Abstract
Oral streptococci have been associated with systemic diseases, including infective endocarditis and neutropenic bacteremia. We analyzed 58 recent oral streptococcal bloodstream isolates, and we obtained clinical and demographic data for source patients. The sodA gene was found to be a better target than the 16S-23S rRNA internal transcribed spacer for DNA sequence-based species identification. Together, Streptococcus mitis and Streptococcus oralis were significantly more likely than the 12 combined remaining species to be isolated from neutropenic patients.
TEXT
Streptococci are the most abundant inhabitants of the human mouth (6, 24), and they gain frequent access to the bloodstream through periodontal lesions or oral abrasions created by routine activities (32). This can lead to serious illnesses, including infective endocarditis (IE) (28) and neutropenic bacteremia (29). The taxonomy of the oral streptococci has long been a source of confusion (4, 9, 20, 22, 31). Sequencing of the 16S rRNA gene has clarified the situation immensely (13); however, this approach lacks the sensitivity required to distinguish certain closely related species, including Streptococcus mitis and Streptococcus oralis (12), or to allow for strain typing or phylogenetic analysis within species. Genes possessing greater variability have therefore been examined, including the 16S-23S rRNA intergenic transcribed spacer (ITS) (2), protein-coding housekeeping genes (1, 5, 7, 10, 12, 14, 15, 19, 23), or both (17, 18). Consensus concerning the gene(s) best suited for these purposes has yet to emerge. Further, although previous studies have identified oral streptococci from clinical blood cultures using definitive sequencing methods (12, 23, 30), none have reported detailed clinical information about underlying disease. We set out to do both.
Bloodstream cultures performed at Virginia Commonwealth University Medical Center Hospital from May 2003 to May 2008 were presumptively identified as containing streptococci by the clinical microbiology laboratory. A computer script was used to exclude nonoral species. To avoid analysis of contaminants, isolation plates were requested only when two or more reports originated from separate cultures of the same patient. One colony from each available isolation plate was grown in broth culture and examined macroscopically and microscopically. If any differences were observed in separate cultures derived from the same patient, both cultures were retained. Otherwise, a single culture from each patient was cryopreserved, and aliquots were removed for PCR amplification.
To determine the species identity and phylogenetic relatedness of each isolate, the 16S-23S ITS was amplified using the 6R and 13BF primers described previously (2). When products were not obtained from some isolates, we noted that the final three nucleotides of the 6R primer did not align with 23S rRNA sequences in GenBank from several species of interest. We therefore shortened and simplified the 6R primer, creating 6R-S, and shortened the 13BF primer to match its annealing temperature (see Table S1 in the supplemental material). A similar modification of the 6R primer was reported recently (18). Using these primers, PCR amplicons were obtained from all isolates and submitted for capillary DNA sequence analysis.
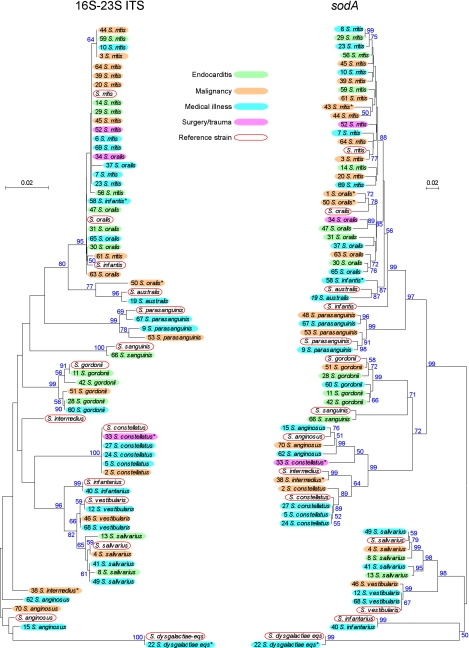
Most DNA sequences contained the complete ITS and portions of the 16S and 23S rRNA genes, which were aligned with ITS sequences from type strains available in GenBank or determined by us from strains obtained from the American Type Culture Collection (ATCC). We found that flanking 16S and 23S sequences, though not retained in most published sequences, facilitated ITS alignment. Indeed, at least four type strain sequences have been published (18) in which a CTAAGG located 78 bp prior to the 3′ end of the 16S rRNA gene appears to have been confused with an identical hexanucleotide sequence defined previously (2) as the beginning of the ITS. Trimmed ITS sequences (2) were then compared and aligned with MEGA 4 software (25) to construct a neighbor-joining phylogenetic tree (Fig. 1).
Fig 1.
Neighbor-joining trees for ITS and sodA sequences. Clinical isolates are indicated by VMC number and final species assignment. Fill colors correspond to patient underlying illness, and red outlines indicate reference strains. The scale indicates the number of base substitutions per site, with distances determined using the maximum composite likelihood method (21). Bootstrap values that were equal to or greater than 50% from 2,000 replicates are indicated in blue text adjacent to the branches. *, isolate for which additional sequences (pfl and/or pyk) were determined.
It has been suggested previously that ITS analysis alone is sufficient for species identification of oral streptococci, including that of the closely related species S. mitis and S. oralis. Conserved single-base deletions at two sites and a total ITS length of 246 bp were suggested to be characteristic of S. oralis, while S. mitis was proposed to lack the deletions and possess an ITS of 248 to 249 bp (2, 3). In our study, we found isolates of both species that possessed both deletions or neither, along with overlapping ITS lengths. Thus, the S. oralis and S. mitis isolates could not be reliably distinguished by these criteria. Another study has suggested that ITS sequence analysis, while not sufficient for distinguishing S. oralis and S. mitis, is sufficient for identification of many other oral streptococcal species, including those belonging to the anginosus group (18). However, we found one isolate of the anginosus group species S. intermedius (VMC38) that could not be classified by ITS (see Table S1 in the supplemental material).
A large number of isolates, particularly for S. mitis and Streptococcus constellatus, also possessed identical sequences, precluding phylogenetic analysis. Identical sequences were also obtained from five pairs of isolates originating from the same patients (data not shown).
Because the ITS analysis was not sufficient for our purposes, we sequenced a second common target—the sodA gene, encoding manganese-dependent superoxide dismutase (1, 5, 10, 12, 14, 19, 27). This allowed for the exclusion of several isolates. The strain pairs mentioned above as originating from the same subjects and having identical ITS sequences also possessed identical sodA sequences, confirming that the isolates were identical, and one isolate from each pair was dropped. Two isolates were dropped due to an apparent mistake in strain handling, and another was not retained because the ITS and sodA sequences indicated that it was a strain of Enterococcus faecalis. A phylogenetic tree derived from the remaining isolates is shown in Fig. 1.
The sodA alignment was superior to that of the ITS for phylogenetic analysis. The only isolates with identical sodA sequences were two S. mitis strains and two strains of Streptococcus vestibularis. The sodA alignment was also more useful for species determination. All reference strains were well separated from one another in the phylogenetic tree. The result was only four ambiguous species assignments for sodA, compared to 15 for the ITS (see Table S1 in the supplemental material). In those cases in which a single species designation was consistent with both the ITS and sodA sequences, that designation was used. Only 3 of the final 58 clinical isolates could not be confidently identified using this method. The VMC1 and VMC43 isolates produced ITS sequences that were too short to be informative, despite repeated sequencing attempts, and VMC58 was inconsistently identified by ITS and sodA sequences (Fig. 1; Table S1). We confirmed the species identity of these isolates utilizing a recently published database containing sequences of seven housekeeping genes from 420 well-characterized streptococcal strains, including sodA, pfl, and pyk (1). Sequences of the pfl and/or pyk genes from all questionable strains were determined and aligned with the sequences in the eMLSA.net database (http://www.emlsa.net/), as were the existing sodA sequences. In all cases, the pfl and pyk species assignments were in agreement with those from sodA. In addition, the sodA alignment indicated that the VMC58 sequence, while divergent from the type strain, fell within a cluster of 13 Streptococcus infantis isolates in the database (data not shown). The consensus species identification of all isolates included in the study are indicated in Fig. 1 and Tables S1 and S2 in the supplemental material.
While these results suggest that sodA analysis alone is sufficient for identifying most strains, we recommend inclusion of at least one additional protein-coding housekeeping gene for all strains. Many oral streptococci are naturally competent, and instances of acquisition of foreign housekeeping genes, including sodA, have been reported (1, 12). This was not apparent in our study, but there were instances of chimeric ITS (VMC38 and VMC50) and sodA (VMC33) sequences. Two recent publications have gone farther, each suggesting sequencing of a different set of seven housekeeping genes for multilocus sequence typing (7) or multilocus sequence analysis (1). These approaches rely on analysis of a larger number of genes to provide increased resolution and to minimize the effects of occasional chimeric or foreign genes (1, 7). These schemes also allow for more sophisticated phylogenetic analyses than are possible with only two or three genes. We occasionally experienced difficulty, however, in amplification and sequencing of pfl and pyk and little success with two of the other recommended genes, map and ppaC (1). Another recent study reported similar difficulties (27). Thus, the choice of additional genes may require empirical testing, but those employed in the seven-gene analyses should be explored first, since large collections of sequences from well-characterized strains are available for comparison (1, 7).
To our knowledge, this is only the second report of a Streptococcus australis (12) or S. infantis (30) blood culture isolate. The only other association of the latter species with any illness comes from two reports of its isolation from the sputum of adults with cystic fibrosis (17, 26). It is therefore interesting that an adult with cystic fibrosis was the source of our S. infantis isolate, VMC58 (see Table S2 in the supplemental material).
Antibiotic susceptibility data are included in Table S2 and are largely in agreement with previous studies (8). Table S2 also provides clinical and demographic data for source patients, categorizing them based on chief underlying illness: medical illness, malignancy, IE, or surgery/trauma. All IE subjects were diagnosed clinically, and all but the VMC56 case (Table S2) were classified as “definite IE” by the modified Duke criteria (16). The lone exception had a history of previous IE and intravenous drug use (IVDU), which in concert with the positive blood cultures would result in categorization as “possible IE.” Data on possible sources of infection, including central lines, gastrointestinal endoscopy, oral mucositis, dental conditions, and IVDU, were also examined. IVDU was found for 9 of the 13 IE patients and for only 1 other patient in the study (Table S2). This difference was statistically significant (P < 0.0001; Fisher's exact test). Thus, although this study was focused on oral species, IVDU was an overwhelming risk factor.
Our phylogenetic analysis uncovered no statistically significant associations between any particular species or clonal type and underlying illness or other clinical parameters, including white blood cell count, highest temperature, vegetation size, valve affected, or patient death. One previous study contained enough information to identify species isolated from neutropenic patients (12). Our results were similar, in that 11 of 12 isolates in that study and 9 of 10 in ours (Table S2) were either S. mitis or the closely related S. oralis. In our study, these two species, taken together, were significantly more likely than the 12 remaining species combined to be isolated from neutropenic patients (P = 0.004; Fisher's exact test). This may reflect the prevalence of these species in the oral cavity. It is interesting, though, that this trend did not carry over to IE. Combining our data with those from the same study (12) for neutropenia and IE, S. oralis and S. mitis were isolated from 20 of 22 neutropenia cases and only 15 of 27 IE cases, a difference that was statistically significant (P = 0.01; Fisher's exact test). As in previous studies (11, 12, 30), sample numbers limited our ability to confidently assess other possible associations between species and particular illnesses or clinical characteristics. However, by pairing definitive species identification with clinical and demographic characteristics for each source patient (Table S2), we have sought to make our results suitable for meta-analysis or other studies utilizing combined data.
DNA sequence accession numbers.
DNA sequences of type strains and clinical isolates determined in this study were deposited in GenBank, under accession numbers JN181256 to JN181394. A whole-genome shotgun sequence of S. sanguinis isolate VMC66 determined at the Baylor College of Medicine for the Human Microbiome Project (S. K. Highlander et al., unpublished data) is available under GenBank accession number NZ_AEVH01000000.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karen Smalara for creating the script to examine electronic records, Yvette Major for strain collection, and Sandra Tallent, Connie Bender, and Gordon Archer for useful discussions.
This work was supported by grants K02AI05490 and R01AI47841 from the National Institutes of Health (T.K.).
Footnotes
Published ahead of print 21 December 11
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Bishop C, et al. 2009. Assigning strains to bacterial species via the internet. BMC Biol. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen CC, Teng LJ, Chang TC. 2004. Identification of clinically relevant viridans group streptococci by sequence analysis of the 16S–23S ribosomal DNA spacer region. J. Clin. Microbiol. 42:2651–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen CC, Teng LJ, Kaiung S, Chang TC. 2005. Identification of clinically relevant viridans streptococci by an oligonucleotide array. J. Clin. Microbiol. 43:1515–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coykendall AL. 1989. Classification and identification of the viridans streptococci. Clin. Microbiol. Rev. 2:315–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Delorme C, Poyart C, Ehrlich SD, Renault P. 2007. Extent of horizontal gene transfer in evolution of streptococci of the salivarius group. J. Bacteriol. 189:1330–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dewhirst FE, et al. 2010. The human oral microbiome. J. Bacteriol. 192:5002–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Do T, et al. 2009. Population structure of Streptococcus oralis. Microbiology 155:2593–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doern GV, Ferraro MJ, Brueggemann AB, Ruoff KL. 1996. Emergence of high rates of antimicrobial resistance among viridans group streptococci in the United States. Antimicrob. Agents Chemother. 40:891–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Facklam R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glazunova OO, Raoult D, Roux V. 2009. Partial sequence comparison of the rpoB, sodA, groEL and gyrB genes within the genus Streptococcus. Int. J. Syst. Evol. Microbiol. 59:2317–2322 [DOI] [PubMed] [Google Scholar]
- 11. Han XY, Kamana M, Rolston KVI. 2006. Viridans streptococci isolated by culture from blood of cancer patients: clinical and microbiologic analysis of 50 cases. J. Clin. Microbiol. 44:160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoshino T, Fujiwara T, Kilian M. 2005. Use of phylogenetic and phenotypic analyses to identify nonhemolytic streptococci isolated from bacteremic patients. J. Clin. Microbiol. 43:6073–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406–408 [DOI] [PubMed] [Google Scholar]
- 14. Kawamura Y, Whiley RA, Shu SE, Ezaki T, Hardie JM. 1999. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology 145(Pt 9):2605–2613 [DOI] [PubMed] [Google Scholar]
- 15. Kiratisin P, Li L, Murray PR, Fischer SH. 2005. Use of housekeeping gene sequencing for species identification of viridans streptococci. Diagn. Microbiol. Infect. Dis. 51:297–301 [DOI] [PubMed] [Google Scholar]
- 16. Li JS, et al. 2000. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin. Infect. Dis. 30:633–638 [DOI] [PubMed] [Google Scholar]
- 17. Maeda Y, et al. 2011. Population structure and characterization of viridans group streptococci (VGS) including Streptococcus pneumoniae isolated from adult patients with cystic fibrosis (CF). J. Cyst. Fibros. 10:133–139 [DOI] [PubMed] [Google Scholar]
- 18. Nielsen XC, Justesen US, Dargis R, Kemp M, Christensen JJ. 2009. Identification of clinically relevant nonhemolytic streptococci on the basis of sequence analysis of 16S–23S intergenic spacer region and partial gdh gene. J. Clin. Microbiol. 47:932–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruoff KL. 1988. Streptococcus anginosus (“Streptococcus milleri”): the unrecognized pathogen. Clin. Microbiol. Rev. 1:102–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 22. Sherman JM. 1937. The streptococci. Microbiol. Mol. Biol. Rev. 1:3–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simmon KE, et al. 2008. Phylogenetic analysis of viridans group streptococci causing endocarditis. J. Clin. Microbiol. 46:3087–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Socransky SS, Manganiello SD. 1971. The oral microbiota of man from birth to senility. J. Periodontol. 42:485–496 [DOI] [PubMed] [Google Scholar]
- 25. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 26. Tazumi A, et al. 2009. Molecular characterization of macrolide resistance determinants [erm(B) and mef(A)] in Streptococcus pneumoniae and viridans group streptococci (VGS) isolated from adult patients with cystic fibrosis (CF). J. Antimicrob. Chemother. 64:501–506 [DOI] [PubMed] [Google Scholar]
- 27. Teles C, Smith A, Ramage G, Lang S. 2011. Identification of clinically relevant viridans group streptococci by phenotypic and genotypic analysis. Eur. J. Clin. Microbiol. Infect. Dis. 30:243–250 [DOI] [PubMed] [Google Scholar]
- 28. Tleyjeh IM, et al. 2007. A systematic review of population-based studies of infective endocarditis. Chest 132:1025–1035 [DOI] [PubMed] [Google Scholar]
- 29. Tunkel AR, Sepkowitz KA. 2002. Infections caused by viridans streptococci in patients with neutropenia. Clin. Infect. Dis. 34:1524–1529 [DOI] [PubMed] [Google Scholar]
- 30. Westling K, et al. 2008. Identification of species of viridans group streptococci in clinical blood culture isolates by sequence analysis of the RNase P RNA gene, rnpB. J. Infect. 56:204–210 [DOI] [PubMed] [Google Scholar]
- 31. Whiley RA, Beighton D. 1998. Current classification of the oral streptococci. Oral Microbiol. Immunol. 13:195–216 [DOI] [PubMed] [Google Scholar]
- 32. Wilson W, et al. 2007. Prevention of infective endocarditis. Guidelines from the American Heart Association. Circulation 116:1736–1754 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.