT for wild-type target sequences or V for variant target sequences. NHDP63 strains with the same sequences as those of the TN strain were considered WT. For HRM cluster verification by sequencing, representative samples of each of the clusters were verified by PCR product sequencing, and the genotypes detected are indicated.
The HRM clustering results were not concordant with expected genotypes for both the rpoB and folP1 genes for strain Airaku-2 (22). VNTR strain typing performed for this strain confirmed that it was indeed not Airaku 2 (44). However, the designation Airaku-2 was retained during the course of the study and in all tables in this report.
The HRM assay for gyrA DRDR separated this strain from the wild type and the other mutants; DNA sequencing results showed a mixed allele (C and T) (see Fig. 2 [gyrA; orange curves] and Fig. 3).
The HRM assay for folP1 and gyrA DRDRs separated this strain from the wild type and the other mutants that share the same genotype; DNA sequencing showed minor mixed alleles at codons 53 and 55 in folP1 and codon 91 in gyrA (see Fig. 2 [blue curve in folP1 panel B and orange curve in gyrA panel B] and Fig. 3).
The reference MFP-LRC strains that were available for this study represented several wild-type strains and two, three, and one unique sequence variant(s) for the folP1, rpoB, and gyrA amplicons, respectively (Table 3). Furthermore, in this panel, one or multiple isolates represented each of these genotypes. The DNA quantitation and HRM cluster classification (wild type [WT] or variant [V]) results are shown in Tables 2 and 3. The HRM cluster classifications were concordant in 8 WT and 10 mutant folp1; 12 WT and 6 mutant rpoB; and 14 WT and 4 mutant gyrA DRDRs (Table 3). Four situations are of note. (i) The Airaku-2 strain was expected to carry mutations in both rpoB and folP1. Repeated HRM assays did not indicate these mutations, as confirmed from the amplicon DNA sequences. Therefore, VNTR strain typing was performed for all MFP-LRC strains to verify strain identity (14, 44). The VNTR strain type of the sample received as Airaku-2 was indeed not consistent with the expected VNTR (44) (data not shown). (ii) For strain Zensho-4, the gyrA amplicon was separated into a different cluster (blue curve in Fig. 2 gyrA panel) compared to other strains that carry the expected mutation type. To verify whether this was due to a sample quality or quantity artifact, the PCR products were sequenced, which confirmed the presence of the mixed alleles at codon 91 (see Fig. 3). (iii) For strain Zensho-5, the HRM clustered the folP1 and gyrA amplicons (blue and orange curves in Fig. 2 folP1 and gyrA panels) as being different from those of strains Kusatsu-6 and NHDP63 with the expected sequence types, respectively. Closer inspection of the sequence detected mixed alleles (in codons 53 and 55 in folP1 and codon 91 in gyrA) as shown in Fig. 3. (iv) For Zensho-9, HRM detected a variant for folP1, which, when sequenced, verified the presence of a folP1 DRDR mutation.
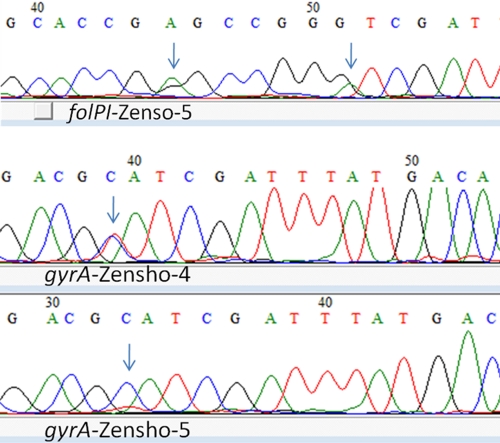
Fig 3.
Sequence chromatograms of samples depicting multiple alleles in gyrA and folP1 DRDR. Arrows indicate nucleotide positions where mixed alleles were detected for samples named in the chromatograms.
Taken together, these results validate that real-time PCR-HRM is suitable for distinguishing wild-type strains from those that carry some of the known mutations in the DRDRs of folP1, rpoB, and gyrA. Furthermore, HRM clustering can be sensitive to the presence of multiple alleles.
Performance of the real-time PCR-HRM DRDR assays on clinical samples.
Having validated that wild-type and several mutation types in the DRDRs can be identified in reference strains, the real-time HRM-PCR assays were tested on clinical samples. For this, 121 DNA samples extracted from clinical biopsy specimens from Philippine patients that had been analyzed previously by conventional PCR-DNA sequence-based mutation surveillance of folP1 and rpoB DRDRs were selected (18). The gyrA DRDR had not been sequenced. This sequence-based survey detected mutations in folP1 but not in rpoB DRDRs (18). The folP1 mutation types found were a codon 55 mutation (CCC→CTC) (n = 1) and a codon 53 mutation (ACC→GTC) (n = 3). The latter mutation type is not represented in the MFP-LRC reference panel (Table 3).
The real-time PCR-HRM results were highly concordant with the sequencing results. All four strains with mutations in folP1, including those with the codon 53 mutation (ACC→GTC), were reidentified (Table 4). Furthermore, a folP1 mutant strain (codon 55 [CCC→CTC]) that was overlooked previously was identified. Thus, the sensitivity for mutation detection was 100% for folP1. Samples that produced HRM variants (i.e., non-WT) clusters were found at 3.3%, 4.9%, and 4.9% for the folP1, rpoB, and gyrA DRDRs. Of these, three appeared as HRM variants in two or more DRDR loci; the amplification curves showed high CT values/and or an early fluorescence plateau indicative of low DNA concentrations and/or PCR inhibition, which can limit the reliability of HRM cluster assignments. When tested in PCR inhibition assays, the addition of these DNA samples to NHDP63 controls increased the CT by 2 to 3 cycles (data not shown). Overall, these results showed a high sensitivity rate for clinical samples and, when combined with a low false-mutation rate, validate that HRM analysis, as established for the mutations tested, is suitable for drug resistance surveillance in the clinic.
Table 4.
Sensitivity and specificity of HRM detection of DRDR mutations in clinical biopsy DNA samples
| Target | Classification | No. of samples of indicated SQ (pg) or % sensitivity or specificitya |
|||||
|---|---|---|---|---|---|---|---|
| Total | <0.1 | 0.1–1 | 1–10 | 10–100 | 100–1,000 | ||
| folP1 | True wild type | 112 | 1 | 16 | 37 | 50 | 8 |
| True mutant | 5 | 2 | 2 | 1 | 0 | 0 | |
| False wild type | 0 | 0 | 0 | 0 | 0 | 0 | |
| False mutant | 4 | 2 | 1 | 1 | 0 | 0 | |
| Total | 121 | 5 | 19 | 39 | 50 | 8 | |
| Sensitivity | 100 | 100 | 100 | 100 | nab | na | |
| Specificity | 96.50 | 33 | 94.10 | 97.40 | 100 | 100 | |
| rpoB | True wild type | 115 | 2 | 12 | 34 | 59 | 8 |
| True mutant | 0 | 0 | 0 | 0 | 0 | 0 | |
| False wild type | 0 | 0 | 0 | 0 | 0 | 0 | |
| False mutant | 6 | 3 | 3 | 0 | 0 | 0 | |
| Total | 121 | 5 | 15 | 34 | 59 | 8 | |
| Sensitivity | na | na | na | na | na | na | |
| Specificity | 95.04 | 40 | 80 | 100 | 100 | 100 | |
| gyrA | True wild type | 115 | 2 | 16 | 36 | 55 | 6 |
| True mutant | 0 | 0 | 0 | 0 | 0 | 0 | |
| False wild type | 0 | 0 | 0 | 0 | 0 | 0 | |
| False mutant | 6 | 4 | 2 | 0 | 0 | 0 | |
| Total | 121 | 5 | 18 | 36 | 55 | 6 | |
| Sensitivity | na | na | na | na | na | na | |
| Specificity | 95 | 20 | 88.90 | 100 | 100 | 100 | |
Sample values represent numbers of samples grouped according to the starting quantity (SQ) in picograms. Sensitivity is defined as number of true mutants/(number of true mutants + number of false wild types). Specificity is defined as number of true wild types/(number of true wild types + number of false mutants).
na, not applicable, as no true mutants were present in the set of samples.
Development of SNP typing assays based on real-time PCR-HRM analysis and assay validation with reference specimens.
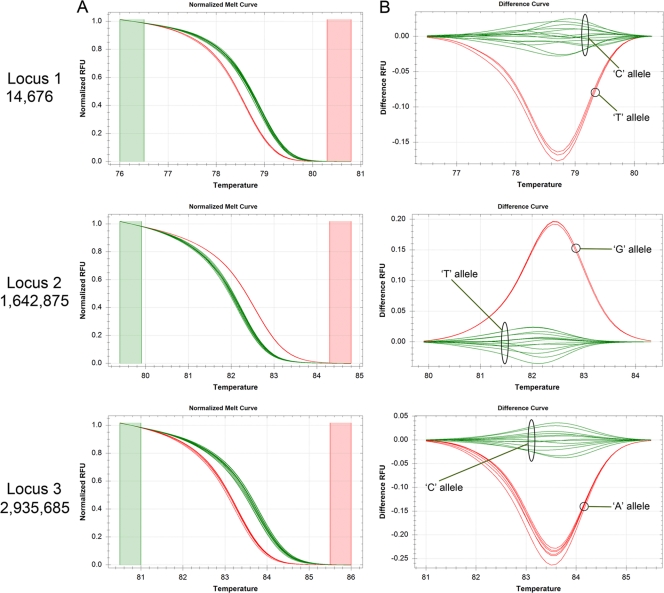
Four lineages of M. leprae (SNP types 1 to 4), distinguishable on the basis of three biallelic SNP loci, have been previously described (28). The PCR conditions for each locus were standardized using NHDP63 (Table 5). ADML strains that represent each of the SNP types and all the 18 MFP reference strains were quantitated using these assays (Table 5). For HRM cluster analysis, NHDP63 DNA, which is of SNP type 3, was used for the reference cluster assignment (Table 6).It was then easy to identify the strains which had alleles that matched or differed from it at each of the three SNP loci (Fig. 4 and Tables 6 and 7). For example, for SNP locus 1, the “C” allele amplicons cluster together with NHDP63 (reference cluster 1), while those with the alternative allele “T” are separate (cluster 2) (Fig. 4). With this HRM cluster approach, the actual SNP haplotype of the M. leprae DNA could then be readily determined.
Table 5.
Comparison of real-time PCR CT values and estimates of starting quantity
| Sample typea | Strain (quantity) | SNPb |
|||||
|---|---|---|---|---|---|---|---|
| Locus 1 |
Locus 2 |
Locus 3 |
|||||
| CT | SQ (pg) | CT | SQ (pg) | CT | SQ (pg) | ||
| ADML | NHDP63 (1 ng) | 21.95 | 1,000.00 | 21.97 | 1,000.00 | 21.01 | 1,000.00 |
| NHDP63 (100 pg) | 25.40 | 100.00 | 25.34 | 100.00 | 24.24 | 100.00 | |
| NHDP63 (10 pg) | 29.09 | 10.00 | 28.73 | 10.00 | 27.76 | 10.00 | |
| NHDP63 (1 pg) | 32.70 | 1.00 | 32.24 | 1.00 | 31.14 | 1.00 | |
| NHDP63 (0.1 pg) | 36.50 | 0.10 | 35.75 | 0.10 | 34.43 | 0.10 | |
| Thai-53 | 27.14 | 26.40 | 27.42 | 20.71 | 26.09 | 29.48 | |
| 3039 | 27.16 | 25.97 | 27.15 | 24.60 | 26.16 | 28.16 | |
| BR4923 | 27.25 | 25.36 | 27.18 | 24.36 | 26.24 | 26.79 | |
| MFP | Airaku-2 | 26.35 | 58.92 | 26.30 | 53.48 | 25.25 | 53.64 |
| Airaku-3 | 27.08 | 37.60 | 27.14 | 30.74 | 25.90 | 34.52 | |
| Amami | 26.13 | 67.50 | 25.85 | 72.29 | 25.01 | 63.49 | |
| Hoshizuka-4 | 29.95 | 6.47 | 29.52 | 6.19 | 28.73 | 5.00 | |
| Kusatsu-3 | 29.17 | 10.40 | 29.32 | 7.11 | 28.00 | 8.22 | |
| Kusatsu-6 | 27.30 | 32.85 | 27.16 | 30.02 | 26.08 | 30.61 | |
| Ryukyu-6 | 28.17 | 19.35 | 27.90 | 18.30 | 26.58 | 21.74 | |
| Zensho-2 | 27.28 | 33.35 | 27.73 | 20.67 | 25.62 | 41.90 | |
| Zensho-4 | 27.70 | 25.73 | 27.26 | 28.14 | 26.41 | 24.31 | |
| Zensho-5 | 27.82 | 23.87 | 27.65 | 21.76 | 27.02 | 16.04 | |
| Zensho-9 | 27.38 | 31.21 | 27.12 | 30.92 | 26.35 | 25.33 | |
| Zensho-15 | 28.68 | 14.11 | 28.45 | 12.79 | 27.64 | 10.52 | |
| Gushiken | 25.90 | 77.88 | 25.96 | 67.15 | 24.39 | 97.04 | |
| Hoshizuka-5 | 28.04 | 20.87 | 28.03 | 16.79 | 26.59 | 21.60 | |
| Indonesia-1 | 27.86 | 23.70 | 27.40 | 25.64 | 25.93 | 33.77 | |
| Korea-3-2 | 27.93 | 22.40 | 28.05 | 16.58 | 26.46 | 23.56 | |
| Thai-53 | 27.42 | 30.50 | 27.32 | 26.91 | 26.19 | 28.33 | |
| Thai-311 | 25.95 | 75.25 | 25.88 | 70.67 | 24.84 | 71.41 | |
ADML, armadillo-derived M. leprae; MFP, mouse footpad-derived M. leprae.
Percent efficiency, correlation of coefficient of determination (R2), and slope are 90.3, 0.994, and 3.579 for locus 1, 91.7, 0.998, and 3.538 for locus 2, and 91.7, 0.999, and 3.539 for locus 3. For starting quantity (SQ) determinations, all DNA templates were tested in triplicate for each target and quantitated according to the NHDP63 DNA standard curve.
Table 6.
Scheme for real-time PCR-HRM assay for M. leprae SNP typing showing the expected real-time PCR-HRM cluster patterns for the three loci which generate four SNP types
| SNP type | Allelea/HRM clusterb |
||
|---|---|---|---|
| Locus 1 | Locus 2 | Locus 3 | |
| 1 | C/1 | G/2 | A/2 |
| 2 | C/1 | T/1 | A/2 |
| 3 | C/1 | T/1 | C/1 |
| 4 | T/2 | T/1 | C/1 |
The SNP alleles are indicated (28).
The NHDP63 allele is assigned to cluster 1 and the alternative allele to cluster 2.
Fig 4.
Real-time PCR-HRM analysis for SNP detection for M. leprae typing (SNP typing assays). Representative real-time PCR-normalized melt curves (A) and differential curves (B) for M. leprae strains analyzed at three SNP loci as indicated beside the panels are shown. The green color was assigned to NHDP63, and the corresponding alleles for each locus are indicated (this is referred to as cluster 1 in Tables 6, 7, and 8). The red curves indicate strains with the alternative allele at each locus (this is referred to as cluster 2 in Tables 6, 7, and 8).
Table 7.
Results of real-time PCR-HRM assay for M. leprae SNP typing of MFP-LRC and armadillo-derived reference strains based on the cluster pattern defined in Table 6
| Sample type | Strain | HRM cluster |
SNP type | ||
|---|---|---|---|---|---|
| Locus 1 | Locus 2 | Locus 3 | |||
| MFP | Airaku-2 | 1 | 2 | 2 | 1 |
| Airaku-3 | 1a | 2a | 2 | 1b | |
| Amami | 1 | 1 | 1 | 3 | |
| Hoshizuka-4 | 1 | 1 | 1 | 3 | |
| Kusatsu-3 | 1 | 1 | 1 | 3 | |
| Kusatsu-6 | 1 | 1 | 1 | 3 | |
| Ryukyu-6 | 1 | 1 | 1 | 3 | |
| Zensho-2 | 1 | 1 | 1 | 3 | |
| Zensho-4 | 1 | 1 | 1 | 3 | |
| Zensho-5 | 1 | 1 | 1 | 3 | |
| Zensho-9 | 1 | 1 | 1 | 3 | |
| Zensho-15 | 1 | 1 | 1 | 3 | |
| Gushiken | 1 | 2 | 2 | 1 | |
| Hoshizuka-5 | 1 | 1 | 1 | 3 | |
| Indonesia-1 | 1a | 2a | 2 | 1b | |
| Korea-3-2 | 1 | 1 | 1 | 3 | |
| Thai-53 | 1 | 2 | 2 | 1 | |
| Thai-311 | 1a | 2a | 2 | 1b | |
| ADML | Thai-53 | 1 | 2 | 2 | 1 |
| 3039 | 1 | 1 | 2 | 2 | |
| BR4923 | 2 | 1 | 1 | 4 | |
| NHDP63 | 1 | 1 | 1 | 3 | |
Sequences of amplicons were verified.
The SNP types are different from those reported in a previous study (22).
While SNP types 1 and 4 can be ascertained by genotyping at just one locus, other types require mapping of at least two loci, as can be seen by the haplotypes shown in Table 6. The real-time PCR-HRM cluster assignment scheme was validated for the MFP-LRC reference samples. The SNP types in these strains had been determined previously by the PCR amplicon sequencing method. In this process, HRM analysis found that the Airaku-3, Indonesia-1, and Thai-311 strains are all actually SNP type 1 and not as previously published (Airaku-3 as SNP type 3 and the other two as type 2) (22).
Performance of the real-time PCR-HRM SNP typing assays on clinical samples.
To verify whether real-time PCR-HRM was suitable for SNP typing of clinical isolates, a set of DNA samples prepared from biopsy specimens from Nepali patients of unknown SNP types were selected. Correct SNP types were readily obtained, as demonstrated by the concordance of the HRM clustering-derived results with those from a previously described PCR-RFLP assay (Table 8). Only SNP types 1 and 2 were identified within this clinical sample set, consistent with the genotypes that are prevalent in this country (28).
Table 8.
Concordance of PCR-RFLP and real-time PCR-HRM methods for M. leprae SNP typing of clinical isolates
| Sample | Sample type | PCR-RFLPa |
SNP type | HRM clusterb |
|||
|---|---|---|---|---|---|---|---|
| Locus 2/CviKI | Locus 3/BstUI | Locus 1 | Locus 2 | Locus 3 | |||
| NP101 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP103 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP108 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP109 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP110 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP111 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP112 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP114 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP116 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP117 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP118 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP119 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP120 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP123 | Clinical | — | — | 1 | 1 | 2 | 2 |
| NP106 | Clinical | — | — | 1 | Vc/1 | 2 | 2 |
| NP113 | Clinical | — | — | 1 | Vc/1 | 2 | 2 |
| NP102 | Clinical | + | — | 2 | 1 | 1 | 2 |
| NP104 | Clinical | + | — | 2 | 1 | 1 | 2 |
| NP115 | Clinical | + | — | 2 | 1 | 1 | 2 |
| NP122 | Clinical | + | — | 2 | 1 | 1 | 2 |
| NP124 | Clinical | + | — | 2 | 1 | 1 | 2 |
| NP105 | Clinical | + | — | 2 | 1 | 1 | 2 |
| NP121 | Clinical | + | — | 2 | 1 | 1 | 2 |
| NP125 | Clinical | NDd | — | 2 | 1 | Vc/1 | 2 |
| NP107 | Clinical | ND | — | 1 | 1 | 2 | 2 |
| NHDP63 | ADML | + | + | 3 | 1 | 1 | 1 |
| Thai53 | ADML | — | + | 1 | 1 | 2 | 2 |
| BR4923 | ADML | + | + | 4 | 2 | 1 | 1 |
The PCR-RFLP assay was performed as previously described (31).
The NHDP63 allele is assigned to cluster 1 and the alternative allele to cluster 2.
V, HRM automatically grouped the three strains NP106, NP113, and 125 into a different cluster (variant). When the melting curves were manually examined, NP113 and NP125 were in the same cluster as NHDP63, while NP106 appeared to belong to a different cluster. Locus 1 amplicons of NP106 was sequenced, and SNP allele C was same as that of the NHDP63 and TN strains.
ND, not determined due to small amount of amplicon.
DISCUSSION
The success of antimicrobial therapies for leprosy is central and critical for preventing morbidities and disabilities and for decreasing the incidence of new cases. The low levels of relapse reported after MDT indicate that this treatment is effective (41). However, the nearly stable incidence rate attests to continuing transmission. Multiple factors can contribute to the emergence and spread of M. leprae drug resistance, particularly in countries of high endemicity. These include unsupervised components in the multidrug therapy, a possible lack of compliance or irregular use, and an absence of standardized tests for cure at release from treatment (RFT) and long-term follow-up. On the other hand, drug resistance may also emerge in countries that have apparently achieved “elimination status” because of scant resources and attention to leprosy.
Drug resistance surveillance and strain typing of M. leprae are useful molecular tools for leprosy control. Traditional PCR and sequencing techniques for these applications are laborious and expensive. In order to improve throughput, reduce costs of molecular tests, and support inclusion of all patients, including new and relapse cases, we explored emerging high-resolution melt technologies combined with real-time PCR. It is now possible to discriminate genetic variants in target loci by post-PCR analysis of the shapes and melting temperatures of amplicon melting curves. Such methods have been utilized for scanning mutations in the tumor suppressor gene TP53 for cancer detection (6), LDL receptor gene for hypercholesterolemia (16), rpoB and inhA genes for rifampin and isoniazid resistance in Mycobacterium tuberculosis strains (2, 36), and the 3′ untranslated region for typing of bronchitis viruses (9). The requirements for real-time PCR-HRM are a compatible thermocycler, a PCR mix containing appropriate enzymes, buffer, and DNA-saturating dyes, and high-resolution melt software. For genotyping by HRM analyses, there are no operator-dependent sample manipulations after the real-time PCR is assembled and no need for additional reagents.
In the context of leprosy molecular epidemiology, we developed and demonstrated real-time PCR-HRM assays for two situations: to detect mutations within drug targets gyrA, rpoB, and folP1 (DRDR assays) and another for strain typing based on SNPs (SNP typing assays). Real-time PCR assays have the added advantage of allowing estimations of the template amount in the clinical DNA samples.
The capability of HRM in separating DNA variants is related to mutation types. As M. leprae is not cultivable in vitro and few laboratories possess facilities and expertise for the propagation of clinical strains in mouse footpad or armadillo animal models, the availability of characterized strains, particularly for testing drug resistance, is limited. Clinically (biopsy or slit skin smear sample) derived DNA containing M. leprae with DRDR mutations is an uncommon resource, precluding availability, sharing, and testing in different laboratories. Furthermore, some DRDR mutations have been reported only once or infrequently. In this context, the feasibility of HRM for DRDR M. leprae mutation screening, in the first study of its kind in leprosy, has been explored by accessing the largest publically shared mouse derived DRDR mutant M. leprae library, maintained at the LRC, Tokyo, Japan. This library, although not comprehensive, contained several mutations in each of three drug targets (rpoB, folP1, and gyrA), allowing standardization and demonstration of proof of concept. Furthermore, a mutation detected when screening clinical strains available in-house was added; all mutant strains within these combined collections could be identified by the HRM assays developed. These include folP1 at codon 53 (ACC→ATC and ACC→GTC) and codon 55 (CCC→CTC). Together, these three mutation types cover 50% of the mutants described worldwide. The DRDR mutation types not tested by HRM are folP1 53 (ACC→GCC) and 55 (CCC→CGC); the former has A→G substitution that is within HRM resolution capability. For rifampin resistance, mutations in rpoB codons 441, 451, 456, and 458 have been reported. We tested HRM for 441 (GAT→TAT), 451 (CAC→TAC), and 456 (TCG→TTG). More than 80% of reported mutants are covered by these three genotypes. Four mutations not included in the current studies are 441(CAC→GAC), 456 (TCG→ATG), 456 (TCG to TTC), and 458 (GTA to GTG). Of these, substitutions in the latter three should be easily detected by HRM due to the level of change expected in Tm values. For gyrA, we tested a mutation in codon 91 (GCA-GTA); this covers more than 90% of the reported mutations. The other mutation is at codon 89 (GGC to TGC); the G→T transition is detectable by HRM. It is of note that for the double mutation (AC→GT) in the folP1 codon 53 ACC to GTC detected in the clinical strains, the Tm change is negligible; the differentiation by HRM was possible only when a small amplicon (52 bp) was generated. There is only one report for this mutation type (24), which incidentally was detected in the same studied population, i.e., leprosy patients in Cebu, Philippines. The identification of multiple cases with this mutation in our study sample was due to transmission within a closely linked community, as captured and described in greater detail by Li et al. by prospective molecular epidemiological approaches (18, 30, 31).
Singh et al. recently reported a method based on TaqMan probe assays for rpoB and folP1 (35). The allele-specific assays are not suitable for unknown or new mutations and require reciprocal testing with both wild-type and mutation-specific primers/probes (35). For these reasons, we find that HRM is convenient for the preliminary screening of DNAs and rapid classification of clinical strains into wild-type or variant clusters. For folP1 and rpoB, three different proven drug resistance mutations were tested; these could be separated from the wild-type strain. Further, for the three variants of each target gene, two different HRM clusters could be detected.
With regard to SNP typing, HRM was robust and straightforward due to the biallelic nature of each of the SNPs. Assignment of SNP types 1, 2, 3, and 4 is based on the finding that only 4 out of 64 possible haplotypes have been detected by mapping SNPs at three loci (28). Thus, SNP types can be assigned by mapping only one or two of the three loci. These four SNP types can be further divided into 16 SNP subtypes; however, these are also restricted within a given region of endemicity. In Philippines, for example, SNP types 1A and 3K have been detected, while in Brazil, 3I and 4P strains are found (29). Thus, typing of SNP types 1 to 4 by real time-HRM analysis provides simple, rapid, and robust classification and is suitable for comparison of M. leprae strains on a global and national level. HRM assays that can be designed based on principles as described herein for the discrimination of the 16 subtypes were beyond the scope of the present study goals and are also restricted by the availability of a representative collection of all strain subtypes. Furthermore, to date, the highest resolution of strains within SNP subtypes has been achieved by VNTR strain typing (7, 15, 30, 31).
Real-time PCR-HRM analysis of various DRDR targets aided in the detection of discrepancies between expected and/or reported DRDR genotypes for strain Airaku-2. Secondary genotyping of VNTR loci clarified that the strain received for HRM testing was indeed not Airaku-2 as reported but could have been Gushiken (44). The HRM analysis was also sensitive in detecting the presence of both wild-type and mutant alleles at the gyrA locus in strain Zensho-4 due to a melt curve that differed from that of the wild type or the expected mutant. VNTR strain typing of Zensho-4 did not show that the sample was contaminated with another wild-type strain. Thus, real-time HRM analysis may enable detection of minor populations of mutant alleles in a wild-type background and emergence of drug resistance. The melt curve of Zensho-5 for gyrA locus, expected to correspond to the wild-type curves, showed the possibility of a variant. Careful reexamination of the sequence chromatogram showed a minor contaminant peak (Fig. 3). Similarly, the melt curves of the folP1 amplicon, although very close to that of the wild type, are slightly different. When VNTR profiles were reviewed, duplicate alleles in several loci were detected. Overall, these data indicate that Zensho-5 DNA was contaminated with another DNA. Strain Zensho-9, previously shown to possess a folP1 wild-type sequence and to be susceptible in MFP assays, is interesting (21, 22). Current data show an folP1 mutation also, which may indicate the emergence of this mutation. The strain type did not show discrepancies by VNTR typing, so sample contamination is not an issue (44). These examples illustrate the potential of real-time PCR-HRM analysis as a sensitive mutation screening tool and for use in quality control, such as when sharing reagents between researchers and for detection of sample contamination.
For each target amplicon, real-time PCR-HRM assays can be accomplished for 26 different samples, each analyzed in triplicate, in a 96-well plate system, resulting in costs of less than $3 per sample and not requiring any other post-PCR reagents or procedures. In contrast, despite multiplexing of the target rpoB, folP, and gyrA DRDRs, which reduced time requirements and template, PCR reagent, and plastic supply costs, the cost of a single sequencing reaction per target for just one direction was $7 to $10 at a subsidized rate. Sequencing is often performed offsite, which separates the PCR from subsequent steps, and adding replicate tests increases costs and labor. Another issue is that in sequence chromatograms, mixtures of wild type and mutant are difficult to delineate above background peak heights and can be missed (as seen in Fig. 3). As a proof of principle, two DNAs with known rpoB and folP1 genotypes (a variant and a wild type) were combined (0% to 100% of each, in 10% increments). Even without a heteroduplex formation step in the PCR program or coamplification at lower denaturation temperature-PCR (COLD-PCR) procedure (17), DNAs in mixtures with as little as 10% of one type of DNA could be separated (data not shown).
Our studies also showed that leprosy clinical DNA samples are amenable for real-time PCR-HRM. The majority of clinical DNA samples that showed real-time PCR CT values of less than 35 were suitable for HRM. This translates to a sensitivity of ∼30 bacilli per PCR. It is highly likely that next-generation parallel-sequencing technologies will advance sample throughput and quantities and qualities of data, including numbers of gene targets and depth of coverage. In the interim, the real-time PCR-HRM assays described here are viable, simple options and can be easily integrated into practice by centralization of tests in a reference laboratory.
ACKNOWLEDGMENTS
These studies were funded by NIH-NIAID grant AI-063457, ARRA supplements to AI-063457, and contract NO1-AI-25469.
We thank Marivic Balagon and all clinicians and staff at CSC-LWM and Anandaban Hospital, Kathmandu, Nepal, for the clinical work and specimen collection and the patients who volunteered to participate in the research studies. We gratefully acknowledge Jason Kasvin-Felton for his generous help and software expertise with generation of the high quality art work. We thank Paul Streng and Kim Petro of Bio-Rad for the RT-PCR training and technical support.
Footnotes
Published ahead of print 14 December 2011
REFERENCES
- 1. Balagon MF, et al. 2009. Long-term relapse risk of multibacillary leprosy after completion of 2 years of multiple drug therapy (WHO-MDT) in Cebu, Philippines. Am. J. Trop. Med. Hyg. 81:895–899 [DOI] [PubMed] [Google Scholar]
- 2. Choi GE, et al. 2010. High-resolution melting curve analysis for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 48:3893–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Colston MJ, Hilson GRF, Banerjee DK. 1978. The ‘proportional bactericidal test’: a method for assessing bactericidal activity of drugs against Mycobacterium leprae in mice. Lepr. Rev. 49:7–15 [PubMed] [Google Scholar]
- 4. dela Cruz E, et al. 1996. Primary dapsone resistance in Cebu, The Philippines; cause for concern. Int. J. Lepr. Other Mycobact Dis. 64:253–256 [PubMed] [Google Scholar]
- 5. Fukuda H, Hiramatsu K. 1999. Primary targets of fluoroquinolones in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:410–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garritano S, et al. 2009. Determining the effectiveness of high resolution melting analysis for SNP genotyping and mutation scanning at the TP53 locus. BMC Genet. 10:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Groathouse NA, et al. 2004. Multiple polymorphic loci for molecular typing of strains of Mycobacterium leprae. J. Clin. Microbiol. 42:1666–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gupta UD, Katoch K, Katoch VM. 2009. Study of rifampicin resistance and comparison of dapsone resistance of M. leprae in pre- and post-MDT era. Indian J. Lepr. 81:131–134 [PubMed] [Google Scholar]
- 9. Hewson KA, Browning GF, Devlin JM, Ignjatovic J, Noormohammadi AH. 2010. Application of high-resolution melt curve analysis for classification of infectious bronchitis viruses in field specimens. Aust. Vet. J. 88:408–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Honore N, Cole ST. 1993. Molecular basis of rifampin resistance in Mycobacterium leprae. Antimicrob. Agents Chemother. 37:414–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Honoré N, Perrani E, Telenti A, Grosset J, Cole ST. 1993. A simple and rapid technique for the detection of rifampin resistance in Mycobacterium leprae. Int. J. Lepr. Other Mycobact. Dis. 61:600–604 [PubMed] [Google Scholar]
- 12. Honoré N, Roche PW, Grosset JH, Cole ST. 2001. A method for rapid detection of rifampicin-resistant isolates of Mycobacterium leprae. Lepr. Rev. 72:441–448 [DOI] [PubMed] [Google Scholar]
- 13. Kai M, et al. 2011. Analysis of drug-resistant strains of Mycobacterium leprae in an endemic area of Vietnam. Clin. Infect. Dis. 52:e127–e132 [DOI] [PubMed] [Google Scholar]
- 14. Kapur V, et al. 1994. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J. Clin. Microbiol. 32:1095–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kimura M, et al. 2009. Rapid variable-number tandem-repeat genotyping for Mycobacterium leprae clinical specimens. J. Clin. Microbiol. 47:1757–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laurie AD, George PM. 2009. Evaluation of high-resolution melting analysis for screening the LDL receptor gene. Clin. Biochem. 42:528–535 [DOI] [PubMed] [Google Scholar]
- 17. Li J, Makrigiorgos GM. 2009. COLD-PCR: a new platform for highly improved mutation detection in cancer and genetic testing. Biochem. Soc. Trans. 37:427–432 [DOI] [PubMed] [Google Scholar]
- 18. Li W, et al. 2011. Transmission of dapsone-resistant leprosy detected by molecular epidemiological approaches. Antimicrob. Agents Chemother. 55:5384–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maeda S, et al. 2001. Multidrug resistant Mycobacterium leprae from patients with leprosy. Antimicrob. Agents Chemother. 45:3635–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Markham NR, Zuker M. 2005. DINA Melt web server for nucleic acid melting prediction. Nucleic. Acids Res. 33:W577–W581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsuoka M. 2010. Drug resistance in leprosy. Jpn. J. Infect. Dis. 63:1–7 [PubMed] [Google Scholar]
- 22. Matsuoka M. 2010. History and characteristics of isolates maintained at the leprosy research center. Nihon Hansenbyo Gakkai Zasshi 79:247–256 [DOI] [PubMed] [Google Scholar]
- 23. Matsuoka M, et al. 2008. A novel method for simple detection of mutations conferring drug resistance in Mycobacterium leprae, based on a DNA microarray, and its applicability in developing countries. J. Med. Microbiol. 57:1213–1219 [DOI] [PubMed] [Google Scholar]
- 24. Matsuoka M, et al. 2007. The frequency of drug resistance mutations in Mycobacterium leprae isolates in untreated and relapsed leprosy patients from Myanmar, Indonesia and the Philippines. Lepr. Rev. 78:343–352 [PubMed] [Google Scholar]
- 25. Matsuoka M, Kashiwabara Y, Namisato M. 2000. A Mycobacterium leprae isolate resistant to dapsone, rifampin, ofloxacin and sparfloxacin. Int. J. Lepr. Other Mycobact. Dis. 68:452–455 [PubMed] [Google Scholar]
- 26. Matsuoka M, et al. 2010. Possible mode of emergence for drug-resistant leprosy is revealed by an analysis of samples from Mexico. Jpn. J. Infect. Dis. 63:412–416 [PubMed] [Google Scholar]
- 27. Molyneux DH. 2004. “Neglected” diseases but unrecognised successes—challenges and opportunities for infectious disease control. Lancet 364:380–383 [DOI] [PubMed] [Google Scholar]
- 28. Monot M, et al. 2005. On the origin of leprosy. Science 308:1040–1042 [DOI] [PubMed] [Google Scholar]
- 29. Monot M, et al. 2009. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat. Genet. 41:1282–1289 [DOI] [PubMed] [Google Scholar]
- 30. Sakamuri RM, et al. 2009. Continuation: study and characterization of Mycobacterium leprae short tandem repeat genotypes and transmission of leprosy in Cebu, Philippines. Lepr. Rev. 80:272–279 [PubMed] [Google Scholar]
- 31. Sakamuri RM, et al. 2009. Population-based molecular epidemiology of leprosy in Cebu, Philippines. J. Clin. Microbiol. 47:2844–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sapkota BR, Ranjit C, Neupane KD, Macdonald M. 2008. Development and evaluation of a novel multiple-primer PCR amplification refractory mutation system for the rapid detection of mutations conferring rifampicin resistance in codon 425 of the rpoB gene of Mycobacterium leprae. J. Med. Microbiol. 57:179–184 [DOI] [PubMed] [Google Scholar]
- 33. Sapkota BR, Ranjit C, Macdonald M. 2006. Reverse line probe assay for the rapid detection of rifampicin resistance in Mycobacterium leprae. Nepal Med. Coll. J. 8:122–127 [PubMed] [Google Scholar]
- 34. Shepard CC. 1967. A kinetic method for the study of the activity of drugs against Mycobacterium leprae in mice. Int. J. Lepr. 35:429–435 [Google Scholar]
- 35. Singh P, et al. 2011. Molecular drug susceptibility testing and genotyping of Mycobacterium leprae from South America. Antimicrob. Agents Chemother. 55:2971–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramirez MV, et al. 2010. Rapid detection of multidrug-resistant Mycobacterium tuberculosis by use of real-time PCR and high-resolution melt analysis. J. Clin. Microbiol. 48:4003–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rozen S, Skaletsky JH. 2000. Primer3 on the WWW for general users and for biologist programmers, p 365–386 In Krawetz S, Misener SA. (ed), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 38. Williams DL, Gillis TP. 2004. Molecular detection of drug resistance in Mycobacterium leprae. Lepr. Rev. 75:118–130 [PubMed] [Google Scholar]
- 39. Williams DL, Spring L, Harris E, Roche P, Gillis TP. 2000. Dihydropteroate synthase of Mycobacterium leprae and dapsone resistance. Antimicrob. Agents Chemother. 44:1530–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Williams DL, Gillis TP. 1999. Detection of drug-resistant Mycobacterium leprae using molecular methods. Indian J. Lepr. 71:137–153 [PubMed] [Google Scholar]
- 41. World Health Organization 2010. Global leprosy situation, 2010. Wkly. Epidemiol. Rec. 85:337–348 [PubMed] [Google Scholar]
- 42. World Health Organization 2009. Drug resistance in leprosy: reports from selected endemic countries. Wkly. Epidemiol. Rec. 84:264–267 [PubMed] [Google Scholar]
- 43. World Health Organization 1982. Chemotherapy of leprosy for control programmes. WHO Technical Report Series 768. World Health Organization, Geneva, Switzerland: [PubMed] [Google Scholar]
- 44. Zhang L, Budiawan T, Matsuoka M. 2005. Diversity of potential short tandem repeats in Mycobacterium leprae and application for molecular typing. J. Clin. Microbiol. 43:5221–5229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]