Abstract
We developed and assessed the diagnostic value of a novel quantitative nested real-time (QNRT) PCR assay targeting the internal transcribed spacer region of ribosomal DNA (rDNA) in a guinea pig model of invasive pulmonary aspergillosis. Groups of 5 immunosuppressed animals that were infected using an aerosol chamber with Aspergillus fumigatus conidia were humanely terminated 1 h postinoculation and at days 3, 5, 7, and 11 postchallenge, and lung tissue, bronchoalveolar lavage (BAL) fluid, whole blood, and serum samples were collected. The QNRT PCR results obtained with the serum and BAL fluid were compared to those achieved with galactomannan and (1→3)-β-d-glucan assays. High fungal burden levels were detected by QNRT PCR in both lung tissue and BAL fluid in all infected animals at each time point, and the sensitivity of each assay in BAL fluid was 100% by day 3 and remained so through the remainder of the study. The sensitivity of detection of fungi in whole blood and serum samples was significantly lower, and some samples remained negative by all three assays despite the advanced stage of the infection. From these data, we can conclude that this novel QNRT PCR method was highly sensitive for the detection of A. fumigatus from different types of samples in this model. In addition, BAL fluid samples appeared to be the most suitable for the early diagnosis of invasive pulmonary aspergillosis. When testing serum, the use of a combination of available assays may increase the possibility of early detection of this opportunistic mycosis.
INTRODUCTION
Invasive aspergillosis represents the most frequent invasive fungal disease in patients with hematological malignancy (20). Conventional diagnostic methods, e.g., culture and cytology/histology, have limited use in this group of immunocompromised patients, where disease progression is rapid and mortality is very high. Moreover, their usefulness depends on the possibility of obtaining deep-tissue samples, which is often impossible in patients with hematologic malignancies (6). Novel non-culture-based approaches focus on the detection of surrogate markers released by the fungus such as surface antigens or DNA. These methods have been developed to hasten the diagnosis and to enable diagnostic driven rather than empirical antifungal treatment strategies. Detection of galactomannan (GM), a cell wall antigen released by Aspergillus spp. and some other fungi in serum, bronchoalveolar lavage (BAL) fluid, or other body fluids, has proven to be an important advance in the detection of invasive pulmonary aspergillosis (1, 10, 11) and has become one of the EORTC/MSG (European Organization for Research and Treatment of Cancer/Mycoses Study Group) criteria for the diagnosis of this invasive mycosis (3). Evidence for its use as a screening method for early detection of invasive aspergillosis is supported by several studies (14). Another cell wall component, (1,3)-β-d-glucan (BG) is released by several clinically important fungal species, including Aspergillus spp., and despite limited evidence compared to GM, it is included in the 2008 update of EORTC/MSG criteria for diagnosis of invasive fungal disease.
Real-time PCR is a powerful technique routinely applied as a diagnostic tool in many different fields, including clinical microbiology (4). However, several limitations of fungal PCR diagnostics exist, including resistance of fungal cells to common DNA isolation protocols; unknown timing of DNA release; the small amount of the fungal target in clinical samples, which reaches the detection limit of real-time PCR assays; and the potential for contamination and colonization to complicate the detection of true disease (2, 9). In the last decade dozens of different PCR assays have been evaluated for the early diagnostics of invasive aspergillosis, but none of them have become generally accepted (5, 12, 25). Proper validation of any novel diagnostic method requires extensive testing of clinical samples, correlation of the results with other well-established laboratory tests, and agreement of the results with clinical data indicating active disease. However, meeting these criteria for novel methods for the diagnosis of invasive aspergillosis is very difficult. The quantity of clinical samples obtained from patients with proven aspergillosis is limited due to the relatively low incidence of the infection, difficulties in obtaining sterile deep tissue samples in patients with hematological disease, and finally, due to the preemptive initiation of antifungal therapy, which may influence the sensitivity of clinically available assays. Contrary to the limited availability of clinical samples, animal models provide a unique source of samples that can be taken at different stages of infection. This makes the use of animal models ideal for the initial evaluation of novel diagnostic methods (21). Animal models in which infection is established via inhalation most closely mimic the clinical pathogenesis of invasive pulmonary aspergillosis in many ways (24, 26), and therefore, samples taken from these models may supplement the clinical samples necessary for the validation of novel laboratory tests.
The aim of this study was to validate a novel, fast, and robust quantitative nested real-time (QNRT) PCR assay designed for the sensitive and specific quantification of Aspergillus fumigatus, as this species is the most common cause of invasive aspergillosis (17), and assess its performance with samples from a guinea pig model of pulmonary disease. The technology uses a short DNA preamplification with a pair of panfungal primers and subsequent quantification with a pair of primers and a TaqMan MGB probe specific exclusively to A. fumigatus (Fig. 1). We tested lung tissue, BAL fluid, whole blood, and serum using this quantitative assay. The results obtained with BAL fluid and serum samples were compared with those achieved using clinically available assays for the detection of GM and BG.
Fig 1.
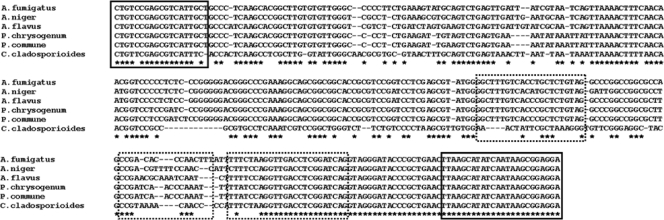
ClustalW multiple-sequence alignment of the region detected by tested QNRT PCR. The primers for the external round (solid lines) and the primers and TaqMan-MGB probe (dashed-lines) are boxed. The GenBank accession numbers for the sequences shown are as follows: A. fumigatus, AY214446; A. flavus, AY214444; A. niger, AY213632; P. chrysogenum, AY213669; P. commune, AY213672; C. cladosporioides, AY213640. The asterisks indicate 100% sequence similarity among all presented species.
MATERIALS AND METHODS
Fungal isolate.
A. fumigatus clinical isolate 293 (AF293), the isolate used in the A. fumigatus genome sequencing project, was grown on potato dextrose agar at 37°C for 7 days (18, 26). Conidia were harvested by washing and scraping of agar surfaces with 0.1% Tween 80 in sterile physiological saline and subsequent filtration for the removal of hyphal fragments. Conidia were concentrated by centrifugation and then resuspended to a target inoculum of ∼1 × 108 conidia/ml by hemocytometer. The number of viable conidia was confirmed by plating dilutions of the inoculum and quantifying CFU.
Animals and immunosuppression.
Two days prior to infection, male Hartley guinea pigs (0.5 kg; Charles River Laboratory, Wilmington, MA) received cyclophosphamide (Cytoxan, 250 mg/kg; Mead Johnson, Princeton, NJ) intraperitoneally and cortisone acetate (250 mg/kg; Sigma, St. Louis, MO) subcutaneously to induce neutropenia. Ceftazidime (100 mg/kg; Glaxo SmithKline Beecham Pharmaceuticals, Philadelphia, PA) was administered daily for prevention of bacterial infections during the study. Additional doses of cyclophosphamide (200 mg/kg) and cortisone acetate (250 mg/kg day) were administered on day 3 postinfection (26). All animal research procedures were approved by the Institutional Animal Care and Use Committee at the University of Texas Health Science Center at San Antonio, and all animals were maintained in accordance with the American Association for Accreditation of Laboratory Animal Care (16).
Inhalational challenge and sample collection.
As previously described, an inexpensive acrylic chamber was used to infect guinea pigs with a nebulized inoculum of 108 A. fumigatus conidia. Twenty-five guinea pigs were exposed to the aerosol mist for 1 h. Confirmation of the conidial delivery of each exposure run was accomplished with the sacrifice of randomly chosen guinea pigs (total n = 5) from each run of the aerosol chamber and determination of lung conidial loads by enumeration of CFU. On days 3, 5, 7, and 11 postinoculation, five guinea pigs were humanely euthanized via anesthesia and exsanguination by cardiac puncture. Blood samples were collected from each animal, and the serum was separated. BAL fluid samples were also collected from each animal, and fungal burdens in the lungs were also determined postmortem using CFU analysis (7). Throughout the course of the study, all animals were monitored daily for any obvious signs of illness or other signs of distress. Any animal that was found to be moribund prior to the study endpoint was humanely euthanized using approved methods. Four uninfected, immunosuppressed controls were also included, and samples (lung tissue, BAL fluid, serum, and whole blood) were collected on day 11 or when they appeared moribund.
DNA isolation.
Samples were stored at −70°C and thawed for 30 min at room temperature before isolation. DNA isolation was performed within a laminar-flow box in a separate laboratory to avoid PCR mixture contamination. Other measures used to avoid external contamination included the use of filtered pipet tips, frequent glove changing, and the use of a UV light in the laminar-flow box when it was not in use. One blank sample (equivalent amount of sterile water (B. Braun, Melsungen, Germany) was isolated with each set of 11 samples in order to exclude the influence of possible external contamination (chemicals, test tubes, etc.). DNA concentration was measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
DNA isolation from whole blood samples.
Twenty-two milliliters of erythrocyte lysis buffer (ELB; 10 mM Tris-Cl [pH 7.6], 5 mM MgCl2, and 10 mM NaCl) was added to 3 ml of whole blood. Samples were gently shaken for 10 min at room temperature and then centrifuged for 10 min at 1,500 × g and 10°C. The supernatant was discarded, and the pellet was washed with 25 ml of ELB. The samples were then gently shaken and centrifuged again, and the washing step was repeated once. Each pellet was resuspended in 1 ml of ELB, transferred to a microcentrifugation tube (Eppendorf), and centrifuged for 10 min at 4,000 × g at room temperature, after which the supernatant was discarded. DNA was extracted from the pellet using a ZR Fungal/Bacterial DNA kit (Zymo Research, Irvine, CA) according to the manufacturer's protocol. The time of sample vortexing with glass beads was extended from 5 to 15 min, and DNA was eluted with 50 μl of elution buffer.
DNA isolation from BAL fluid samples.
BAL fluid (0.8 to 2 ml, depending on the available volume) was centrifuged for 10 min at 5,000 × g at room temperature. The supernatant was discarded, and DNA was extracted from the pellet using a ZR Fungal/Bacterial DNA kit as described above.
DNA isolation from lung tissue and serum samples.
Lung tissue samples were homogenized in sterile saline for 20 s with an IKA T8 Ultra Turrax disperser (IKA Labortechnik, Staufen, Germany). Two hundred microliters of homogenate was added to the ZR BashingBead lysis tubes with 750 μl of the lysis solution (both from the ZR Fungal/Bacterial DNA kit) and incubated at room temperature overnight. The next day, the tubes were vortexed for 15 min and DNA was isolated according to the manufacturer's protocol using 50 μl of elution buffer. DNA was isolated from 400 μl of serum with the QIAamp DNA Blood mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol.
PCR amplification.
Five microliters of DNA from each sample was amplified in a 25-μl reaction mixture containing 1×ABsoluteQPCR Mix (Thermo Scientific) and 400 nM each primer (ASP-F, 5′-CTGTCCGAGCGTCATTGCT-3′; ITS-R, 5′-TCCTCCGCTTATTGATATGCTTAA-3′) (22) using an amplification profile of 95°C for 15 min, followed by 20 cycles of 95°C for 15 s and 60°C for 1 min. One microliter of the initial PCR product was used as a template in the second round of amplification in a 25-μl reaction mixture containing 1×ABsoluteQPCR Mix, 400 nM each primer (forward primer, 5′-GGCTTTGTCACCTGCTCTGTAG-3′; reverse primer, 5′-CTGATCCGAGGTCAACCTTAGAAA-3′), and 200 nM TaqMan-MGB probe (5′–6-carboxyfluorescein–CCGACACCCAACTTT-MGBNFQ-3′) (sequences of the all oligonucleotides: international patent application PCT/CZ2010/000052, published under no. WO 2010/121578, granted Czech patent no. CZ 302670). The amplification profile of the second PCR was 95°C for 15 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min using a Rotorgene 6000 (Corbett Research, Mortlake, New South Wales, Australia). Each sample was run in duplicate. Sterile water (B. Braun) was used as the no-template control in each run. Results were analyzed using Rotorgene 6000 software version 1.7. Quantification was made using a standard curve with 5 μl of the PCR product cloned into plasmid pCR 2.1 (Invitrogen, Grand Island, NY) in 10-fold dilutions ranging from 2 × 106 to 2 copies of plasmid DNA/5 μl. In order to reduce artificial differences in fungal loads caused by dissimilar efficiencies of DNA isolation from the individual samples, all PCR results are presented as numbers of copies per microgram of DNA used for PCR amplification (number of copies detected by QNRT PCR × (1,000/DNA concentration [in ng/μl] × 5). The amount of free DNA in serum is very low and almost unmeasurable. Therefore, real-time PCR results obtained with this fluid are presented as numbers of copies per milliliter of serum. Any sample in which a PCR product was detected following QNRT PCR was considered positive.
Test of cross-reactivity with other fungal species.
Cross-reactivity of the A. fumigatus-specific assay was tested by amplification of DNA isolated directly from fungal strains obtained from the Czech Collection of Microorganisms or clinical isolates. DNA was isolated from 100 μl of a fungal suspension using a protocol for the isolation of DNA from BAL fluid samples as described above. Five microliters of isolated DNA was used as the template for the second round of PCR as described above.
GM detection.
Both serum and BAL fluid GM concentrations were determined by the Platelia Aspergillus kit (Bio-Rad, Marnes-la-Coquette, France) according to the manufacturer's instructions. BAL fluid samples were centrifuged at 1,000 rpm for 10 min, and 300 μl of the supernatant of each sample was used for GM detection. A sample with a GM index of ≥0.5 was considered positive.
BG detection.
Both serum and BAL fluid BG concentrations were measured with the Fungitell test kit (Associates of Cape Cod, East Falmouth, MA) as recommended by the manufacturer. BAL fluid specimens were centrifuged at 1,000 rpm for 10 min, and 5 μl of the supernatant of each sample was used for BG detection. All specimens were tested in duplicate. A BG level of ≥80 pg/ml was considered positive.
Statistical analysis.
The Mann-Whitney test was used to determine if differences in fungal burdens detected in samples and on different collection days were significant (http://faculty.vassar.edu/lowry/utest.html). Spearman's rank correlation coefficient was calculated to confirm the correlation between the different assays (http://www.wessa.net/) (27). Fisher's exact test was used to determine if significant differences existed between the numbers of samples positive by each assay with different sample types.
RESULTS
Sensitivity and specificity of QNRT PCR assay.
The primers and TaqMan-MGB probe used in the second round of PCR were designed to selectively amplify A. fumigatus, not other Aspergillus species or related airborne fungi. Cross-reactivity with wild-type strains was not found with any of the species listed in Table 1. All clinical and environmental isolates were verified by DNA sequencing.
Table 1.
Species used to test assay specificity
| Species | Source or strain |
|---|---|
| Aspergillus flavus | Clinical isolate |
| Aspergillus niger | Clinical isolate |
| Aspergillus nidulans | CCMF-266 |
| Aspergillus vitricola | Environmental isolate |
| Aspergillus versicolor | Environmental isolate |
| Aspergillus sydowii | Environmental isolate |
| Cladosporium cladosporioides | CCMF-348 |
| Penicillium comunne | CCMF-327 |
| Penicillium brevicompactum | CCM8040 |
| Penicillium olsonii | Environmental isolate |
| Penicillium chrysogenum | CCM 8034 |
| Fusarium oxysporum | Clinical isolate |
| Fusarium proliferatum | Clinical isolate |
| Fusarium solani | CCM8014 |
| Rhizopus microsporus | Clinical isolate |
| Rhizopus oryzae | Clinical isolate |
| Mucor racemosus | CCM8190 |
| Rhizomucor pusillus | Clinical isolate |
| Absidia corymbifera | CCM8077 |
| Candida albicans | Clinical isolate |
| Candida glabrata | Clinical isolate |
| Candida tropicalis | Clinical isolate |
| Candida parapsilosis | Clinical isolate |
| Candida krusei | Clinical isolate |
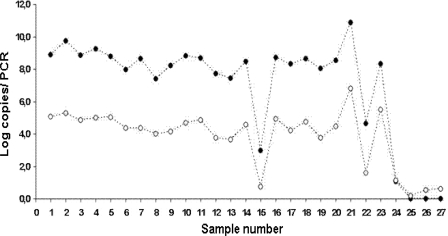
The analytical sensitivity of the assay is presented in Table 2. Both single-round and nested reverse transcription PCRs reproducibly detected 0.01 fg of DNA, corresponding to 2 plasmid copies per PCR. The use of nested PCR did not increase sensitivity but did result in lower cycle threshold (CT) values and better performance and reproducibility of the assay. The delta CT between the two reactions remained stable, reflecting linear amplification of all dilutions. We used BAL fluid samples to test the difference between the fungal loads detected after single-round PCR (45 cycles with A. fumigatus-specific oligonucleotides) and nested PCR (20 cycles of amplification with panfungal primers plus 45 cycles with A. fumigatus-specific oligonucleotides) (Fig. 2). Use of fungal DNA preamplification increased the detected copy numbers without affecting assay performance and efficiency. As inhibition of amplification due to a large amount of template was not observed, we can conclude that this ultrasensitive approach might be useful for sample testing.
Table 2.
Representative amplification of serial dilutions of plasmid DNA with cloned internal transcribed spacer region detected in second round of PCRa
| Amt of DNA | Mean CT ± SD (log no. of copies/reaction) |
ΔCT | |
|---|---|---|---|
| Single-round PCR | Nested real-time PCR | ||
| 10 pg | 16.6 ± 0.49 (6.3) | 3.09 ± 0.2 (10.8) | 13.5 |
| 1 pg | 19.9 ± 0.74 (5.3) | 7.12 ± 0.3 (9.5) | 12.8 |
| 0.1 pg | 23.4 ± 0.61 (4.3) | 9.8 ± 0.3 (8.7) | 13.6 |
| 10 fg | 27.5 ± 1.13 (3.3) | 12.6 ± 0.4 (7.8) | 14.9 |
| 1 fg | 30.5 ± 1.46 (2.3) | 15.5 ± 0.2 (6.9) | 15.0 |
| 0.1 fg | 33.9 ± 1.88 (1.3) | 18.3 ± 0.3 (6.0) | 15.6 |
| 0.01 fg | 35.8 ± 1.86 (0.3) | 20.3 ± 0.8 (5.4) | 15.5 |
Both single-round and nested PCRs detected 0.01 fg of DNA, corresponding to 2 copies of plasmid DNA per reaction.
Fig 2.
Comparison of fungal loads detected in BAL fluid samples with single-round and nested PCRs. Filled circles denote nested PCR data. Empty circles denote single-round PCR data.
Detection of A. fumigatus in lung tissue.
The results from individual guinea pigs, as well as the mean and standard deviations for each assay at each time point, are reported in Table 3. QNRT PCR detected high fungal burdens in 25 (100%) of 25 tissue samples from infected animals, while all uninfected controls were PCR negative. Initially, the high load of DNA measured 1 h postinfection (mean, log10 33.55 ± 18.80 copies/μg of DNA) was decreased at day 3 (13.56 ± 8.56 copies/μg of DNA) but was then increased on days 5 and 7. Two of five animals originally assigned to the day 11 group died prematurely on day 5, and the variability in the remaining 3 at this later time point was too large to determine if the fungal burdens were increasing further. Semiquantitative cultures (CFU) were able to detect Aspergillus conidia as early as 1 h postinfection, and as observed with QNRT PCR, fungal burdens measured by this assay decreased until day 3 and remained stable at the other time points. These results are consistent with previously reported data in this model (26).
Table 3.
Comparison of PCR, GM enzyme immunoassay, and BG results obtained with samples from A. fumigatus-infected guinea pigsa
| Day p.i.b or parameter | Fungal burden |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Lung tissue |
BAL fluid |
Serum |
Whole blood, PCR | ||||||
| Culture | PCR | PCR | GM | BG | PCR | GM | BG | ||
| 0 | 4.50 | 31.04 | 72.52 | 0.14 | 79 | 0 | 0.11 | 15 | 37.69 |
| 0 | 4.47 | 60.22 | 98.20 | 0.17 | 20 | 0 | 0.09 | 18 | 0.00 |
| 0 | 4.47 | 41.17 | 85.61 | 0.23 | 61 | 0.1 | 0.09 | 29 | 0.00 |
| 0 | 4.43 | 25.78 | 96.07 | 0.17 | 35 | 0.1 | 0.05 | 8 | 7.44 |
| 0 | 4.28 | 9.52 | 90.15 | 0.2 | 70 | 3.99 | 0.32 | 15 | 18.54 |
| Mean (SD) | 4.43 (0.09) | 33.5 (18.8) | 88.5 (10.2) | 0.18 (0.03) | 53.0 (24.7) | 0.84 (1.76) | 0.13 (0.11) | 17.0 (7.65) | 12.7 (15.9) |
| 3 | 3.43 | 6.52 | 115.51 | 3.3 | 570 | 0 | 0.11 | 13 | 60.72 |
| 3 | 3.62 | 8.42 | 148.74 | 1.32 | 160 | 1.64 | 1.08 | 20 | 0.00 |
| 3 | 3.25 | 7.70 | 137.81 | 1.07 | 125 | 4.1 | 0.11 | 20 | 47.81 |
| 3 | 3.53 | 19.43 | 108.90 | 1.62 | 273 | 0.75 | 0.42 | 2 | 42.82 |
| 3 | 3.58 | 25.75 | 95.65 | 1.96 | 622 | 0 | 0.11 | 10 | 96.63 |
| Mean (SD) | 3.48 (0.15) | 13.6 (8.56) | 121.3 (21.6) | 1.85 (0.87) | 350 (232) | 1.30 (1.71) | 0.37 (0.42) | 13.0 (7.55) | 49.6 (34.8) |
| 5 | 3.23 | 47.62 | 118.66 | 6.44 | 1243 | 0 | 0.45 | 0 | 0.00 |
| 5 | 2.96 | 25.66 | 103.40 | 1.69 | 1295 | 0 | 0.23 | 7 | 42.40 |
| 5 | 3.41 | 69.32 | 81.10 | 1.6 | 420 | 0 | 0.24 | 62 | 36.77 |
| 5 | 3.48 | 78.44 | 109.30 | 7.45 | 1352 | 0 | 0.38 | 25 | 0.00 |
| 5 | 2.79 | 40.35 | 22.06 | 4.04 | 1100 | 0 | 0.43 | 0 | 0.00 |
| Mean (SD) | 3.17 (0.29) | 43.4 (24.6) | 86.9 (38.8) | 4.24 (2.68) | 1082 (382) | 0 (0) | 0.35 (0.10) | 18.8 (26.2) | 15.8 (21.8) |
| 7 | 3.51 | 73.00 | 78.02 | 12.6 | 1356 | 4.35 | 3.49 | 30 | 63.54 |
| 7 | 3.66 | 44.02 | 91.32 | 12.26 | 1588 | 4.28 | 10.97 | 220 | 0.00 |
| 7 | 2.62 | 33.99 | 99.46 | 6.18 | 1476 | 0 | 0.15 | 0 | 53.72 |
| 7 | 3.99 | 79.58 | 92.03 | 12.55 | 1530 | 4.13 | 12.49 | 1020 | 0.00 |
| 7 | 2.76 | 78.35 | 113.72 | 4.95 | 1510 | 0.4 | 0.13 | 0 | 0.00 |
| Mean (SD) | 3.31 (0.59) | 61.8 (21.2) | 94.9 (13.0) | 9.71 (3.81) | 1492 (86.2) | 2.63 (2.23) | 5.45 (5.92) | 254 (438) | 23.4 (32.3) |
| 11 | 3.60 | 125.59 | 81.86 | 12.13 | 1612 | 0.49 | 5.09 | 126 | 50.86 |
| 11 | 2.68 | 25.53 | 56.86 | 3.91 | 1150 | 0.7 | 0.62 | 30 | 14.75 |
| 11 | 3.53 | 19.45 | 72.10 | 12.11 | 1530 | 0 | 8.16 | 312 | 17.72 |
| 11d | 3.38 | 6.96 | NDc | ND | ND | ND | ND | ND | ND |
| 11d | 3.04 | 35.29 | ND | ND | ND | ND | ND | ND | ND |
| Mean (SD) | 3.27 (0.51) | 56.9 (59.6) | 70.3 (12.6) | 9.38 (4.74) | 1431 (246) | 0.40 (0.36) | 4.62 (3.79) | 156 (143) | 27.8 (20.0) |
| 11e | ND | 0.00 | 10.62 | 0.24 | 51 | 3.51 | 11.82 | 60 | 0.00 |
| 11e | ND | 0.00 | 0.00 | 0.14 | 35 | 0 | 0.12 | 0 | 0.00 |
| 11e | ND | 0.00 | 0.00 | 0.16 | 26 | 1.05 | 0.09 | 0 | 0.00 |
| 11e | ND | 0.00 | 0.00 | 0.11 | 75 | 0 | 0.11 | 0 | 0.00 |
Units used: culture, log no. of CFU/g of tissue; PCR for lung tissue, BAL fluid, or whole blood, log10 no. of copies/μg of DNA; PCR for serum, no. of copies/ml of serum; GM, index of positivity; BG, pg/ml of BAL fluid or whole blood. Positive results are in bold.
p.i., postinfection.
ND, not done.
Dead on day 5.
Uninfected.
Detection of A. fumigatus in BAL fluid.
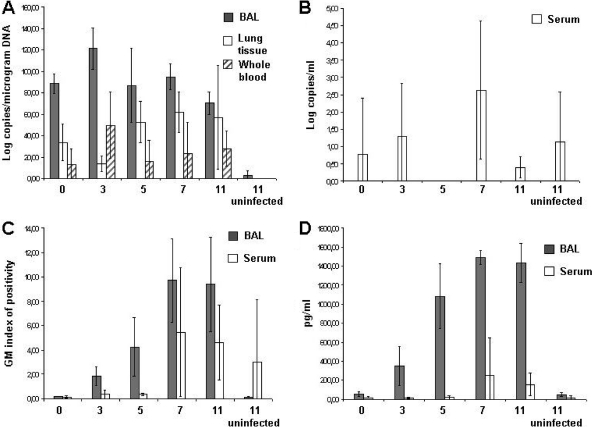
The QNRT PCR assay detected a high fungal burden in BAL fluid samples beginning at 1 h postinfection (mean, log10 88.51 ± 10.22 copies/μg of DNA). Detection at this early time point most likely represents inhaled conidia. However, the values measured at the later time points following germination of the conidia into hyphae (mean range for days 3, 5, 7, and 11, log10 70.27 to 121.32 copies/μg of DNA) remained elevated. One sample from an uninfected control animal was positive. Although the fungal burden was much lower than in other samples, the serum from this animal was positive for GM and BG.
BAL fluid was negative for GM and BG at 1 h postinfection, but all samples became positive on day 3 and levels steadily increased throughout the study (Fig. 3). None of the BAL fluid samples on days 3 to 11 of the study were negative by either of these assays, which were also 100% specific, as none of the uninfected controls were positive (Table 3).
Fig 3.
Time courses of fungal burdens as detected by all three methods. Results are presented as means ± standard deviations. (A) QNRT PCR detection in BAL fluid (gray), lung tissue (white), and whole blood (dashed boxes). (B) QNTR-PCR detection in serum. (C) GM detection in BAL fluid (gray) and serum (white). (D) BG detection in BAL fluid (gray) and serum (white).
Detection of A. fumigatus in serum and whole blood.
By QNRT PCR, fungal loads were detected in 10 (43.5%) of 23 serum samples from infected animals throughout the course of infection and in 9 (50%) of 18 samples collected on days 3 to 11. All samples collected on day 5 were negative. Two animals from the uninfected control group were also positive.
GM was positive in 7 (30.4%) of 23 samples from all infected animals (7 [38.8%] of 18 on days 3 to 11). Except for one sample collected on day 3, all positive results occurred on either day 7 or day 11. One sample from the uninfected control group was also positive for GM. Only 4 (17.4%) of 23 serum samples from infected animals were positive for BG (4 [22.2%] of 18 on days 3 to 11). Similar to GM, BG was detected beginning on the seventh day of infection. In whole blood, QNRT PCR detected fungal DNA in (60.9%) 14 of 23 samples from infected animals throughout the course of the study and in 11 (61.1%) of 18 samples on days 3 to 11. As shown in Table 3 and Fig. 3, detection of fungal DNA within this fluid was highly variable. However, no samples from the uninfected control group were positive.
Compared to serum, the number of samples of BAL fluid from infected guinea pigs that were positive, either throughout the course of infection or on days 3 to 11, was significantly higher for the QNRT PCR, GM, and BG assays (P < 0.01 for each comparison). The mean values within the BAL fluid samples for most time points for each assay were also significantly greater than those detected in the serum. Similarly, with BAL fluid, the number of samples positive by QNRT PCR and the mean value at each time point were significantly higher than with whole blood (P < 0.05 for each comparison).
DISCUSSION
In this study, our QNRT PCR assay proved to be highly sensitive and specific for the detection of A. fumigatus in various samples collected from guinea pigs. This approach combines the advantages of nested PCR, which is highly sensitive, and real-time PCR, which may allow the monitoring of disease progression. Our system is highly adaptable, and we have designed primers and probes that specifically target other Aspergillus species (data not shown in this study). These assays use the same primers for the first round of PCR, and species-specific oligonucleotides are used in the second round. These primers and probes were not evaluated in the present study, where the causative agent of infection was known. Traditionally discussed problems with nested PCR, i.e., higher rates of false-positive results due to contamination, were solved by DNA isolation and PCR setup in laminar-flow boxes in separate rooms, isolation of water samples with each batch of samples, and use of the undiluted PCR product as the template for the second round of PCR amplification. Moreover, the design of the highly A. fumigatus-specific oligonucleotides used in the internal round prevented cross-reactivity with airborne fungi.
Generally, the amount of fungal DNA present in clinical samples is very low. Even in patients with proven aspergillosis, DNA copy numbers may be low or PCR CT values may be very high and can be at the detection limit of a real-time PCR. In addition, fungal DNA is amplified with a high background of host DNA, which may make the detection of fungal DNA difficult. With our nested PCR assay, the first round of amplification with panfungal primers enriches the fungal DNA fraction and improves the fungal versus nonfungal DNA ratio. Specific oligonucleotides used in the second round can therefore easily find their targets, allowing for more-efficient PCRs (19). The preamplification step enables the detection of very low copy numbers in blood samples. The lower CT values observed with this technique make the PCR assay more robust and reproducible, which is very important in routine diagnostics.
In this study, QNRT PCR detected fungal DNA in all lung tissue samples from infected animals as early as 1 h postinfection; therefore, we conclude that our DNA isolation method is applicable for isolation of fungal DNA from inhaled conidia. GM and BG were not detected at this time point, as the conidia had not yet begun to germinate and form hyphae at this early time point (13, 15). Due to this fact, data obtained from day 0 were excluded from analysis and were not used to calculate sensitivity. At day 3, there was a significant decrease in the fungal burden (P < 0.05). Similar results were also observed in other studies where a drop of the fungal burden occurred between day 0 and day 3 when measured by semiquantitative culture (23, 26, 28). This may be due to partial removal of the infecting organism by a residual innate immune system response. At day 5, the fungal burden increased again and remained relatively stable until day 7. Because only 3 samples were taken on day 11 (two animals died prematurely on day 5) and there was high interindividual variability, we cannot assess if the fungal burden increased further or remained stable. However, the relative sensitivity and specificity of the QNRT PCR assay in lung tissue were both 100%.
The samples most suitable for early invasive aspergillosis diagnostics in this study proved to be BAL fluid samples, as they were positive in all infected animals with each assay (QNRT PCR, GM, and BG) beginning on day 3 (Table 3). The relative sensitivity of QNRT PCR, GM, and BG in BAL fluid was therefore 100%, and the specificities were 75%, 100%, and 100%, respectively. Beginning on day 3, the GM data correlated with the BG results (Fig. 3 C and D) (Spearman rank correlation, rho 0.93; P < 0.0001), which is in concordance with the results of other studies (8, 23). Although fungal burdens determined by QNRT PCR were highly variable, among different animals they remained elevated throughout the duration of the study.
Results from serum and whole blood were more heterogeneous. The sensitivities of the QNRT PCR, GM, and BG in serum were 50%, 39% and 22%, respectively, while specificities ranged from 50% to 100%. PCR detected fungal burdens in all but one sample that were either GM or BG positive. The only serum sample that was negative by PCR but positive by serological methods was from one animal with a very low fungal burden in lung tissue. Two of four serum samples taken from uninfected animals at day 11 were positive by PCR but at very low levels compared to those observed in the infected animals. One of these samples was also positive by GM. Although the cause of this false positive is unknown, it may have been due to cross-contamination during sample collection. The measurement of fungal DNA in individual serum and whole blood samples was at the detection limit of real-time PCR, and there was no correlation between the fungal loads detected in serum and whole blood. Unfortunately, data concerning the health condition of the animals at the time of death were not collected; therefore, we cannot correlate the results of these assays with the physical state of the guinea pigs.
In conclusion, this novel QNRT PCR method is highly sensitive and specific for detection of A. fumigatus in different types of samples. With serum and whole blood, QNRT PCR was more sensitive than GM and BG. However, despite a high fungal burden in lung tissue and BAL fluid, serum and whole blood might be negative with the various assays even in advanced stages of disease. Therefore, we conclude that when testing serum, a combination of assays may increase the possibility of early detection of invasive pulmonary aspergillosis, and the results of all methods may not be positive at the same time due to potential differences in the kinetics of antigen and DNA release. With BAL fluid, all three methods were positive throughout the course of the infection, and therefore, BAL fluid samples seem to be the most suitable for early detection of invasive pulmonary aspergillosis. It is highly probable that similar interpatient variability may occur with clinical samples, and this may contribute to the ambiguous interpretation of PCR results and subsequently restrict standardization and use fungal PCR diagnostics in routine clinical practice.
ACKNOWLEDGMENTS
This study was supported in part by grants from the Ministry of Health of the Czech Republic (NS10442-3/2009, NS10441-3/2009), the Ministry of Industry and Trade of the Czech Republic (FR-TI2/254), and NIH/NIAID contract N01-AI-30041.
Footnotes
Published ahead of print 21 December 2011
REFERENCES
- 1. Becker MJ, et al. 2003. Galactomannan detection in computerized tomography-based broncho-alveolar lavage fluid and serum in haematological patients at risk for invasive pulmonary aspergillosis. Br. J. Haematol. 121:448–457 [DOI] [PubMed] [Google Scholar]
- 2. Christie JD. 2003. Diagnosis of invasive mold infection. Is PCR the answer? Am. J. Clin. Pathol. 119:38–44 [DOI] [PubMed] [Google Scholar]
- 3. De Pauw B, et al. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 46:1813–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Espy MJ, et al. 2006. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev. 19:165–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ferns RB. 2006. Evaluation of the role of real-time PCR in the diagnosis of invasive aspergillosis. Leuk. Lymphoma 47:15–20 [DOI] [PubMed] [Google Scholar]
- 6. Francesconi A, et al. 2006. Characterization and comparison of galactomannan enzyme immunoassay and quantitative real-time PCR assay for detection of Aspergillus fumigatus in bronchoalveolar lavage fluid from experimental invasive pulmonary aspergillosis. J. Clin. Microbiol. 44:2475–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graybill JR, Kaster SR. 1984. Experimental murine aspergillosis. Comparison of amphotericin B and a new polyene antifungal drug, SCH 28191. Am. Rev. Respir. Dis. 129:292–295 [PubMed] [Google Scholar]
- 8. Khan ZU, Ahmad S, Theyyathel AM. 2008. Diagnostic value of DNA and (1→3)-d-glucan detection in serum and bronchoalveolar lavage of mice experimentally infected with Fusarium oxysporum. 47301-0. J. Med. Microbiol. 57:36–42 [DOI] [PubMed] [Google Scholar]
- 9. Klingspor L, Loeffler J. 2009. Aspergillus PCR formidable challenges and progress. Med. Mycol. 47(Suppl. 1):S241–S247 [DOI] [PubMed] [Google Scholar]
- 10. Maertens J, et al. 2009. Bronchoalveolar lavage fluid galactomannan for the diagnosis of invasive pulmonary aspergillosis in patients with hematologic diseases. Clin. Infect. Dis. 49:1688–1693 [DOI] [PubMed] [Google Scholar]
- 11. Meersseman W, et al. 2008. Galactomannan in bronchoalveolar lavage fluid: a tool for diagnosing aspergillosis in intensive care unit patients. Am. J. Respir. Crit. Care Med. 177:27–34 [DOI] [PubMed] [Google Scholar]
- 12. Mengoli C, Cruciani M, Barnes RA, Loeffler J, Donnelly JP. 2009. Use of PCR for diagnosis of invasive aspergillosis: systematic review and meta-analysis. Lancet Infect. Dis. 9:89–96 [DOI] [PubMed] [Google Scholar]
- 13. Mennink-Kersten MASH, Ruegebrink D, Wasei N, Melchers WJG, Verweij PE. 2006. In vitro release by Aspergillus fumigatus of galactofuranose antigens, 1,3-{beta}-d-glucan, and DNA, surrogate markers used for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 44:1711–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miceli MH, et al. 2008. Strong correlation between serum aspergillus galactomannan index and outcome of aspergillosis in patients with hematological cancer: clinical and research implications. Clin. Infect. Dis. 46:1412–1422 [DOI] [PubMed] [Google Scholar]
- 15. Morton CO, et al. 2010. Dynamics of extracellular release of Aspergillus fumigatus DNA and galactomannan during growth in blood and serum. J. Med. Microbiol. 59:408–413 [DOI] [PubMed] [Google Scholar]
- 16. National Academy of Sciences 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC [Google Scholar]
- 17. Neofytos D, et al. 2009. Epidemiology and outcome of invasive fungal infection in adult hematopoietic stem cell transplant recipients: analysis of Multicenter Prospective Antifungal Therapy (PATH) Alliance registry. Clin. Infect. Dis. 48:265–273 [DOI] [PubMed] [Google Scholar]
- 18. Nierman WC, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 19. Nogva HK, Rudi K. 2004. Potential influence of the first PCR cycles in real-time comparative gene quantifications. Biotechniques 37:246–248, 250–253 [DOI] [PubMed] [Google Scholar]
- 20. Pagano L, Caira M, Valentini CG, Posteraro B, Fianchi L. 2010. Current therapeutic approaches to fungal infections in immunocompromised hematological patients. Blood Rev. 24:51–61 [DOI] [PubMed] [Google Scholar]
- 21. Patterson TF. 2005. The future of animal models of invasive aspergillosis. Med. Mycol. 43(Suppl. 1):S115–S119 [DOI] [PubMed] [Google Scholar]
- 22. Schabereiter-Gurtner C, Selitsch B, Rotter ML, Hirschl AM, Willinger B. 2007. Development of novel real-time PCR assays for detection and differentiation of eleven medically important Aspergillus and Candida species in clinical specimens. J. Clin. Microbiol. 45:906–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sheppard DC, et al. 2006. Comparison of three methodologies for the determination of pulmonary fungal burden in experimental murine aspergillosis. Clin. Microbiol. Infect. 12:376–380 [DOI] [PubMed] [Google Scholar]
- 24. Sheppard DC, et al. 2004. Novel inhalational murine model of invasive pulmonary aspergillosis. Antimicrob. Agents Chemother. 48:1908–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tuon FF. 2007. A systematic literature review on the diagnosis of invasive aspergillosis using polymerase chain reaction (PCR) from bronchoalveolar lavage clinical samples. Rev. Iberoam. Micol. 24:89–94 [PubMed] [Google Scholar]
- 26. Vallor AC, et al. 2008. Assessment of Aspergillus fumigatus burden in pulmonary tissue of guinea pigs by quantitative PCR, galactomannan enzyme immunoassay, and quantitative culture. Antimicrob. Agents Chemother. 52:2593–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wessa P. 2011. Free statistics software, Office for Research Development and Education, version 1.1.23-r7. http://www.wessa.net/
- 28. Zhao Y, et al. 2010. Detection of Aspergillus fumigatus in a rat model of invasive pulmonary aspergillosis by real-time nucleic acid sequence-based amplification. J. Clin. Microbiol. 48:1378–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]