Abstract
The non-long-terminal-repeat (non-LTR) retrotransposons (also called long interspersed repetitive elements [LINEs]) are among the oldest retroelements. Here we describe the properties of such an element from a primitive protozoan parasite, Entamoeba histolytica, that infects the human gut. This 4.8-kb element, called EhLINE1, is present in about 140 copies dispersed throughout the genome. The element belongs to the R4 clade of non-LTR elements. It has a centrally located reverse transcriptase domain and a restriction enzyme-like endonuclease (EN) domain at the carboxy terminus. We have cloned and expressed a 794-bp fragment containing the EN domain in Escherichia coli. The purified protein could nick supercoiled pBluescript DNA to yield open circular and linear DNAs. The conserved PDX12-14D motif was required for activity. Genomic sequences flanking the sites of insertion of EhLINE1 and the putative partner short interspersed repetitive element (SINE), EhSINE1, were analyzed. Both elements resulted in short target site duplications (TSD) upon insertion. A common feature was the presence of a short T-rich stretch just upstream of the TSD in most insertion sites. By sequence analysis an empty target site in the E. histolytica genome, known to be occupied by EhSINE1, was identified. When a 176-bp fragment containing the empty site was used as a substrate for EN, it was prominently nicked on the bottom strand at the precise point of insertion of EhSINE1, showing that this SINE could use the LINE-encoded endonuclease for its insertion. The nick on the bottom strand was toward the right of the TSD, which is uncommon. The lack of strict target site-specificity of the restriction enzyme-like EN encoded by EhLINE1 is also exceptional. A model for retrotransposition of EhLINE1/SINE1 is presented.
The advent of large-scale genome sequencing will make it possible to understand the origin and evolution of transposable elements and their dynamic relationship with the resident genome. In this respect the study of ancient lineages of eukaryotes is particularly beneficial. Several primitive eukaryotes, for example, the amitochondrial human pathogens Giardia lamblia (1, 7) and Entamoeba histolytica (26), have recently been shown to harbor non-long-terminal-repeat (non-LTR) retrotransposons. These retrotransposons (also called long interspersed repetitive elements [LINEs]) are considered more primitive than the LTR retrotransposons (13, 35). In keeping with this contention, it is interesting that non-LTR elements are found abundantly in parasitic protozoa while there is only one report so far of an LTR element in these organisms (3).
Data from whole-genome sequencing efforts have revealed the presence of multiple LINE families in E. histolytica (30). Of these, the most abundant seems to be the 4.8-kb E. histolytica retrotransposon-like element (EhRLE), a LINE family from E. histolytica that had been described previously (26). In addition, short repetitive sequences of about 0.5 kb that are abundantly transcribed have also been identified (10, 32, 33). These show striking sequence conservation at the 3′ ends with partner LINEs (3, 30, 33) and might, therefore, be considered short interspersed repetitive elements (SINEs) (24). Sequence analysis of the LINE EhRLE shows the presence of well-conserved functional domains in the open reading frame. Sequence comparison and phylogeny based on the reverse transcriptase (RT) domain (3, 30) places EhRLE closest to the R4 clade (belonging to the R2 group of non-LTR elements) (12, 21), which includes the R4 element, which inserts into the 28S rRNA gene of nematodes (6); the Dong element, which inserts into TAA repeats that may be found in the nontranscribed spacer of insect ribososmal DNA and sometimes outside it (36); and Rex6 elements of vertebrates, some of which insert into TAA repeats, while for others the target preferences are not clear (31). In common with the R2 group, EhRLE contains an endonuclease (EN) domain downstream of a centrally located RT domain. The EN domain has the highly conserved CCHC, PDX12-14D, RHD, and KXXXY motifs found in the R2 group and partly shared with type IIS restriction enzymes (37). However, while most known members of the R2 group insert in a sequence-specific manner, this does not appear to be the case with EhRLE. The element is found on all chromosomes of E. histolytica, is not telomerically located, and is found close to protein-coding genes (26). The putative partner SINE of EhRLE (IE/Ehapt2) is also widely dispersed throughout the genome (10, 30, 33).
The mobilization of SINEs by LINEs would have a great impact on genome evolution, especially because SINEs are generally very abundantly transcribed. Several pairs of LINEs and SINEs in which the 3′ tails of each pair share a common sequence have been reported from a diversity of organisms (5, 19, 23). The involvement of this tail in SINE mobilization has recently been demonstrated for the eel LINE/SINE pair (19). Although sequence conservation of the 3′ tail of the E. histolytica LINE/SINE pair is suggestive of SINE mobilization by LINE-encoded enzymes, there is no experimental evidence yet in favor of this hypothesis. Nothing is known about the mechanism of retrotransposition in this primitive parasite. The general model that has served to explain LINE retrotransposition is target-primed reverse transcription, proposed by Eickbush and coworkers from their work on the silk moth element R2Bm (20). According to this model, the LINE-encoded endonuclease nicks the bottom strand of the target site, generating a 3′-OH group that primes reverse transcription of the element RNA.
In order to understand the mode of transmission of EhRLE in the E. histolytica genome, we have cloned and expressed the EN domain in Escherichia coli and studied its properties with respect to target site specificity of nicking in vitro. Here we show that the enzyme is not site specific and that it nicks a variety of sequences that fall into a loose consensus. This is the first clear example of an element belonging to the R2 group that does not insert in a site-specific manner. Some Rex6 elements of vertebrates also probably lack strict target site specificity, but this has yet to be clearly defined (31). We also show that the enzyme can nick an empty target site, known to be occupied by IE/Ehapt2 in vivo, at the precise point of insertion. This provides strong functional evidence that IE/Ehapt2 is a SINE element that utilizes the enzymatic machinery of EhRLE for its insertion. For the sake of uniformity in nomenclature, we henceforth refer to EhRLE as EhLINE1 and to IE/Ehapt2 as EhSINE1.
MATERIALS AND METHODS
Cloning of EhLINE1 EN.
The clones from the shotgun library used for E. histolytica genome sequencing (accession numbers AZ669903 and AZ541056) were selected for cloning the EN domain. These were a kind gift from B. Loftus, The Institute for Genomic Research (TIGR). A 276-bp EcoRI-NdeI fragment from AZ669903 and a 600-bp NdeI-BamHI fragment from AZ541056 were ligated and cloned into pBS [pBluescript II KS(+); Stratagene]. For expression of EhLINE1 EN in E. coli, the EcoRI-NotI fragment from pBS was cloned into the EcoRI-NotI site of pET30b (Novagen) and transformed into E. coli BL21(DE3) to yield pET-EN. The PDX14D-to-PAX14D mutation was generated by a QuikChange site-directed mutagenesis kit (Stratagene) using primers 5′-CAATATCACAAAATACCAGCCAAATACGTATTAAATAAAAAG-3′ and 5′-TTTATTTAATACGTATTTGGCTGGTATTTTGTGATATTGTCC-3′. The sequences of the wild-type (pET-EN) and mutant (pET-ENM) constructs were confirmed by sequencing.
Expression and purification of EhLINE1 EN.
E. coli cells (BL21; 200 ml) containing pET-EN were grown at 30°C in Luria-Bertani medium containing 30 μg of kanamycin/ml to an A600 of 0.7. For induction of His6-tagged EN and mutated EN (ENM) proteins, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM and the cells were further incubated for 3 h. Recombinant proteins were purified by Ni2+-nitrilotriacetic acid-agarose affinity chromatography as described by the supplier (Qiagen). The cells were harvested by centrifugation and then resuspended in 10 ml of buffer A (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 20 mM imidazole [pH 8.0], 10 mM β-mercaptoethanol, 0.5% Triton X-100, and 2 mM phenylmethylsulfonyl fluoride). These were then lysed by three cycles of freeze-thawing, followed by sonication on ice. The lysate was centrifuged at 12,000 × g for 30 min at 4°C. The supernatant was passed through a 0.45-μm-pore-size filter (Millipore) and incubated with 1 ml of preequilibrated Ni2+-nitrilotriacetic acid-agarose (Qiagen) for 2 h at 4°C with gentle mixing. It was then packed in a C10/10 column (Amersham Pharmacia Biosciences) and washed several times with buffer A containing 50 mM imidazole. The recombinant protein was eluted with an imidazole gradient (50 to 500 mM). The EhLINE1 EN protein was found to elute at an imidazole concentration of 150 to 200 mM. Fractions containing EhLINE1 EN were identified by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and then pooled and concentrated by dialyzing against buffer B (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 20 mM MgCl2, 2 mM dithiothreitol [DTT], 20% sucrose). The sucrose was removed by dialyzing against buffer C (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 20 mM MgCl2, 2 mM DTT, 10% glycerol). The purified protein was stored in aliquots of 2 to 5 μg in 30% glycerol at −20°C.
Immunoblotting.
Proteins were separated on an SDS-10% polyacrylamide gel and transferred to Hybond ECL nitrocellulose paper (Amersham Pharmacia Biosciences). After transfer, the membrane was blocked with 5% bovine serum albumin in TBS-T (20 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20 [pH 7.6]) overnight and subsequently incubated with the anti-His tag antibody (Amersham Pharmacia Biosciences) for 1 h at room temperature. The membrane was washed with TBS-T three times and incubated for 1 h at room temperature with a horseradish peroxidase-conjugated secondary antibody. Subsequently, the membrane was washed three times with TBS-T. The proteins were visualized with an enhanced chemiluminescence kit (Amersham Pharmacia Biosciences).
Nicking assay.
Supercoiled plasmid DNA was purified by using a Qiagen-tip100 plasmid purification kit. The EhLINE1 EN reaction mixture contained 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 20 mM MgCl2, 1 mM DTT, 0.2 μg of supercoiled pBS DNA, and 80 ng of purified protein in a total reaction volume of 30 μl. The reaction was carried out at 37°C for 1 h and was stopped by addition of 25 mM EDTA. Products were separated on a 0.8% agarose gel containing 0.5 μg of ethidium bromide/ml.
Growth of E. histolytica and genomic DNA purification.
E. histolytica strain HM-1:IMSS was maintained in TYI-S-33 medium (11) at 36°C with appropriate antibiotics. Total DNA was purified from cells grown to late-log phase, as described previously (2).
Mapping of nicked sites.
The primer (50 pmol) was end labeled in a 20-μl reaction mixture by using 50 μCi of [γ-32P]ATP (Amersham Pharmacia Biosciences) and T4 polynucleotide kinase (New England Biolabs). The reaction was stopped by incubation at 65°C for 20 min, and the labeled primer was purified by being passed through a Sephadex G-25 column (Amersham Pharmacia Biosciences) (25). The DNA substrates were generated by PCR with E. histolytica genomic DNA as the template and a combination of one end-labeled primer and one unlabeled primer. The sequences of the primers used are indicated in the figure legends. After PCR the products were separated on a 6% native polyacrylamide gel, and the DNA band corresponding to the full-length product was excised from the gel. The DNA was recovered by the “crush-and-soak” method (25).
The DNA substrate (100 ng) was incubated with 40 ng of protein in a 10-μl reaction mixture for 1 h at 37°C. The protein was inactivated as mentioned above. For denaturing electrophoresis on 6% polyacrylamide gels, a 2-μl aliquot of the reaction product was mixed with 8 μl of formamide gel loading dye (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol FF), boiled for 5 min, and chilled on ice before loading. The parallel sequencing reaction was carried out by using a Thermo Sequenase cycle sequencing kit (Amersham Pharmacia Biosciences). Template DNA (100 to 150 ng) and 1 to 2 pmol of primer were used for each sequencing reaction. Electrophoresis was carried out at 65 W for 1 to 3 h with the gel temperature maintained at 45 to 50°C. The gels were fixed, dried, and exposed to X-ray film.
Phylogenetic analysis.
Sequences used for phylogenetic analysis were taken from the work of Burke et al. (7). The sequences of EhLINE1, EhLINE2, and EhLINE3 were assembled from the following entries in the Genome Sequence Survey (GSS) database: EhLINE1, accession no. AZ535823, AZ684839, AZ542852, AZ669999, AZ687050, AZ533295, AZ546488, AZ678960, AZ669903, and AZ541065; EhLINE2, accession no. AZ547248, BH152719, AZ687791, AZ534852, AZ546599, BH135183, AZ676959, AZ688833, BH164129, AZ534795, AZ542853, AZ545537, and BH146735; EhLINE3, accession no. AZ532904, BH135862, AZ687777 (removed stop codon), AZ544710, AZ692964, AZ542119, AZ679161, AZ550903, AZ530263, AZ550894 (corrected frameshift), and AZ549664. The RT and EN domains were aligned by using the Clustal X alignment program (29). The rooted phylogenetic tree was derived by the neighbor-joining method using the PAM250 matrix of PHYLIP (14) and maximum-parsimony heuristic options in PAUP (27). Bootstrapping was carried out by using PAUP, with 100 data sets.
RESULTS
Cloning and purification of the EN polypeptide.
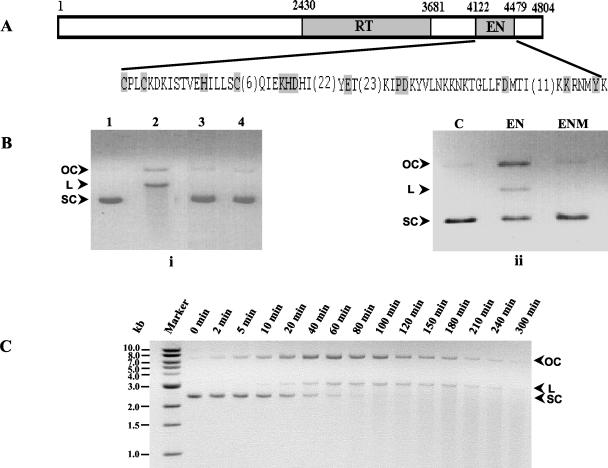
From nucleotide sequence analysis of EhLINE1, the conserved motifs of the EN domain were localized to nucleotide positions 4122 to 4479 (Fig. 1A). Two GSS entries encompassing this region and lacking any stop codons were used to clone the EN domain. The clones corresponding to these entries (accession no. AZ669903 and AZ541065) were a kind gift from TIGR. The 794-bp EcoRI-BamHI fragment (nucleotide positions 4010 to 4804) containing the EN domain was cloned into the E. coli expression vector pET30b. The expressed protein contained a His tag, and together with other vector sequences at the amino terminus, it was 309 amino acids long, with an expected molecular mass of 35.5 kDa. It was purified by nickel-agarose chromatography, and its identity was confirmed by using an anti-His tag antibody. As a control, the PDX14D motif in the EN domain, thought to be essential for enzyme activity, was mutated (37). The ENM motif was changed to PAX14D and expressed in E. coli. The recombinant EN protein purified from E. coli was stable for at least 6 months in 30% glycerol at −20°C.
FIG. 1.
Properties of the recombinant EN protein. (A) The nucleotide positions of the RT and EN domains in EhLINE1 are given above the diagram. The amino acid sequence of the EN domain is given below; conserved motifs are shaded. Numbers in parentheses indicate the number of amino acids between adjacent motifs. (B) The EhLINE1 EN protein was expressed in the E. coli expression vector pET30b (pET-EN) and purified as described in Materials and Methods. The endonucleolytic activity of the EN protein was measured with pBS DNA as the substrate. (i) Supercoiled pBS DNA (200 ng) was incubated with 80 ng of protein at 37°C for 1 h. Lane 1, untreated pBS DNA; lanes 2 to 4, pBS DNA incubated with protein purified from E. coli containing either pET-EN induced with IPTG (lane 2), uninduced pET-EN (lane 3), or induced pET30b (lane 4). (ii) pBS DNA (200 ng) was either left untreated (lane C) or incubated with 80 ng of either wild-type EN or ENM at 37°C for 30 min. DNA was electrophoresed through 0.8% agarose at 5 V/cm for 4 h and visualized by ethidium bromide staining. OC, open circular, L, linear, SC, supercoiled. (C) The time course of the endonucleolytic cleavage of pBS with EN protein was determined by incubating pBS DNA (100 ng) for the indicated times with 40 ng of EN protein. The reaction was terminated by addition of EDTA. DNA was electrophoresed through 0.8% agarose at 5 V/cm for 4 h and visualized by ethidium bromide staining.
Lack of a strict target-site consensus sequence for insertion of EhLINE1 and EhSINE1.
To design a suitable substrate for testing the endonucleolytic activity of the EN protein, we looked at the flanking genomic sequences at the sites of insertion of EhLINE1 and its putative partner SINE, EhSINE1. Sequences were obtained from the GSS and contig databases and were aligned using ClustalW. The 5′ and 3′ ends of the elements were defined as follows. Both elements started with 5′-AGATC, after which the sequence of the EhLINE1 family diverged from that of the EhSINE1 family, but within each family the sequence was highly conserved. The 3′ ends of the two families shared a 74-bp sequence, which ended with CTTTTTATTT-3′, with minor variations. The sequences upstream and downstream of EhLINE1 and EhSINE1 showed that these elements do not insert in a strictly site-specific manner. However, a T-rich stretch of 15 to 20 nucleotides was almost universally present about 13 to 19 nucleotides upstream of the insertion site of both elements.
The EN protein can cleave a nonspecific substrate, pBS.
The lack of a strict target site consensus for insertion of EhLINE1 prompted us to check if the EN protein could utilize a nonspecific substrate, pBS, as found for the human L1 endonuclease (15). Supercoiled pBS DNA was efficiently nicked by the purified EN protein to yield open circular and linear DNAs (Fig. 1B). An equal amount of ENM had very low activity, while no activity was found in extracts from uninduced and pET30b-induced cells. In a time course of endonucleolytic activity, it was evident that supercoiled pBS DNA was first nicked to yield open circular DNA, which was then converted to linear DNA (Fig. 1C). Since the amount of linear DNA produced at early time points was very small compared with that of open circular DNA, the enzyme appears to make predominantly single-strand nicks and not double-strand breaks. On further incubation, the linear DNA gave a smear of low-molecular-weight molecules. This (and experiments reported below) shows that the enzyme can use both circular and linear DNAs as substrates. The slow appearance of smears, and the presence of detectable levels of high-molecular-weight linear DNA even after 240 min, indicates that smears are probably due to closely spaced nicks on the two DNA strands, although a low level of exonucleolytic activity cannot be ruled out. The absence of exonuclease activity in our enzyme preparation is also borne out by the experiments described below in which end-labeled linear DNAs were used as substrates. The nicks produced by the endonuclease had 5′-PO4 and 3′-OH ends, since open circular pBS DNA generated by the enzyme could be converted to the covalently closed circular form upon incubation with T4 DNA ligase (data not shown).
To look for any possible hot spots where the enzyme nicks preferentially, pBS DNA was treated with EN protein for 10 min, following which the enzyme was denatured and DNA was linearized with EcoRI. The fragments generated were detected by Southern hybridization. The appearance of discrete bands along with a background smear showed that the enzyme did have a preference for some sites (data not shown).
Nicking activity of EN at the target sites occupied by EhLINE1 and EhSINE1.
To assay the activity of EN on its natural substrate, we looked for sites of insertion of EhLINE1/SINE1 in the E. histolytica genome, which may be unoccupied. Since E. histolytica is thought to be polyploid (n = 4) (9), it may be possible to find homologous regions of chromosomes, some of which harbor these elements while others are unoccupied. To look for unoccupied sites, it is necessary to have contigs containing the full-length element, from which flanking sequences can be obtained. Since the assembly of the E. histolytica genome sequences is still in progress, we undertook this exercise only for the 0.55-kb EhSINE1 at this point. The GSS database was adequate for this purpose, since it contains, on average, 0.8 kb of sequence available from either end of a clone. Thirty-eight entries containing the full-length EhSINE1 could be retrieved from the database. Sequences flanking the ends of the element in each entry were stitched together and used to search the database for unoccupied sites. Only one unoccupied site was found (AZ669709). The corresponding EhSINE1-containing sequence was AZ550231. The infrequent occurrence of unoccupied sites may be a limitation of the database, or it may be a consequence of the transposition mechanism, which does not leave unoccupied sites even in a polyploid genome. Both EhSINE1 and EhLINE1 insertions are accompanied by short target site duplications (TSDs), as seen from sequence analysis. In the case of AZ550231, EhSINE1 insertion resulted in a 22-bp TSD, which is present only once in AZ669709.
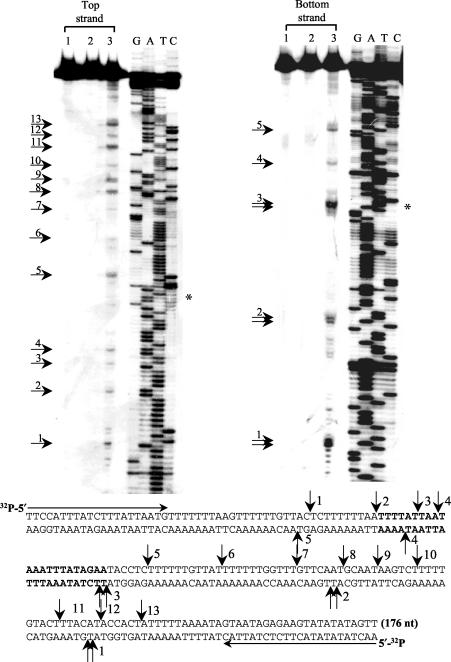
A 176-bp fragment containing the unoccupied site was obtained by PCR amplification of E. histolytica genomic DNA using primers designed from the sequence in AZ669709 (Fig. 2). The fragment was sequenced to confirm its identity with the sequence of the GSS entry. The fragment was then labeled either in the bottom strand or in the top strand by PCR amplification using the corresponding end-labeled primer. It was incubated with the endonuclease, and the resulting products were denatured and separated on a sequencing gel along with a sequencing reaction of the same template with each primer. The sites of nicking by the endonuclease on each strand were identified from the parallel sequencing run. The substrate in which the bottom strand was labeled showed three major nicking sites in which the products were doublets (Fig. 2, double arrows 1 to 3) and two less-preferred sites in which the products were primarily single bands (Fig. 2, arrows 4 and 5). Of these, bands 1 and 3 were the brightest. Band 3 corresponded with the exact site of insertion of EhSINE1. The pattern of nicks on the top strand was different from that on the bottom strand. The enzyme acted on many sites, but none was as strongly preferred as the sites in bands 1 and 3 on the bottom strand. In addition, most of the bands generated from the top strand appeared to be singlets (Fig. 2). The sequences surrounding the nicks on the two strands are listed in Table 1. From this it appears that the enzyme nicks at some preferred sequences and not randomly, since clear patterns could be discerned. The differences between the nicking patterns of the top and bottom strands could, in part, be due to the fact that the top strand was T rich while the bottom strand was A rich. The property of this endonuclease to nick at one of two consecutive phosphodiester bonds has also been observed with human L1 endonuclease (15) and R2Bm endonuclease (34). We were not able to repeat this experiment with more unoccupied sites of EhSINE1, since only one such site could be identified in the database used for this study.
FIG. 2.
Nicking profile of EN protein on a 176-bp substrate containing an unoccupied insertion site of EhSINE1. This site was found in a GSS database entry (accession no. AZ669709). The fragment containing the site was obtained by PCR amplification of E. histolytica genomic DNA. The sequence of the fragment used is shown at the bottom, and the primers used for PCR are marked at the two ends by horizontal arrows. Either the top strand or the bottom strand of the substrate was radiolabeled by using the appropriate primer end labeled with [γ-32P]ATP. The radiolabeled 176-bp fragment was gel purified, and 100 ng was incubated with 40 ng of EN protein at 37°C for 1 h (lane 3). Controls used were protein purified from pET-EN uninduced cells (lane 2) and a reaction mix with no added protein (lane 1). After enzyme digestion, the products were denatured by boiling in formamide and were separated through a 6% polyacrylamide gel at 60 W for 2 h. A sequencing reaction using the same primers was run in parallel. Asterisk marks the point of insertion of EhSINE1. Numbered arrows mark the positions of prominent nicks in the two strands. The 22-bp sequence that is duplicated upon EhSINE1 insertion is boldfaced.
TABLE 1.
Sequences surrounding the nicking site of the EN proteina
| Band | Sequence surrounding the following site:
|
|||||
|---|---|---|---|---|---|---|
| Unoccupied site
|
EhSINE1 insertion
|
EhLINE1 insertion
|
||||
| Bottom strand | Top strand | 3′ end | 5′ end | 3′ end | 5′ end | |
| 1 | 5′-GTATG | 5′-TTACT | 5′-GTATG | 5′-GGAGA | 5′-GATAAT | 5′-GTATT |
| ⇈ | ↑ | ⇈ | ↑ | ↑ | ⇈ | |
| 2 | 5′-GCATT | 5′-TTAAT | 5′-GCATT | 5′-GAGTC | 5′-GTATA | 5′-GCATT |
| ⇈ | ↑ | ⇈ | ↑ | ↑ | ⇈ | |
| 3 | 5′-GTATT | 5′-TTATT | 5′-GTATT | 5′-GTGTT | 5′-GCAAT | |
| ⇈ | ↑ | ⇈ | ↑ | ↑ | ||
| 4 | 5′-TAATA | 5′-TTAAT | 5′-GTATT | 5′-GCCACC | 5′-GTATT | |
| ↑ | ↑ | ⇈ | ⇈ | ↑ | ||
| 5 | 5′-GAGTA | 5′-CCTCT | ||||
| ↑ | ↑ | |||||
| 6 | 5′-TTATT | |||||
| ↑ | ||||||
| 7 | 5′-GTTTG | |||||
| ↑ | ||||||
| 8 | 5′-CAATG | |||||
| ↑ | ||||||
| 9 | 5′-CAATA | |||||
| ↑ | ||||||
| 10 | 5′-GTCTT | |||||
| ↑ | ||||||
| 11 | 5′-TACTT | |||||
| ↑ | ||||||
| 12 | 5′-ACATA | |||||
| ↑ | ||||||
| 13 | 5′-CACTA | |||||
| ↑ | ||||||
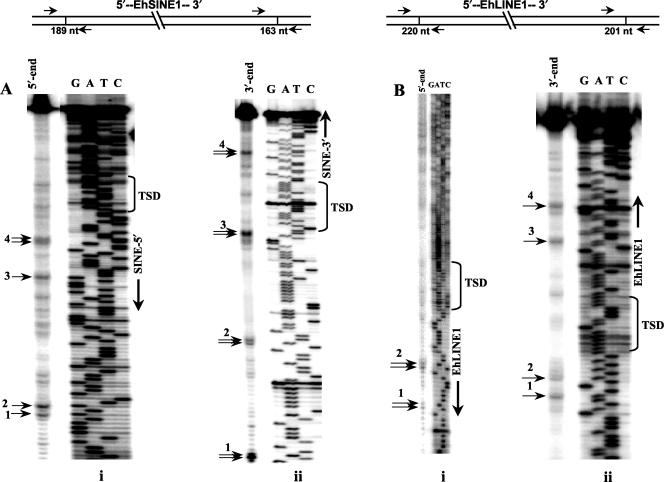
To further understand the nicking preferences of the endonuclease, we used as substrates fragments containing the boundary regions of sites where EhSINE1 and EhLINE1 had inserted. For EhSINE1 the GSS entry AZ550231 contained the entire element. Primers were designed to PCR amplify two fragments (189 and 163 bp, respectively), one from the 5′ end and one from the 3′ end, containing a part of the element and adjoining genomic sequence (Fig. 3A). E. histolytica genomic DNA was used for PCR amplification. To obtain radioactively labeled fragments, the bottom strand primer was end labeled in each case. After incubation with the endonuclease, the resulting products were separated on a sequencing gel as described for Fig. 2. Several bands of various intensities were observed with both fragments. Of these, the four most prominent bands are marked (Fig. 3A). In the fragment from the 3′ end, there was a prominent nick at the 3′ boundary with the TSD (band 3), which corroborates the data for the unoccupied site in Fig. 2. The sequences at the nicking sites are listed in Table 1.
FIG. 3.
(A) Nicking profile of EN protein on DNA fragments derived from the two ends of an EhSINE1 insertion. The sequences of the element and flanking regions were obtained from GSS entries AZ550231 and AZ542529 and were used to design primers for PCR amplification of genomic DNA in order to obtain fragments of the indicated sizes from the 5′ end (i) and the 3′ end (ii). Both fragments were labeled in the bottom strand and used as substrates for nicking by EN protein. Reaction conditions and electrophoretic analysis were carried out as described in the legend to Fig. 2. Electrophoresis was done at 60 W for 1.5 h. Numbered arrows indicate positions of the most prominent nicks. The TSD sequence is marked. In the fragment from the 3′ end, the alignment of band 1 with the sequencing run was slightly skewed. (B) Nicking profile of EN protein on DNA fragments derived from the two ends of an EhLINE1 insertion. Genomic sequences flanking EhLINE1 were obtained from the E. histolytica genome database at the Sanger Centre (contig 8160). Fragments of the indicated sizes were amplified from the two ends and treated with EN protein as described above.
The same experiment was repeated for EhLINE1 by using contig 8160 from the E. histolytica genome database at the Sanger Centre. This contig contains the entire EhLINE1. Again, the boundary fragments were obtained by PCR amplification, and the bottom strand was labeled. The nicking pattern is shown in Fig. 3B, and the nicked sequences are listed in Table 1. Here, too, the enzyme nicked the bottom strand very close to the 3′ end of the TSD in the fragment derived from the 3′ end (band 2). However, unlike the corresponding band in the EhSINE1 insertion site, this band was not the most prominent (Fig. 2 and 3A). It is likely that mutations occurred after the original transposition event took place, making the sequence in contig 8160 a less favorable substrate. The fragment from the 5′ end did not give very prominent nicks.
Once better assembly of the E. histolytica genome sequence is available, it may be possible to find some unoccupied sites of EhLINE1 and use them as substrates. Although the EhLINE1-encoded endonuclease lacks site specificity, it is clear from Table 1 that it nicks at some preferred sites. For example, T was the most common nucleotide found both 3′ and 5′ of the nick, followed by A. Another common feature was the presence of a G 3 to 4 nucleotides upstream of the nick in a large number of sites.
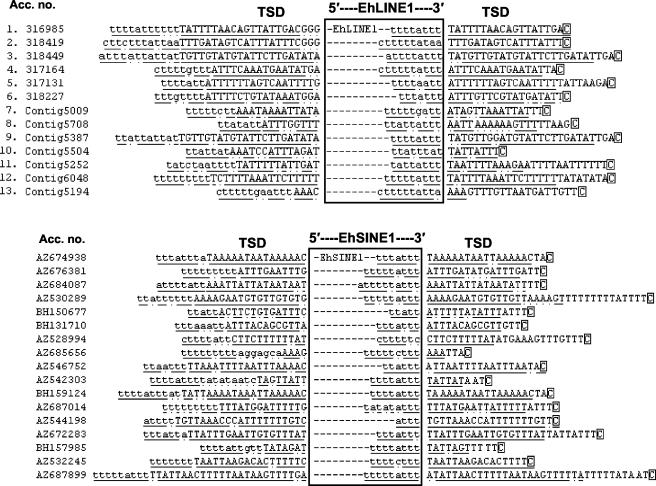
To gain further insight into the possible mode of retrotransposition of EhLINE1/SINE1, we analyzed the sequences flanking the inserted element, as available in the database. We selected entries containing full-length EhLINE1 and EhSINE1 (Fig. 4). The TSD could be readily identified in most cases and ranged from 3 to 22 bp. Two important features that emerged from this analysis are as follows. First, of 13 entries containing full-length EhLINE1 and 17 entries containing full-length EhSINE1 shown in Fig. 4, 21 of 30 contained a C (G in the bottom strand) within 0 to 5 nucleotides from the TSD 3′ end, while the average occurrence of C in a 60-nucleotide stretch surrounding the TSDs in these entries was calculated to be 8%. Since a large number of nicks made by the endonuclease lay close to a 5′ G residue (Table 1), this finding strengthens the possibility that the G in the bottom strand may be involved in target site recognition and nicking by the endonuclease, leading to insertion of the element. Second, the TSD upstream of the element was almost always immediately preceded by a T-rich stretch. This stretch showed a very good sequence match with the 3′ ends of EhLINE1 and EhSINE1 (underlined in Fig. 4). Taking into account the features observed, we propose a model for retrotransposition of these elements (Fig. 5).
FIG. 4.
Conserved sequence features of EhLINE1 and EhSINE1 insertion. EhLINE1 sequences were obtained from the E. histolytica genome database at TIGR (sequences 1 to 6) and the Sanger Centre (sequences 7 to 13), while EhSINE1 sequences were from the GSS database. Boxes show the 3′-most sequence of the element. The TSDs at the two ends are uppercased. Regions of identity between them are underlined, while nonidentical nucleotides are marked by dots. The sequences immediately upstream of the TSD and at the 3′ end of the element, which share close identity, are lowercased. Their regions of identity are underlined, and nonidentical nucleotides are marked by dots. The nearest C flanking the 3′ TSD at its 3′ end is boxed.
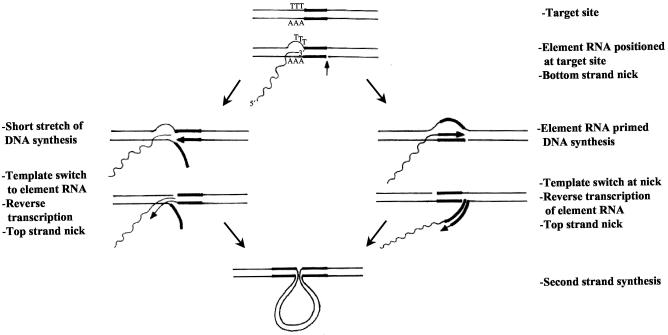
FIG. 5.
Model for EhLINE1/SINE1 retrotransposition. The part of the target site that will be duplicated upon retrotransposition (TSD) is boldfaced. The element RNA is shown as a wavy line, and its 3′ end is complementary to the bottom strand in the T-rich stretch upstream of the TSD (see Fig. 4). The position of the bottom strand nick is inferred from Fig. 2.
DISCUSSION
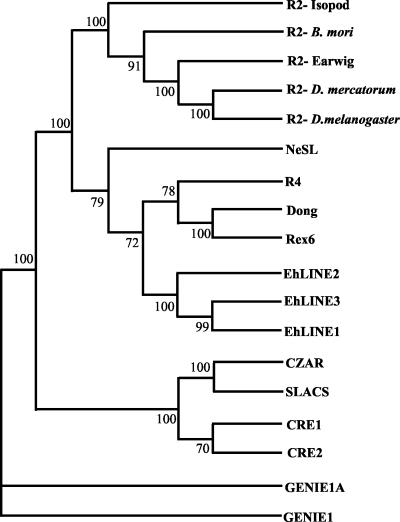
The non-LTR retrotransposons described so far can be classified into two broad categories based on the natures of the endonucleases encoded by the elements. One class encodes the apurinic endonuclease (APE), while the other encodes a restriction enzyme-like endonuclease (EN). Based on RT phylogeny, all the elements of the latter class belong to the R2 group, which is considered to be of ancient origin. The EN domain has been included in phylogenetic analysis, along with the RT domain to improve the resolution (7). Using this approach we show that EhLINE1 (as also EhLINE2 and -3) can be grouped in the R4 clade along with Rex6, Dong, R4, and NeSL (Fig. 6). Excluding some members such as Genie2 of Giardia, which is highly degenerate (7), and some vertebrate Rex6 elements where target sites have not been well defined (31), all well-characterized members of the R2 group show some level of site specificity and insert into defined sites (28S rRNA gene, spliced leader gene, TAA repeats, telomere). EhLINE1 is a clear exception. It is widely dispersed in the genome of its host. In addition, the properties of the EN domain of EhLINE1 reported here show that the enzyme is not strictly site specific and nicks a variety of related target sequences in vitro.
FIG. 6.
Phylogenetic positions of EhLINEs based on RT and EN domains. The analysis was performed essentially as described elsewhere (7). The entire sequence from the RT domain to the C terminus of the element (∼600 amino acid residues) was included in each case. The tree was rooted on the Genie sequence. Numbers indicate bootstrap values at each node. The organisms to which the elements belong are as follows: GENIE1 and -1A, G. lamblia; CRE1 and CRE2, Crithidia fasciculata; SLACS, Trypanosoma brucei; CZAR, Trypanosoma cruzi; Rex6, Oryzias latipas; Dong, B. mori; R4, Ascaris lumbricoides; NeSL, Caenorhabditis elegans; R2-D. melanogaster, Drosophila melanogaster; R2-D. mercatorum, Drosophila mercatorum; R2-Earwig, Forficula auricularia; R2-Isopod, Porcellio scaber.
It is generally believed that target site selection is determined by the element-encoded endonuclease (28) and that consequently, elements with the restriction enzyme-like (EN) type of endonuclease would be site specific. However, the dispersed insertion pattern of EhLINE1 shows that other factors might be involved. This was also suggested by studies on elements belonging to the APE group, where most members are not site specific but many are known to insert at defined target sites (DRE, Tdd-3, and related elements in Dictyostelium discoideum, Zepp elements of Chlorella spp., R1 of Bombyx mori) (13). Although these elements inserted at defined sites, the endonucleases encoded by them were not strictly site specific. It was therefore proposed that apart from the endonuclease, other important factors, such as local chromatin organization and interaction with the transcriptional apparatus of particular genes, might be involved in target site selection (16). It would be interesting to know which factors may have led to the evolution of a loose target specificity of the EhLINE1 endonuclease.
Several studies have shown that SINEs, which do not encode their own transposition functions, exist in partnership with specific LINEs and utilize the enzymatic machinery of the latter for their own transposition (5, 22, 23). The consensus sequences flanking the site of insertion of human and rodent SINEs agreed very well with the experimentally observed hot spots of nicking by the human L1 endonuclease, suggesting that the latter may be involved in SINE mobilization (15, 18). The most compelling evidence in this regard has been the direct in vivo demonstration that eel LINE-encoded functions provided in trans could mobilize a partner SINE which shared the same 3′ tail (19). Here we show the LINE/SINE functional linkage by using a different approach, namely, the ability of the LINE-encoded endonuclease to nick an empty target site, known to be occupied by the partner SINE in vivo. Although the EhLINE1 endonuclease is not strictly site specific and nicked the DNA substrate at several sites, it made a very prominent nick exactly at the point where EhSINE1 had inserted into the genome (Fig. 2), making it highly likely that this SINE element is mobilized by EhLINE1.
The nicking specificity of EhLINE1 EN provided important clues about the possible mode of transposition of this element. The endonuclease nicked the bottom strand at the right end of the TSD (Fig. 2 and 3), implying that the top strand nick would be toward the left of the TSD, resulting in 5′ overhangs. This nicking orientation is also found for the endonuclease encoded by R2Bm (12, 37), whereas the APE enzymes encoded by L1 (15, 18), Tx1L (8), and R1Bm (16) nick the two DNA strands in the opposite orientation to give 3′ overhangs. Due to this difference in nicking orientation, the model proposed for insertion of L1 (18) and R1Bm (16) is not directly applicable to EhLINE1. We propose a speculative model (Fig. 5) to account for TSDs with an enzyme that nicks to produce 5′ overhangs. This model takes into account the conserved feature found in most EhLINE1/SINE1 insertions, namely, the presence of a T-rich stretch just upstream of the TSD, as shown in Fig. 4. According to this model, the EhLINE1/SINE1 transcript finds the target site by virtue of its T-rich 3′ tail. This could happen either concurrently with or after the nicking of the bottom strand by the endonuclease. The 3′-OH of the RNA is then used by the RT (or a host enzyme) to copy the bottom strand, up to the nick. This disruption is followed by a template shift, such that the 3′-OH of the nick is used to prime reverse transcription of the RNA template, which now has a short DNA extension (the TSD). Alternatively, the RT uses the 3′-OH of the nick in the bottom strand to copy the DNA template for a short stretch (the TSD) until it can begin to reverse transcribe the element RNA by switching its template. The importance of the T-rich stretch upstream of the TSD, and that of other aspects of our model, needs to be experimentally verified. In the context of the RT switching its template, it is interesting that the RT encoded by R2Bm has been shown to jump from the 5′ end of one RNA template to the 3′ end of another (4). We have also noted the presence of a G upstream of the nicking site in the bottom strand (Table 1 and Fig. 4). Its involvement, if any, in the endonucleolytic cleavage reaction has to be tested by using appropriate oligonucleotides as substrates. This G, if essential, may have evolved to limit the number of insertion sites of EhLINE1/SINE1 in the AT-rich E. histolytica genome (17). In conclusion, the EhLINE1 element is a novel member of the R4 clade, which underscores the mechanistic diversity that may be encountered as more members of these lineages are discovered.
Acknowledgments
This work was supported by grants to S.B. from the Council of Scientific and Industrial Research and the Department of Science and Technology, Government of India. P.K.M. is a recipient of a research fellowship from CSIR, New Delhi, India.
REFERENCES
- 1.Arkhipova, I. R., and H. G. Morrison. 2001. Three retrotransposon families in the genome of Giardia lamblia: two telomeric, one dead. Proc. Natl. Acad. Sci. USA 98:14497-14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharya, S., A. Bhattacharya, and L. S. Diamond. 1988. Comparison of repeated DNA from strains of Entamoeba histolytica and other Entamoeba. Mol. Biochem. Parasitol. 27:257-262. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya, S., A. Bakre, and A. Bhattacharya. 2002. Mobile genetic elements in protozoan parasites. J. Genet. 81:73-86. [DOI] [PubMed] [Google Scholar]
- 4.Bibillo, A., and T. H. Eickbush. 2001. The reverse transcriptase of the R2 non-LTR retrotransposon: continuous synthesis of cDNA on non-continuous RNA templates. J. Mol. Biol. 316:459-473. [DOI] [PubMed] [Google Scholar]
- 5.Boeke, J. D. 1997. LINEs and Alus—the polyA connection. Nat. Genet. 16:6-7. [DOI] [PubMed] [Google Scholar]
- 6.Burke, W. D., F. Muller, and T. H. Eickbush. 1995. R4, a non-LTR retrotransposon specific to the large subunit rRNA genes of nematodes. Nucleic Acids Res. 23:4628-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke, W. D., H. S. Malik, S. M. Rich, and T. H. Eickbush. 2002. Ancient lineages of non-LTR retrotransposons in the primitive eukaryote, Giardia lamblia. Mol. Biol. Evol. 19:619-630. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, S., G. Pont-Kingdon, and D. Carroll. 2000. Target specificity of the endonuclease from the Xenopus laevis non-long terminal repeat retrotransposon, Tx1L. Mol. Cell. Biol. 20:1219-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, C. G., M. Espinosa Cantellano, and A. Bhattacharya. 2000. Entamoeba histolytica: an overview of the biology of the organism, p. 1-45. In J. I. Ravdin (ed.), Amebiasis. Imperial College Press, London, United Kingdom.
- 10.Cruz-Reyes, J., T. Ur-Rehman, W. M. Spice, and J. P. Ackers. 1995. A novel transcribed repeat element from Entamoeba histolytica. Gene 166:183-184. [DOI] [PubMed] [Google Scholar]
- 11.Diamond, L. S., D. R. Harlow, and C. Cunnick. 1978. A new medium for axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431-432. [DOI] [PubMed] [Google Scholar]
- 12.Eickbush, T. H. 2002. R2 and related site-specific non-long terminal repeat retrotransposons, p. 813-835. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 13.Eickbush, T. H., and H. S. Malik. 2002. Origin and evolution of retrotransposons, p. 1111-1144. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 14.Felsenstein, J. 1993. PHYLIP (phylogeny inference package), version 3.55. Department of Genetics, University of Washington, Seattle.
- 15.Feng, Q., J. V. Moran, H. H. Kazazian, Jr., and J. D. Boeke. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905-916. [DOI] [PubMed] [Google Scholar]
- 16.Feng, Q., G. Schumann, and J. D. Boeke. 1998. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc. Natl. Acad. Sci. USA 95:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gelderman, A. H., I. L. Bartgis, D. B. Keister, and L. S. Diamond. 1971. A comparison of genome sizes and thermal denaturation-derived base composition of DNAs from several members of Entamoeba (histolytica group). J. Parasitol. 57:912-916. [PubMed] [Google Scholar]
- 18.Jurka, J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retrotransposons. Proc. Natl. Acad. Sci. USA 94:1872-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajikawa, M., and N. Okada. 2002. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell 111:433-444. [DOI] [PubMed] [Google Scholar]
- 20.Luan, D. D., M. H. Korman, J. L. Jakubczak, and T. H. Eickbush. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- 21.Malik, H. S., W. D. Burke, and T. H. Eickbush. 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16:793-805. [DOI] [PubMed] [Google Scholar]
- 22.Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis, J. D. Boeke, and H. H. Kazazian, Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917-927. [DOI] [PubMed] [Google Scholar]
- 23.Moran, J. V., and N. Gilbert. 2002. Mammalian LINE-1 retrotransposons and related elements, p. 836-869. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 24.Ohshima, K., M. Hamada, Y. Terai, and N. Okada. 1996. The 3′ ends of tRNA-derived short interspersed repetitive elements are derived from the 3′ ends of long interspersed repetitive elements. Mol. Cell. Biol. 16:3756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Sharma, R., A. Bagchi, A. Bhattacharya, and S. Bhattacharya. 2001. Characterization of a retrotransposon-like element in Entamoeba histolytica. Mol. Biochem. Parasitol. 116:45-53. [DOI] [PubMed] [Google Scholar]
- 27.Swofford, D. L. 1999. PAUP v 4.0. Laboratory of Molecular Systematics, Smithsonian Institution, Washington, D.C.
- 28.Takahashi, H., and H. Fujiwara. 2002. Transplantation of target site specificity by swapping the endonuclease domains of two LINEs. EMBO J. 21:408-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.VanDellen, K., J. Field, Z. Wang, B. Loftus, and J. Samuelson. 2002. LINEs and SINE-like elements of the protist Entamoeba histolytica. Gene 297:229-239. [DOI] [PubMed] [Google Scholar]
- 31.Volff, J.-N., C. Korting, A. Froschauer, K. Sweeney, and M. Schartl. 2001. Non-LTR retrotransposons encoding a restriction enzyme-like endonuclease in vertebrates. J. Mol. Evol. 52:351-360. [DOI] [PubMed] [Google Scholar]
- 32.Wilhoeft, U., H. Bub, and E. Tannich. 1999. Analysis of cDNA expressed sequence tags from Entamoeba histolytica: identification of two highly abundant polyadenylated transcripts with no overt open reading frames. Protist 150:61-70. [DOI] [PubMed] [Google Scholar]
- 33.Wilhoeft, U., H. Bub, and E. Tannich. 2002. The abundant polyadenylated transcript 2 DNA sequence of the pathogenic protozoan parasite Entamoeba histolytica represents a nonautonomous non-long-terminal-repeat retrotransposon-like element which is absent in the closely related nonpathogenic species Entamoeba dispar. Infect. Immun. 70:6798-6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong, Y., and T. H. Eickbush. 1988. Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell 55:235-246. [DOI] [PubMed] [Google Scholar]
- 35.Xiong, Y., and T. H. Eickbush. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9:3353-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong, Y., and T. H. Eickbush. 1993. Dong, a non-long terminal repeat retrotransposable element from Bombyx mori. Nucleic Acids Res. 21:1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, J., H. S. Malik, and T. H. Eickbush. 1999. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. USA 96:7847-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]