Abstract
Resistance against β-lactam antibiotics is a growing challenge for managing severe bacterial infections. The rapid and cost-efficient determination of β-lactam resistance is an important prerequisite for the choice of an adequate antibiotic therapy. β-Lactam resistance is based mainly on the expression/overexpression of β-lactamases, which destroy the central β-lactam ring of these drugs by hydrolysis. Hydrolysis corresponds to a mass shift of +18 Da, which can be easily detected by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). Therefore, a MALDI-TOF MS-based assay was set up to investigate different enterobacteria for resistance against different β-lactam antibiotics: ampicillin, piperacillin, cefotaxime, ceftazidime, ertapenem, imipenem, and meropenem. β-Lactamases are enzymes that have a high turnover rate. Therefore, hydrolysis can be detected by MALDI-TOF MS already after a few hours of incubation of the bacteria to be tested with the given antibiotic. The comparison of the MS-derived data with the data from the routine procedure revealed identical classification of the bacteria according to sensitivity and resistance. The MALDI-TOF MS-based assay delivers the results on the same day. The approved routine procedures require at least an additional overnight incubation.
INTRODUCTION
The growing number of antibiotic-resistant microorganisms is an increasing health care problem (7). The rapid detection and determination of the type of resistance are important prerequisites for the adequate choice of an efficient antibiotic therapy. Resistance can be based on different mechanisms. These include altered influx/efflux systems preventing the antibiotic agents from acting in the cells (6, 14), prevention of the interaction of the antibiotic agent with the target by alteration of the target binding site (10), and enzymatic modification or destruction of the activity of the antibiotic agent (19).
A large group of antibiotics is the group of β-lactam antibiotics. These comprise different antibiotics, such as penicillin derivatives (e.g., ampicillin and piperacillin), cephalosporins with different spectra, monobactams, and carbapenems. Because of their minimal side effects and their good physiological compatibility, they are frequently administered. As a result, the development of resistance against these drugs is a major problem (4). The shared molecular structure of these antibiotics is the central β-lactam ring. Targets of β-lactam antibiotics are the periplasmic bacterial peptidoglycan transpeptidases (9), which are irreversibly destroyed by formation of a dead-end complex impairing cell wall synthesis. A common resistance mechanism developed by different microorganisms is the inactivation of β-lactam antibiotics by expression/overexpression of β-lactamases, which destroy the β-lactam ring by hydrolysis (2, 9). The hydrolyzed form is unable to interact with the transpeptidases and thereby is inactivated.
Usually, resistance is analyzed on the phenotypic level by disk diffusion assays, the Etest, or automated microbiology systems such as the Phoenix (BD Diagnostic Systems, Heidelberg, Germany) or the Vitek 2 (bioMérieux, Nürtingen, Germany), which determine the MIC by monitoring the growth of the bacteria (2, 18). Starting from a pure culture, the disk diffusion assay and Etest take at least one work day before a result can be stated because an overnight incubation step is required. Some automated systems can provide an antimicrobial susceptibility test (AST) report after a few hours. In the case of Bacteroides fragilis, it was recently shown that the differentiation between cfiA-negative and cfiA-positive isolates (cfiA encodes a class B metallo-beta-lactamase) was possible directly from matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) profile spectra (13, 20). However, such a clonal differentiation of resistant and susceptible clones cannot be expected for the majority of bacteria. Further, a novel MALDI-TOF MS-based assay for the analysis of β-lactamase activity has been described (3, 8, 11). For the β-lactam antibiotics, resistance can be easily monitored by mass spectrometry because hydrolysis of the β-lactam ring by β-lactamases results in disappearance of the original mass peak through a molecular mass shift of +18 Da of the antibiotic agent. In many cases, the hydrolysis is directly followed by a decarboxylation of the hydrolyzed product, resulting in a further mass shift of −44 Da from the hydrolyzed form. Since mass spectrometry is designed to determine such mass differences of molecules, a MALDI-TOF MS-based assay was set up to analyze the hydrolysis reactions of different β-lactam antibiotics.
MATERIALS AND METHODS
Bacterial strains and cultivation.
Escherichia coli strain DH5α (ATCC, Germany) was used as a β-lactamase-negative control strain. Clinical isolates of E. coli (Max von Pettenkofer-Institute, Munich, Germany) classified by standard methods as β-lactamase-producing strains were employed as positive controls (5 strains). The extended-spectrum β-lactamase (ESBL) phenotype indicated by automated microbiology systems (Phoenix and Vitek) was further corroborated by a confirmatory ESBL test using the Oxoid combination disk method (5). Clinical isolates of Klebsiella pneumoniae (Max von Pettenkofer-Institute) classified as carbapenemase-nonproducing or -producing strains by standard laboratory methods (e.g., Hodge test) as well as by PCR (kpc gene) were used as negative (1 strain) or positive (2 strains) controls, respectively. See the supplemental material for a detailed characterization of the isolates.
Bacteria were cultivated on Columbia blood agar plates (Heipha, Germany) without selective agents overnight at 37°C. Fresh overnight cultures were employed for the tests.
Hydrolysis assay with plated microorganisms.
Ampicillin (Sigma-Aldrich, Germany; 10 mg/ml in H2O), piperacillin (Sigma-Aldrich, Germany; 1 mg/ml in H2O), ceftazidime (Sigma-Aldrich, Germany; 0.25 mg/ml in 10 mM ammonium hydrogen citrate, pH 7.0 [Fluka, Germany]), cefotaxime (Sigma-Aldrich, Germany; 0.5 mg/ml in H2O), imipenem (Sigma-Aldrich, Germany; 0.5 mg/ml in 10 mM ammonium hydrogen citrate, pH 7.0 [Fluka, Germany]), ertapenem (MSD, Munich, Germany; 0.5 mg/ml in 10 mM ammonium hydrogen citrate, pH 7.0 [Fluka, Germany]), and meropenem (AstraZeneca, Wedel, Germany; 0.5 mg/ml in 10 mM ammonium hydrogen citrate, pH 7.0 [Fluka, Germany]) were tested with the different E. coli strains and the different K. pneumoniae strains. For inhibition of hydrolysis, the tests were performed in the presence of clavulanic acid (Sigma, Germany; 0.05 mg/ml), tazobactam (Sigma, Germany; 0.05 mg/ml), or 3-aminophenylboronic acid (APBA) (Aldrich, Germany; 2 mg/ml). The amount of bacteria filling a 1-μl inoculation loop was resuspended in 10 μl of the antibiotic solution and incubated at 37°C under agitation for 3 h or for the indicated time. Subsequently, the tubes were centrifuged for 2 min at 13,000 × g at room temperature. The cell-free supernatant was analyzed by MALDI-TOF MS.
Preparation of cells for hydrolysis assay from fresh blood cultures.
Blood culture bottles (BD Bactec Plus+ Aerobic/F; Becton Dickinson, Germany) were inoculated with a carbapenemase-sensitive K. pneumoniae strain and two carbapenemase-resistant K. pneumoniae strains. Incubation was performed in a Bactec blood culture instrument (Becton Dickinson, Heidelberg, Germany) until the culture was flagged as positive. One milliliter of blood culture fluid was taken from the culture bottle with a syringe, and cells were isolated using the MALDI Sepsityper kit (Bruker Daltonik GmbH, Germany). Briefly, the blood cells were lysed with solution 1+. After centrifugation, the cells were washed with washing solution once and subsequently washed with water-ethanol. The last steps of the protocol were modified. Washing with the washing solution from the Sepsityper kit was performed twice instead of once. Subsequently, the isolated bacteria were directly resuspended in 10 μl of the antibiotic solution instead of water-ethanol as advised in the kit manual. The following incubation and preparation were performed as described above.
MALDI-TOF MS analysis.
One microliter of the supernatant from the hydrolysis assay was directly spotted onto a polished steel MALDI target plate. Dried spots were overlaid with MALDI matrix (10 mg/ml of α-cyano-4-hydroxy-cinnamic acid [α-HCCA] in 50% acetonitrile–2.5% trifluoroacetic acid; Bruker Daltonik, Bremen, Germany). After drying of the matrix, MALDI-TOF MS measurements were performed with a Microflex LT benchtop mass spectrometer (Bruker Daltonik GmbH, Germany) equipped with a 60-Hz nitrogen laser. Parameter settings have been optimized for the low mass range (ion source 1 [IS1], 20 kV; IS2, 17.5 kV; lens, 6.5 kV; detector gain, 2,650 V; gating, none). Spectra were recorded in the positive linear mode in the mass range of 100 Da to 1,000 Da with the maximum laser frequency. For calibration, an external standard consisting of bradykinin(1-5) (Sigma-Aldrich, Germany) and bradykinin(1-7) (Sigma-Aldrich, Germany) was prepared in the same way as the antibiotic samples. The HCCA peaks [M + H]+ at 190.05 Da and [2 M + H]+ at 379.02 Da, the bradykinin(1-5) peak [M + H]+ at 573.31 Da, and the bradykinin(1-7) peak [M + H]+ at 757.40 Da were employed for calibration.
Data analysis.
The acquired MALDI-TOF MS spectra were analyzed with the software Flexanalysis 3.3 (Bruker Daltonik GmbH, Germany). Spectra were smoothed (algorithm, SavitzkyGolay; width, 0.2 m/z; cycles, 1) and baseline subtracted (algorithm, TopHat). Peaks were manually selected with the following parameter settings: peak detection algorithm, centroid; signal-to-noise threshold, 2; relative intensity threshold, 0%; minimum intensity threshold, 100; peak width, 0.2 m/z; height, 80%; baseline subtraction, TopHat. Only peaks belonging to the corresponding antibiotic drug and the respective degradation products were labeled. Since no automated data evaluation was available at this time, the classification of susceptible, intermediate, and resistant was done by estimation. For this, the peak intensities of the nonhydrolyzed forms were compared to the intensities of the hydrolyzed forms. Strains were classified as presumably susceptible if the intensity distributions of the nonhydrolyzed and the hydrolyzed forms were similar to those for the negative control. Strains were classified as resistant if the intensities of the hydrolyzed forms represented 80% or more of the intensities of the nonhydrolyzed and the hydrolyzed forms. Strains showing an intensity distribution between that of the negative control and the 80% distribution were appraised as presumably intermediate. The number of analyzed strains was not sufficient to draw more precise cutoffs.
Routine assay.
The MICs of the antibiotics were determined by employing the Epsilometer test. In brief, Etest strips (bioMérieux, Nürtingen, Germany) were placed on 85-mm Mueller-Hinton agar plates (BD Diagnostics Systems) which had been inoculated with 0.5 McFarland standard suspensions of test isolates. All plates were incubated at 35°C for 20 to 24 h before being examined. The MIC was determined to be the value at which the elliptical growth margin intersected the Etest strip. The evaluation of resistance was performed according to the current CLSI guideline M100-S21 for aerobic bacteria (5a).
RESULTS
Analysis of penicillin derivatives with different E. coli strains.
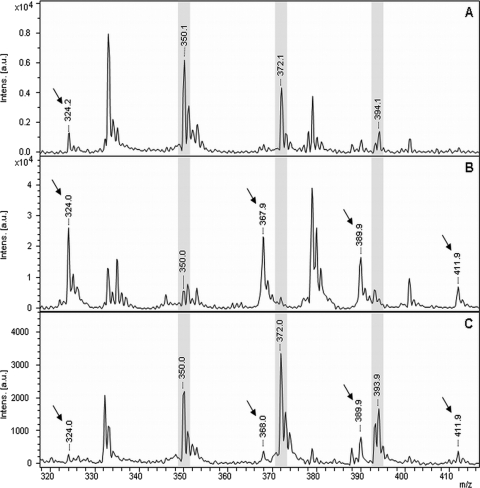
The MS spectrum corresponding to the supernatant derived from E. coli DH5α after incubation with ampicillin for 3 h revealed the molecular peak of ampicillin [M + H]+ at 350.1 Da and the corresponding sodium adducts [M + Na]+ at 372.1 Da and [M + 2Na]+ at 394.1 Da. In contrast, the spectra of the supernatants of the β-lactamase-producing strains revealed clearly decreased intensities of the molecular peak of ampicillin and the corresponding adducts. Additional peaks at 367.9, 389.9, 411.8, and 324.0 Da appeared to correspond to the hydrolyzed form of ampicillin, its sodium adducts, and the hydrolyzed, decarboxylated form of ampicillin, respectively. The different ampicillin forms and the appropriate masses are listed in Table 1. Comparison of the spectra revealed a slight spontaneous hydrolysis of ampicillin also for E. coli DH5α. Performing the hydrolysis assay with β-lactamase-producing E. coli strains in the presence of the β-lactamase inhibitor clavulanic acid demonstrated the inhibition of hydrolysis and the conservation of the nonhydrolyzed forms of ampicillin. Figure 1 shows representative spectra of the hydrolysis and the inhibition of hydrolysis of ampicillin for the ampicillin-sensitive E. coli strain DH5α and an ampicillin-resistant strain, respectively. Except for the control strain E. coli DH5α, all E. coli strains tested showed resistance against the penicillin derivative ampicillin. The K. pneumoniae control strain 32199 showed intermediate hydrolysis behavior. The other K. pneumoniae strains showed the resistance pattern.
Table 1.
Calculated masses and corresponding molecular forms defining the sensitivity and resistance patterns for different antibiotics
| Antibiotic | Mol wt (g/mol) | Sensitivity pattern (Da) |
Resistance pattern (Da) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [M + H]+ | [M + Na]+ | [M + K]+ | [M + 2 Na]+ | [M + Na + K]+ | [M + 3 Na]+ | [M-Xa + H]+ | [Mhydr. + H]+ | [Mhydr. + Na]+ | [Mhydr. + 2 Na]+ | [Mhydr. + Na + K]+ | [Mhydr./decarb. + H]+ | [Mhydr.-X + H]+ | [Mhydr./decarb.-X + H]+ | [Mhydr./decarb. + Na]+ | [Mhydr./decarb. + K]+ | ||
| Ampicillin | 349.4 | 350.4 | 372.4 | 394.4 | 368.4 | 390.4 | 412.4 | 324.4 | |||||||||
| Piperacillin | 517.5 | 518.5 | 540.5 | 562.5 | 536.5 | 558.5 | 580.5 | (492.5) | |||||||||
| Cefotaxime | 455.5 | 456.5 | 478.5 | 396.5 | 414.5 | 370.5 | |||||||||||
| Ceftazidime | 546.6 | 547.6 | 468.6 | 486.6 | 442.6 | ||||||||||||
| Ertapenem | 475.5 | 476.5 | 498.5 | 514.5 | 520.5 | 536.5 | 542.5 | 494.5 | 516.5 | 538.5 | 554.5 | 450.5 | 472.5 | 488.5 | |||
| Imipenem | 299.4 | 300.4 | |||||||||||||||
| Meropenem | 383.4 | 384.5 | 406.5 | 428.5 | |||||||||||||
X, acetyl for cefotaxime and pyridine for ceftazidime.
Fig 1.
(A and B) MALDI-TOF MS spectra of ampicillin after incubation with the β-lactamase-negative E. coli strain DH5α (A) and a β-lactamase-producing strain (B). (C) Inhibition of hydrolysis by a β-lactamase-producing strains was performed in the presence of clavulanic acid. Peaks corresponding to the nonhydrolyzed form of ampicillin are highlighted in gray. Peaks corresponding to the hydrolyzed form of ampicillin are indicated with an arrow.
Comparable results were achieved for the hydrolysis of piperacillin and the inhibition of hydrolysis by the β-lactamase inhibitor tazobactam. In contrast to ampicillin, piperacillin more strongly tends to form sodium adducts, resulting in an almost undetectable molecular peak [M + H]+ for piperacillin (517.9 Da) and in the [M + Na]+ peak (539.9 Da) as the main form detected for the nonhydrolyzed forms. Incubation of piperacillin with β-lactamase-producing strains resulted in the detection of the hydrolyzed forms [Mhydr. + H]+ at 535.9 Da, [Mhydr. + Na]+ at 557.9 Da, and [Mhydr. + 2Na]+ at 557.9 Da. Compared to ampicillin, the hydrolyzed form of piperacillin seems to be more stable, and only a very small peak for the hydrolyzed decarboxylated form [Mhydr./decarb. + H]+ at 492.0 Da was detectable. Again, a spontaneous hydrolysis was also observed for the control strain E. coli DH5α and in the inhibition setups employing tazobactam (Fig. 2). Table 1 gives a summary of the peaks and the corresponding molecular structures. The analysis results for the different strains tested are listed in Table 2. Additionally, the corresponding results of the routine assay are shown. For ampicillin, no difference was observed between the MALDI-TOF MS analysis and the routine Etest. Similar results were obtained for piperacillin and the inhibition of its hydrolysis by tazobactam by the two different methods. An exception was observed for one E. coli strain, which was classified as “presumptive intermediate” by MALDI-TOF MS and “resistant” by the routine procedure, and for the K. pneumoniae control strain which was classified as “presumably intermediate” by MALDI-TOF MS and “sensitive” by the Etest.
Fig 2.
(A and B) MALDI-TOF MS spectra of piperacillin after incubation with the β-lactamase-negative E. coli strain DH5α (A) and a β-lactamase-producing strain (B). (C) Inhibition of hydrolysis by a β-lactamase-producing strains was performed in the presence of tazobactam. Peaks corresponding to the nonhydrolyzed form of piperacillin are highlighted in gray. Peaks corresponding to the hydrolyzed form of piperacillin are indicated with an arrow.
Table 2.
Comparison of MALDI-TOF MS results and results from the routine procedure
| Species | Strain | Test | Resulta for: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | Ampicillin/clavulanic acid | Piperacillin | Piperacillin-tazobactam | Cefotaxime | Cefotaxime-clavulanic acid | Ceftazidime | Ceftazidime-clavulanic acid | Ertapenem | Ertapenem-APBA | Imipenem | Imipenem-APBA | Meropenem | Meropenem-APBA | |||
| E. coli | DH5α | MALDI-TOF MS | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| 901200230 | MALDI-TOF MS | R | S | R | S | R | S | R | S | S | S | S | S | S | S | |
| Etest/MIC (μg/ml) | R/>256 | S/8 | R/>256 | S/6 | R/>256 | S/0.125 | I/8 | S/0.19 | S/0.016 | ND | S/0.125 | ND | S/0.023 | ND | ||
| 901200284 | MALDI-TOF MS | R | S | R | S | R | S | R | S | S | S | S | S | S | S | |
| Etest/MIC (μg/ml) | R/>256 | S/6 | R/>256 | S/1.5 | R/12 | S/0.064 | R/16 | S/0.125 | S/0.023 | ND | S/0.125 | ND | S/0.016 | ND | ||
| 901200288 | MALDI-TOF MS | R | S | R | S | R | S | R | S | S | S | S | S | S | S | |
| Etest/MIC (μg/ml) | R/>256 | S/6 | R/>256 | S/6 | R/>256 | S/0.125 | R/16 | S/0.25 | S/0.023 | ND | S/0.19 | ND | S/0.032 | ND | ||
| 901200370 | MALDI-TOF MS | R | S | R | S | R | S | R | S | S | S | S | S | S | S | |
| Etest/MIC (μg/ml) | R/>256 | S/6 | R/>256 | S/2 | R/>256 | S/0.125 | R/48 | S/0.38 | S/0.125 | ND | S/0.19 | ND | S/0.047 | ND | ||
| 100329-02 | MALDI-TOF MS | R | S | I | S | R | S | S | S | S | S | S | S | S | S | |
| Etest/MIC (μg/ml) | R/>256 | S/6 | R/>256 | S/1 | R/16 | S/0.047 | S/1.5 | S/0.125 | S/0.016 | ND | S/0.125 | ND | S/0.016 | ND | ||
| 100329-03 | MALDI-TOF MS | R | S | R | S | R | S | S | S | S | S | S | S | S | S | |
| Etest/MIC (μg/ml) | R/>256 | S/4 | R/>256 | S/2 | R/>256 | S/0.125 | S/1 | S/0.125 | S/0.008 | ND | S/0.19 | ND | S/0.016 | ND | ||
| Klebsiella pneumoniae | 32199 | MALDI-TOF MS | I | S | I | S | S | S | S | S | S | S | S | S | S | S |
| Etest/MIC (μg/ml) | I/24 | S/1.5 | S/6 | S/2 | S/0.032 | NA/0.047 | S/0.125 | NA/0.125 | S/0.008 | ND | S/0.125 | ND | S/0.016 | ND | ||
| 6391 | MALDI-TOF MS | R | R | R | R | R | R | R | R | R | S | R | R | R | R | |
| Etest/MIC (μg/ml) | R/>256 | R/>256 | R/>256 | R/>256 | R/>256 | R/>4 | R/>256 | R/>4 | R/>32 | ND | R/>32 | ND | R/>32 | ND | ||
| 6737 | MALDI-TOF MS | R | R | R | R | R | R | R | R | R | S | R | R | R | S | |
| Etest/MIC (μg/ml) | R/>256 | R/>256 | R/>256 | R/>256 | R/>256 | R/>4 | R/>256 | R/>4 | R/8 | ND | R/6 | ND | R/6 | ND | ||
For the MALDI-TOF MS results, S, R, and I are the sensitivity, resistance, and intermediate patterns, respectively, according to Table 1. ND, not determined; NA, not applicable.
Analysis of cephalosporins with different E. coli and K. pneumoniae strains.
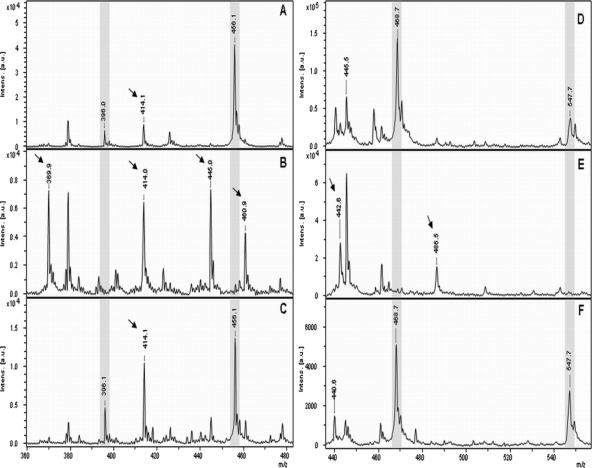
The hydrolysis of the cephalosporins cefotaxime and ceftazidime by different E. coli and K. pneumoniae strains was analyzed. The molecular peak of cefotaxime ([M + H]+ 456.1 Da) was detectable in the spectrum derived from E. coli DH5α. For cefotaxime, no sodium adducts were found in the spectra. Analyzing the lower mass range of the spectra revealed that cefotaxime seems to be very labile under MALDI-TOF conditions. At the distance of −60 Da, a peak was observed at 396.0 Da, corresponding to the elimination of the acetyl group of cefotaxime. Additionally, a small peak at +18 Da (414.1 Da) from the deacetylated form was detectable, indicating a slight spontaneous hydrolysis of the deacetylated form. Incubation of cefotaxime with resistant strains resulted in the disappearance of the molecular peak at 456.1 Da, but no hydrolysis product of the whole cefotaxime corresponding to a peak of +18 Da (474.1 Da) could be observed. Two additional peaks at 445.0 Da and 460.9 Da appeared in the spectra derived from the cephalosporin-resistant bacteria. The corresponding molecular structures have not been identified so far. Further, all spectra derived from resistant bacteria revealed a clear disappearance of the peak at 396.0 Da (the deacetylated, nonhydrolyzed form) and an increase of the peak at 414 Da corresponding to the hydrolyzed, deacetylated form of cefotaxime. An additional peak at 369.9 Da was observed, which represents the deacetylated, hydrolyzed, and decarboxylated form of cefotaxime. Inhibition of hydrolysis of cefotaxime was performed in the presence of clavulanate. The peaks at 369.9 Da, 445.0 Da, and 460.9 Da, which are products/indicators of hydrolysis, disappeared or were clearly reduced. In contrast, the peaks at 456.1 Da and 396.1 Da were detectable again. Representative spectra are shown in Fig. 3A to C. The different forms of cefotaxime and the corresponding masses are listed in Table 1. Considering the observed peak pattern, a clear differentiation between cefotaxime-resistant and -sensitive bacteria was possible. The results obtained by MALDI-TOF MS were concordant with the results obtained by the Etest (Table 2).
Fig 3.
MALDI-TOF MS spectra of cefotaxime (A to C) and ceftazidime (D to F) after incubation with the β-lactamase-negative E. coli strain DH5α (A and D) and a β-lactamase producing strain (B and E). Inhibition of hydrolysis by a β-lactamase-producing strains was performed in the presence of clavulanic acid (C and F). Peaks corresponding to the nonhydrolyzed form of the respective antibiotic are highlighted in gray. Peaks corresponding to the hydrolyzed form of the respective antibiotic are indicated with an arrow.
The analysis of ceftazidime ([M + H]+ 547.6 Da) after incubation with sensitive and resistant bacteria revealed similar results (Fig. 3D to F). After incubation of ceftazidime with sensitive bacteria, only the molecular peak of ceftazidime (547.7 Da), and no sodium adducts, was detectable. In the lower mass range at a distance of −79 Da, a peak at 468.7 Da was visible, corresponding to the elimination of the pyridine ring of ceftazidime. After incubation of ceftazidime with resistant bacteria, no hydrolysis product of the complete ceftazidime molecule could be detected, but hydrolysis of the pyridine-eliminated molecule was detectable at [M + H]+ 486.6 Da. Further, the additional elimination of the carboxyl group (−44 Da of the pyridine-eliminated, hydrolyzed form) was detectable at [M + H]+ 442.6 Da. Inhibition of the β-lactamase activity by clavulanic acid resulted in the preservation of the peaks at 468.7 Da and 547.7 Da. Table 1 gives a summary of the different forms of ceftazidime and the corresponding masses. From the characteristic peak pattern, a differentiation between ceftazidime-sensitive and ceftazidime-resistant bacteria was possible. The comparison of the results obtained with the MALDI-TOF MS measurements with the results obtained with the approved routine method revealed an almost complete match (Table 2). An exception was observed for one E. coli strain, which was classified as “resistant” by MALDI-TOF MS and as “intermediate” by the Etest.
Analysis of carbapenems with different K. pneumoniae strains.
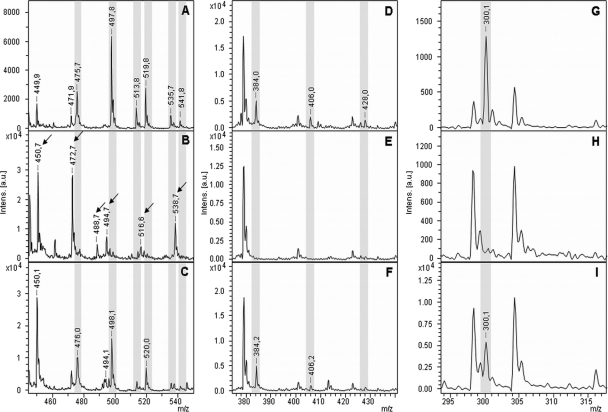
The hydrolysis of the carbapenems ertapenem, meropenem, and imipenem by different K. pneumoniae strains was investigated. After incubation of ertapenem ([M + H]+ 476.5 Da) with a carbapenem-sensitive strain, the molecular peak at 476.8 Da and different adduct peaks were detectable. The main peak of the spectrum at 498.7 Da corresponds to the single sodium adduct of ertapenem. The peaks at 514.7 Da, 520.7 Da, 536.8 Da, and 542.7 Da correspond to [M + K]+, [M + 2Na]+, [M + Na + K]+, and [M + 3Na]+, respectively. Carbapenem-resistant K. pneumoniae strains completely hydrolyzed ertapenem. The resulting peak pattern corresponds to the hydrolyzed form [Mhydr. + H]+ at 494.7 Da, the hydrolyzed sodium adduct [Mhydr. + Na]+ at 516.6 Da, the hydrolyzed double sodium adduct [Mhydr. + 2Na]+ at 538.7 Da, the hydrolyzed, decarboxylated form [Mhydr./decarb. + H]+ at 450.7, its sodium adduct [Mhydr./decarb. + Na]+ at 472 Da, and the hydrolyzed, decarboxylated potassium adduct [Mhydr./decarb. + K]+ at 488 Da (Fig. 4A and B). The completely different peak patterns resulting from carbapenem-sensitive and carbapenem-resistant strains facilitated the unambiguous identification of carbapenemase-positive strains (Table 1). Analysis of the strains with the approved routine Etest revealed an identical evaluation (Table 2). Inhibition of the carbapenemase activity was demonstrated by performing the assay in the presence of APBA (Fig. 4C).
Fig 4.
MALDI-TOF MS analysis of carbapenems after incubation with a carbapenem-sensitive K. pneumoniae strain (A, D, and G), with a carbapenem-resistant K. pneumoniae strain (B, E, and H), and with a carbapenem-resistant strain in the presence of the carbapenemase inhibitor APBA (C, F, and I). (A, B, and C) Conversion of ertapenem. (D, E, and F) Reaction of meropenem. (G, H, and I) Disappearance of the molecular peak of imipenem after incubation with a carbapenemase-resistant K. pneumoniae strain and the conservation of this peak in the presence of APBA. Incubation times were 20 min for ertapenem, 15 min for meropenem, and 10 min for imipenem. Peaks corresponding to the nonhydrolyzed form of the respective antibiotic are highlighted in gray. Peaks corresponding to the hydrolyzed forms of the respective antibiotic are indicated with an arrow.
The incubation of meropenem in the presence of a carbapenemase-sensitive strain resulted in the detection of the molecular peak [M + H]+ at 384.5 Da and the sodium adducts [M + Na]+ at 406.5 Da and [M + 2Na]+ at 428.5 Da (Table 1). Additional peaks at 401.4 Da and 423.4 Da could not be assigned to a known molecular structure of meropenem (Fig. 4D). After incubation of meropenem with carbapenemase-resistant K. pneumoniae strains, the peaks corresponding to the nonhydrolyzed forms of meropenem disappeared, but no hydrolysis products (expected [Mhydr. + H]+ at 402.5 Da, [Mhydr. + Na]+ at 424.5 Da, and [Mhydr. + 2Na]+ at 446.5 Da) were detectable under the conditions employed (Fig. 4E). For meropenem, the disappearance of the molecular peaks at [M + H]+ 384.5 Da, [M + Na]+ 406.5 Da, and [M + 2Na]+ 428.5 Da correlates with the presence of a carbapenem-resistant bacterial strain. Applying this criterion, the MALDI-TOF MS-based evaluation matched the results of the approved routine assay (Table 2). The inhibition set up with APBA conserved the sensitivity pattern, verifying the presence of a carbapenemase (Fig. 4F).
Incubation of imipenem in the presence of a K. pneumoniae carbapenemase-sensitive strain resulted in a spectrum containing only the molecular peak for imipenem ([M + H]+ 300.4 Da) (Table 1). No further salt adducts were detectable. After hydrolysis by carbapenemase-positive strains, this peak disappeared, and no additional peaks appeared under the conditions employed (expected [Mhydr. + H]+ at 318.4 Da). Also, the lower mass range did not contain any peaks corresponding to a fragment of the hydrolyzed form of imipenem (Fig. 4G and H). The presence of a carbapenemase was confirmed by the inhibition of the hydrolysis reaction by APBA (Fig. 4I). Similar to the case for meropenem, the identification of carbapenemase-positive strains was based only on the disappearance of the molecular peak of imipenem. Nevertheless, the MALDI-TOF MS-based assay and the Etest delivered the same classification of carbapenemase-negative and carbapenemase-positive strains (Table 2).
Identification of carbapenemase-positive K. pneumoniae strains from fresh positive blood cultures.
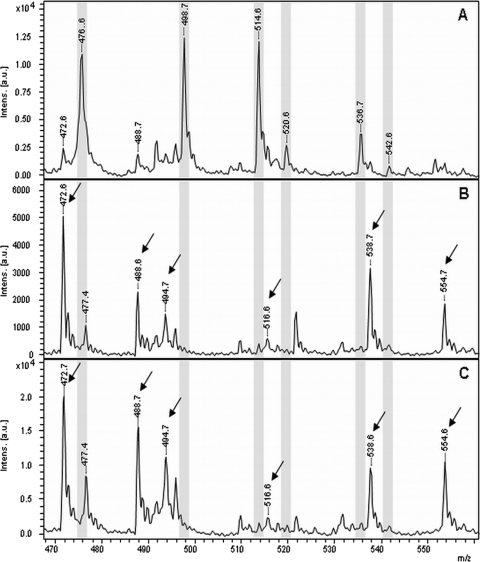
The rapid detection of β-lactam-resistant bacteria from positive blood cultures is an important issue because the decision for an adequate therapy is urgently needed for patients with sepsis. The hydrolysis of ertapenem by different K. pneumoniae strains derived from positive blood cultures was investigated. In the case of a resistant K. pneumoniae strain, the amount of bacteria isolated from 1 ml of fresh blood culture was sufficient for hydrolyzing ertapenem within 3 h of incubation. Figure 5A to C represent the spectra derived from the incubation of ertapenem with a carbapenem-sensitive K. pneumoniae strain and two carbapenem-resistant K. pneumoniae strains isolated from a fresh blood culture, respectively. The characteristic peak pattern, which was described above for the discrimination of carbapenemase-positive and carbapenemase-negative plated bacteria, was also found for bacteria from fresh positive blood cultures. An exception was the peak at 450.5 Da, which was detectable in all spectra. This peak corresponds to the mass of the hydrolyzed, decarboxylated form of ertapenem. In the case of the isolation of bacteria from fresh positive blood cultures, the spectrum of the carbapenem-sensitive strain also comprised a peak at this mass position. Therefore, it was not considered for the classification of spectra derived from blood cultures. The spectrum of the sensitive strain comprises the [M + H]+ molecular peak of ertapenem at 476.6. Da and the corresponding adduct peaks at 498.7, 514.6, 520.6, 536.7, and 542.6 Da for the single sodium adduct, the single potassium adduct, the double sodium adduct, the potassium sodium adduct, and the triple sodium adduct, respectively. These peaks were not detectable in the spectra derived from carbapenem-resistant bacteria. In contrast, the peaks representing the different hydrolysis products were found at 472.6, 488.6, 494.7, 516.6, 538.7, and 554.7 Da, corresponding to the hydrolyzed decarboxylated sodium adduct, the hydrolyzed decarboxylated potassium adduct, the hydrolyzed product, the hydrolyzed single sodium adduct, the hydrolyzed double sodium adduct, and the hydrolyzed potassium sodium adduct, respectively. The peak at m/z 450.5 Da in spectra from positive blood cultures probably resembles an unspecific background signal derived from the blood culture.
Fig 5.
MALDI-TOF MS spectra after incubation of ertapenem with different K. pneumoniae strains isolated from fresh blood cultures. (A) Carbapenem-sensitive strain. (B and C) Carbapenem-resistant strains. Peaks representing the nonhydrolyzed form of the respective antibiotic are highlighted in gray. Peaks corresponding to the hydrolyzed forms of the respective antibiotic are indicated with an arrow.
DISCUSSION
The MALDI-TOF MS-based analysis of bacteria according to their presumable resistance/susceptibility against β-lactam antibiotics is a novel approach, which has recently been applied for the analysis of the hydrolysis of ampicillin, ertapenem, and meropenem by different bacterial strains (3, 8, 11). In contrast to the approved routine assays, which determine the bacterial growth in relation to the exposure to the antibiotic, the MALDI-TOF MS-based approach directly analyzes the underlying enzymatic reaction of the β-lactamase on the molecular level. Therefore, this assay is much faster, and interpreted results are already available after about 4 h. The results of the hydrolysis of the carbapenems demonstrate that this time can be reduced. The optimal incubation time will have to be determined after standardization of the assay. Preliminary experience indicates that an incubation time of 1 h might be sufficient, delivering interpreted results already after 2 h. A prerequisite for the applicability of this assay is the presence of a β-lactamase, which is responsible for the resistance. Other mechanisms, such as changed porins or upregulation of the efflux pumps, are not detected.
The measurements of the small antibiotic molecules required an optimization of the instrument settings to obtain sufficient resolution and sensitivity. For calibration of the spectra, a special calibration standard was developed to cover the mass range of interest. Here, it has to be considered that during the MALDI process a proton is added to the molecules, resulting in an increase of the molecular mass by 1 Da. The large deviation of the observed mass of meropenem (382.975 Da) and its adducts from the expected mass (meropenem [M + H]+ = 384.5 Da), found by Hrabak et al. (8), might be explained by insufficient calibration. The data of Burckhardt and Zimmermann (3) also showed poor calibration, which might be due to the one-point calibration employing the matrix peak at 379 Da, which is located outside the mass range of interest (ertapenem mass [M + H]+ 476.5 Da). The exact calibration of the mass spectrometer applied in this study enabled a measurement with a mass accuracy of less than 0.5 Da.
Our data demonstrate that this approach is applicable for the analysis of different classes of β-lactam antibiotics and β-lactamase inhibitors. The hydrolysis of the central β-lactam rings of the β-lactam antibiotics leads to a molecular mass shift of +18 Da, which is a huge mass difference in the mass range of the antibiotics and can be easily detected by mass spectrometry. Further molecular reactions can lead to additional defined molecular mass peaks specific for the respective antibiotic. Depending on the antibiotic drug, different molecular forms and adduct formations are detectable in the spectra. The minimal change in the mass spectrum after hydrolysis of a β-lactam antibiotic was the disappearance of the molecular peak and its salt adducts. On the other hand, to make the analysis more reliable, it is preferable not only to monitor the disappearance of a mass peak but also to observe the corresponding products which are generated through hydrolysis or by subsequent reactions.
For the penicillin derivatives ampicillin and piperacillin, the single sodium adducts (+22 Da, through replacement of a proton [−1 Da] by sodium [+23 Da]) and the double sodium adducts (+44 Da) of both the molecular peak and the hydrolyzed form were detected. The hydrolyzed form of ampicillin was labile and tended to decarboxylate, resulting in a mass shift of −44 Da calculated from the mass of the hydrolysis product. The decarboxylated form did not form any sodium adducts. The corresponding peak of the hydrolyzed, decarboxylated form of piperacillin was very small. In the case of the cephalosporins, the adduct formation was weak (cefotaxime) or not detectable (ceftazidime). Further, the molecular structures of the cephalosporins were not stable under the MALDI-TOF MS conditions used, resulting in a partial elimination of an acetyl group and a pyridine group for cefotaxime and ceftazidime, respectively. The hydrolysis products of the cephalosporin immediately eliminated the mentioned groups. The resulting fragments did not form any salt adducts but tended to eliminate a carboxyl group, detectable by a mass shift of −44 Da. The carbapenem ertapenem showed spectra comparable to those of the penicillin derivatives. In contrast to the penicillin derivatives, ertapenem also formed potassium and sodium/potassium adducts. After hydrolysis, ertapenem formed single sodium and double sodium adducts. The hydrolyzed form of ertapenem showed strong decarboxylation and subsequent single sodium and potassium adduct formation. Imipenem did not form any adducts, resulting in the detection only of the molecular peak after incubation with carbapenem-sensitive bacteria. The incubation with carbapenem-resistant bacteria led to the complete disappearance of any imipenem-derived peak. The reason for this could be that the hydrolysis products were very labile, and/or the hydrolysis products and the resulting fragments might be difficult to be ionized, which is a prerequisite for MALDI-TOF MS. Further, the hydrolysis products and possible fragments might bind the cellular structures and therefore not be detectable in the cellular supernatant. Similar results were obtained for meropenem under the conditions used. In contrast to that of imipenem, the molecular structure of meropenem formed single and double sodium adducts. These findings are in concordance with recently published data (8).
The interpretation and evaluation of the spectra require expert knowledge, but after the assignment of peaks and the definition of a sensitivity pattern and a resistance pattern, classification of bacteria becomes quite easy. Additional efforts to suppress the strong adduct formation could further improve the assay, because these adducts reduce the sensitivity of the measurements and result in complex spectra.
For standardization and automation of assay evaluation, different approaches depending on the molecular products appearing after hydrolysis of the β-lactam ring might be useful. For the detection of ampicillin-resistant strains, the quotient of the sum of all peak areas belonging to the nonhydrolyzed forms and of the sum of all peak areas belonging to the hydrolyzed forms was >1 for all sensitive strains and <1 for all resistant strains (data not shown). For imipenem and meropenem this evaluation approach was not applicable because no hydrolysis products could be detected. Another approach for standardization might be the quantitation of the nonhydrolyzed and hydrolyzed forms by employing a spiked standard and the subsequent determination of a calibration equation (1).
Comparison of the classification results obtained by the MALDI-TOF MS-based approach and the approved routine method revealed identical results. The concentrations necessary for the MALDI-TOF MS detection are about 1,000-fold above the concentrations used for the routine assay. Nevertheless, this has no impact on the classification results. The reason for this might be that the amount of antibiotic per bacterial cell is important and not the absolute concentration. The routine assay usually employs about 104 cells per plate. For the MALDI-TOF MS-based assay, at least 107 cells per 10 μl were used (the exact cell numbers have not been determined). Further, it is not necessary for the MALDI-TOF MS-based assay that the cells stay alive, as long as the β-lactamase is active.
Verification and classification of β-lactam resistance are usually done by the inhibition of growth of resistant bacteria caused by the β-lactamase inhibitors. The format of the MALDI-TOF MS assay allowed for the simple testing of inhibition of the β-lactamase activity. Inhibition of ESBL activity was demonstrated for the antibiotic/inhibitor combinations ampicillin/clavulanic acid, piperacillin/tazobactam, cefotaxime/clavulanic acid, and ceftazidime/clavulanic acid. Using the respective antibiotic/inhibitor combinations, the described sensitivity patterns of the corresponding antibiotics were observed for resistant bacteria. Thereby, the presence of a β-lactamase of the ESBL type was confirmed. Inhibition of carbapenemases was performed with APBA. The use of the combinations ertapenem/APBA, imipenem/APBA, and meropenem/APBA inhibited the hydrolysis of ertapenem, imipenem, and meropenem by carbapenem-resistant bacteria, respectively.
Additionally, the MALDI-TOF MS-based assay was also applicable for the detection of resistant cells from fresh positive blood cultures. After an optimized isolation of the bacterial cells from the blood culture, the hydrolysis of ertapenem by carbapenem-resistant bacteria was shown. In contrast to the spectra derived from plated bacteria, the spectra derived from carbapenem-sensitive K. pneumoniae strains from blood cultures comprised a peak at 450 Da. A peak at this position also appeared in the resistance pattern. No other peaks belonging to the resistance pattern were detected. Therefore, it can be assumed that the peak at 450 Da in the spectrum from a blood culture-derived analysis was a contamination from the blood culture and should not be considered for classification. This has to be kept in mind when an algorithm for automated data analysis is created. Especially, for positive blood cultures it is very important to quickly identify the microorganisms and to get information about possible resistances. MALDI-TOF MS provides quick identification (12, 15–17), and the MALDI-TOF MS-based resistance assay facilitates the rapid detection of β-lactamase-mediated resistance. Using the respective antibiotics/inhibitor combinations, differentiation and confirmation of the underlying β-lactamase also are possible. This will support the early start of an appropriate antibiotic therapy.
The MALDI-TOF MS-based β-lactamase assay might also be useful in the pharmaceutical area. Since the incubation time is drastically reduced, results regarding β-lactamase activity are available after a few hours. Therefore, this assay may speed up the development of novel β-lactam antibiotics and β-lactamase inhibitors and may save costs.
Further validation of this MALDI-TOF MS-based resistance assay will be necessary before it is applied for diagnostic use. Particularly, different bacterial species with different types of β-lactamases should be investigated. Additionally, strains with only low expression levels of β-lactamase have to be investigated, and the classification results should be carefully compared with the results of the approved routine procedures.
Supplementary Material
ACKNOWLEDGMENTS
Katrin Sparbier and Markus Kostrzewa are employed at the mass spectrometry company Bruker Daltonik GmbH.
This study was supported by a grant from the Bayerische Forschungsstiftung (Forschungsverbund “FORPROTECT—Infektionsprotektion durch neue Diagnostikverfahren und Therapieansäze”) to S.S. and M.K.
This article is dedicated to the memory of Dr. Ulrich Weller, a bright scientist and reliable colleague.
Footnotes
Published ahead of print 28 December 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Baechle D, et al. 2007. Towards stable diagnostic setups in clinical proteomics: absolute quantitation of peptide biomarkers using MALDI-TOF-MS. Proteomics 1:1280–1284 [Google Scholar]
- 2. Bou G. 2007. Minimum inhibitory concentration (MIC) analysis and susceptibility testing of MRSA. Methods Mol. Biol. 391:29–49 [DOI] [PubMed] [Google Scholar]
- 3. Burckhardt I, Zimmermann S. 2011. Using matrix-assisted laser desorption ionization–time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 49:3321–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bush K. 2010. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 13:558–564 [DOI] [PubMed] [Google Scholar]
- 5. Carter MW, Oakton KJ, Warner M, Livermore DM. 2000. Detection of extended-spectrum beta-lactamases in klebsiellae with the Oxoid combination disk method. J. Clin. Microbiol. 38:4228–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a. Clinical and Laboratory Standards Institute 2011. M100-S21. Performance standards for antimicrobial susceptibility testing: 21st informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Delcour AH. 2009. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 1794:808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gottlieb T, Nimmo GR. 2011. Antibiotic resistance is an emerging threat to public health: an urgent call to action at the Antimicrobial Resistance Summit 2011. Med. J. Aust. 194:281–283 [DOI] [PubMed] [Google Scholar]
- 8. Hrabak J, Walkova R, Studentova V, Chudackova E, Bergerova T. 2011. Carbapenemase activity detection by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:3222–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koch AL. 2000. Penicillin binding proteins, beta-lactams, and lactamases: offensives, attacks, and defensive countermeasures. Crit. Rev. Microbiol. 26:205–220 [DOI] [PubMed] [Google Scholar]
- 10. Lambert PA. 2005. Bacterial resistance to antibiotics: modified target sites. Adv. Drug Deliv. Rev. 57:1471–1485 [DOI] [PubMed] [Google Scholar]
- 11. Ledeboer NA, Hodinka RL. 2011. Molecular detection of resistance determinants. J. Clin. Microbiol. 49:S20–S24 [Google Scholar]
- 12. Mellmann A, et al. 2009. High interlaboratory reproducibility of matrix-assisted laser desorption ionization–time of flight mass spectrometry-based species identification of nonfermenting bacteria. J. Clin. Microbiol. 47:3732–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagy E, Becker S, Soki J, Urban E, Kostrzewa M. 2011. Differentiation of division I (cfiA-negative) and division II (cfiA-positive) Bacteroides fragilis strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Med. Microbiol. 60:1584–1590 [DOI] [PubMed] [Google Scholar]
- 14. Poole K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3:255–264 [PubMed] [Google Scholar]
- 15. Sauer S, Kliem M. 2010. Mass spectrometry tools for the classification and identification of bacteria. Nat. Rev. Microbiol. 8:74–82 [DOI] [PubMed] [Google Scholar]
- 16. Schubert S, et al. Novel, improved sample preparation for rapid, direct identification from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. J. Mol. Diagn. 13:701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seng P, et al. 2010. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 5:1733–1754 [DOI] [PubMed] [Google Scholar]
- 18. Woodford N, et al. 2010. Comparison of BD Phoenix, Vitek 2, and MicroScan automated systems for detection and inference of mechanisms responsible for carbapenem resistance in Enterobacteriaceae. J. Clin. Microbiol. 48:2999–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wright GD. 2005. Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv. Drug Deliv. Rev. 57:1451–1470 [DOI] [PubMed] [Google Scholar]
- 20. Wybo I, et al. 2011. Differentiation of cfiA-negative and cfiA-positive Bacteroides fragilis isolates by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:1961–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.