Abstract
Human adenoviruses (HAdVs) are ubiquitous double-stranded DNA viruses that cause a wide array of diseases in humans including pharyngitis, pneumonia, gastroenteritis, hemorrhagic cystitis, and keratoconjunctivitis. They also cause life-threatening opportunistic infections in immunocompromised individuals and are responsible for outbreaks in certain populations. Diagnosis is traditionally by cell culture or antigen detection methods. However, some HAdVs can take up to 4 weeks to isolate, and diarrheagenic types 40 and 41 will not grow in routine cell culture. Therefore, the goal of this study was to design a rapid, real-time PCR assay to detect all known 57 HAdV types from multiple different specimen sources. Primers and fluorescence resonance energy transfer hybridization probes were designed to target a 185-bp region of the penton base gene of HAdV. The analytical sensitivity was determined to be 10 copies/μl for HAdV types showing exact primer/probe homology in specimen matrix. Using whole-virus strains, the analytical sensitivity for representative HAdV types ranged from 10−1 to 103 50% tissue culture infective dose (TCID50)/ml. The assay demonstrated 100% sensitivity and 99% specificity. This real-time PCR assay provides a rapid method for the detection of all 57 known HAdV types from respiratory specimens, blood, stool, urine, and ocular swabs.
INTRODUCTION
Human adenoviruses (HAdVs) are ubiquitous double-stranded DNA viruses that cause a wide array of diseases in humans, including pharyngitis, pneumonia, gastroenteritis, hemorrhagic cystitis, and keratoconjunctivitis (23). Adenoviruses may also cause life-threatening opportunistic infections in immunocompromised individuals and are responsible for outbreaks in certain populations such as day care attendees and military recruits (23).
Adenoviruses are classified in the genus Mastadenovirus, which contains 7 known HAdV species (HAdV-A to HAdV-G). Historically, HAdVs were further classified by hemagglutination and serum neutralization reactions into 51 serotypes (29). New adenovirus types have since been identified by several authors based on genomic data, including several emerging and recombinant viruses, for a total of 57 unique HAdV types (1, 14, 16–18, 27, 29, 32, 33). Most recently, a primate adenovirus from New World monkeys was detected which crossed the species barrier to infect humans (2).
Diagnosis is traditionally by cell culture or antigen detection methods. Most HAdVs grow readily in cell culture and can be detected in a variety of specimen types including nasopharyngeal, throat and conjunctival swabs, urine, cerebrospinal fluid (CSF), and stool. However, some types can take up to 4 weeks to isolate, and diarrheagenic types 40 and 41 will not grow in routine cell culture. Direct antigen detection from clinical specimens may be used for diagnosis of acute infections but provides markedly low sensitivity compared to culture (30). Less commonly, serology is used to detect a rise in adenovirus-specific antibodies following infection (23).
Real-time PCR offers a rapid and sensitive alternative to culture, antigen detection, and serology (31). Due to the limitations of conventional diagnostic methods for HAdV, the aim of this study was to develop and validate a real-time PCR assay for all 57 known HAdV types from multiple different specimen sources, including respiratory specimens, blood, stool, urine, and conjunctival swabs.
(This study was presented in part at the General Meeting of the American Society of Microbiology, 2007, Toronto, Canada.)
MATERIALS AND METHODS
Clinical specimens.
Selected patient specimens (n = 763) submitted to the clinical virology laboratory for routine viral culture from January 2006 through December 2008 were concurrently tested by PCR, conventional tube culture, and shell vial culture. Specimens included upper and lower respiratory specimens (bronchoalveolar lavage [BAL], bronchial washings, tracheal secretions, induced or expectorated sputum, throat swabs, nasal/nasopharyngeal swabs), body fluids (pleural fluid and peritoneal fluid), fresh tissue, ocular swab specimens (conjunctival and corneal specimens collected on a swab), urine, and stool. The Mayo Foundation institutional review board approved the use of all clinical specimens.
In addition to prospectively obtained specimens, 28 archived culture-positive and -negative specimens (lower respiratory, nasal, and throat swabs; unfixed tissue; ocular swab specimens; urine; and stool; ≤1 year from date of collection; stored at −20°C) were tested by the PCR assay and compared to the original culture result.
Culture of clinical specimens.
Adenovirus was isolated in conventional tube culture using rhesus monkey kidney and MRC-5 cell lines and in shell vial (MRC-5) culture (LabCorp, Viromed, Minneapolis, MN). Virus identification was confirmed by the presence of characteristic cytopathic effects (CPEs) and characteristic staining patterns with specific monoclonal antibodies (Millipore, Light Diagnostics, Billerica, MA).
Specimen processing and nucleic acid extraction.
Specimens collected on a swab were agitated and expressed into a tube containing 3 ml of M5 viral transport medium (Remel, Lenexa, KS) for subsequent extraction. Given the potential viscous nature of liquid respiratory specimens and body fluids, these were first processed by adding 500 μl of raw specimen and 100 μl of proteinase K (Roche Applied Sciences, Indianapolis, IN) into a 2.0-ml tube containing 0.1-mm silica glass beads and 2.4-mm zirconia beads (Biospec Products, Bartlesville, OK), followed by incubation at 55°C for 15 min on a Thermomixer instrument (Eppendorf, Hamburg, Germany) for mixing at 1,400 rpm and subsequent heating on a 95°C heat block for 5 min.
To process soft or formed stools for DNA extraction, a sterile cotton-tipped swab was dipped into various locations of the specimen and then agitated and expressed into a 2.0-ml tube containing 1 ml of sterile water. For liquid stools, 100 μl of specimen was placed directly into 1 ml of sterile water. Tubes were then briefly vortexed and subjected to DNA extraction.
Fresh tissue specimens were processed by placing a small piece of tissue (approximately 0.5 cm3) into a 1.5-ml tube containing 400 μl of 1× Tris-EDTA (Sigma-Aldrich, St. Louis, MO), 100 μl of proteinase K (Roche Applied Sciences), and 50 μl of 10% sodium dodecyl sulfate (Sigma-Aldrich). The samples were vortexed briefly and placed on the Thermomixer for at least 4 h at 55°C with a mixing speed of 500 rpm.
Following the processing steps above, 200 μl of each specimen underwent extraction on the MagNA Pure Compact or 1.2 instrument (Roche Applied Sciences) using the total nucleic acid isolation kit, with a final elution volume of 100 μl.
Real-time PCR.
The real-time PCR assay was developed on the LightCycler (LC) 2.0 instrument (Roche Applied Sciences). Primers and fluorescence resonance energy transfer (FRET) hybridization probes were designed to target a 185-bp region of the penton base gene. Primers and probes were synthesized by TIB MolBiol (primer-probe set 261; Adelphia, NJ), and their sequences are provided in Table 1. Chosen primer sequences had complete homology with 41 of 46 HAdV-type targeted gene sequences examined and 2 mutations in the remaining 5 target gene sequences examined (types 6, 57, 2, 1, and 5). The donor probe was labeled with fluorescein, and the acceptor probe was labeled with Red 640. The probes were designed to anneal to the targeted nucleotide sequences with 2 bases between donor and acceptor probes, across which energy transfer occurred. The probes had 0 to 3 mutations in the targeted region, resulting in differences in the melting temperatures produced (Table 2). The PCR assay was performed using the LC FastStart DNA master hybridization probe kit (Roche Applied Sciences) with reaction mixtures consisting of 2 mM MgCl2, 1× LightCycler FastStart mix, 0.7 μM each primer, 0.2 μM fluorescein-labeled probe, and 0.4 μM LC Red 640-labeled probe. The total reaction volume was 20 μl (15 μl master mix plus 5 μl extracted nucleic acid). PCR amplification with real-time detection was performed using the following cycling parameters: template denaturing (1 cycle) at 95°C for 10 min, followed by 45 amplification cycles of 95°C for 10 s, 55°C for 15 s, and 72°C for 15 s. Following amplification, melting temperature (Tm) analysis was performed by measuring the fluorescent signal during the following cycling profile: 95°C for 0 s, 59°C for 20 s, 45°C for 20 s with a 0.2°C/s transition, and 85°C for 0 s with a 0.2°C/s transition. Generation of an acceptable melting peak (demonstrating ≥0.02 fluorescent height) was used for the determination of a positive or negative result.
Table 1.
Primer-probe sequences used in experiments for the detection of the penton base gene of HAdVa
| Name | Sequence (5′ to 3′) |
|---|---|
| ADV 1 | CGT CTT CAA YCG CTT |
| ADV 2 | TGT AGA CGT AGG GAC AGG |
| ADV 3 | CCG TCA GTG AAA ACG TGC-FL |
| ADV 4 | Red 640-GCT CTC ACA GAT CAC GGG-P |
Kit no. 261 (TIB MolBiol).
Table 2.
HAdV types with corresponding LoD, Tm range, and mutations in probe region
| HAdV typeb | LoD for representative type of whole virus (TCID50/ml)b | Tm range | Fluorescein probe areaa | LC Red 640 probe areaa |
|---|---|---|---|---|
| 8, 9, 10, 13, 15, 17, 19, 20, 22, 23, 25-30, 32, 33, 36-39, 42-47, 49, 51, 53, 56 | 10−1 (type 19), 100 (type 9) | 61.1–62.0 | CCG TCA GTG AAA ACG TGC | GCT CTC ACA GAT CAC GGG |
| 24 | Not performed | 58.2 | CGG TCA GTG AAA ACG TGC | GCT CTC ACA GAT CAC GGG |
| 31 | Not performed | 56.4 | CCG GCA GTG AAA ACG TTC | GCT CTC ACA GAT CAC GGG |
| 1, 2, 3, 4-7, 11, 14, 21, 34, 35, 50, 55, 57 | 100 (type 4), 101 (types 1, 3) | 55.3–56.7 | CCG TCA GTG AAA ACG TTC | GCT CTC ACA GAT CAC GGG |
| 18, 40, 41, 52 | 10−1 | 55.9–56.6 | CCG TCA GTG AAA ACG TTC | GCC CTC ACA GAT CAC GGA |
| 48, 54 | Not performed | 52.0 | CCG TTA GTG AAA ACG TGC | GCT CTC ACA GAT CAC GGG |
| 16 | 10−1 | 50.9 | CCA TCA GTG AAA ACG TTC | GCT CTC ACA GAT CAC GGG |
| 12 | 103 | 49.9 | CCG TCA GTG AAA ACG TTC | GCC CTT ACA GAT CAC GGG |
Mutations in the region targeted by the probes are in bold. There are 2 intervening nucleotides between the donor (fluorescein) probe and acceptor (LC Red 640) probe across which energy transfer occurs.
Bolded HAdV types were used to determine the limit of detection for each Tm-based grouping shown above (see Table S3 in the supplemental material for virus strains used). Whole-virus strains were quantitated using the 50% tissue culture infective dose (TCID50).
Generation of a positive control plasmid.
Human adenovirus type 9 (VR1086; American Type Culture Collection [ATCC]) was used to construct a positive control plasmid using the pCR2.1-TOPO TA cloning kit (Invitrogen Corporation, Carlsbad, CA) following the manufacturer's instructions. The plasmid construct was then purified using the High Pure plasmid isolation kit (Roche Applied Sciences) according to the manufacturer's instructions. The expected size of the recombinant plasmid was confirmed with 2% agarose gel electrophoresis. Subsequent Sanger sequencing confirmed the insert to be the targeted region of the penton base gene derived from HAdV type 9.
Analytical sensitivity and specificity.
The analytical sensitivity of the assay was determined by testing a dilution series of the HAdV positive control plasmid. A 10-fold dilution (104 copies/μl serially diluted to 100 copies/μl) of the plasmid was performed in Tris-EDTA (TE) buffer, and each dilution was then tested in triplicate. The limit of detection (LoD) in buffer was determined to be the highest dilution whereby all three replicates were positive. This LoD was then confirmed in each specimen matrix group to by spiking 30 analyte-negative specimens from each specimen group at the approximate LoD determined in TE buffer. The specimen matrices tested were upper and lower respiratory specimens (bronchoalveolar lavage, bronchial washings, tracheal secretions, induced or expectorated sputum, throat swabs, nasal/nasopharyngeal swabs), body fluids (pleural, peritoneal, and synovial), fresh tissue, ocular swab specimens (conjunctival and corneal), urine, and stool. After spiking, specimens underwent extraction and amplification with the novel HAdV PCR. The limit of detection was also obtained for representative HAdV types by making serial dilutions of quantitated whole virus and testing each dilution in triplicate.
The analytical specificity was determined by performing a BLAST search of each primer, each probe, and the entire amplicon sequence using the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). In addition, a panel of nucleic acid from 113 microorganisms, including fungi (n = 23), bacteria (n = 60), mycobacteria (n = 10), viruses (n = 19), and human DNA (human diploid fibroblasts, Medical Research Council-5 cells; Viromed, Minneapolis, MN) (n = 1) was tested (see Tables S1 and S2 in the supplemental material). Panel organisms were chosen based on their ability to cause similar diseases, their potential for being found in the specimen source as a pathogen or normal flora, and/or their genetic similarity to the HAdV penton base target. Amplification and Sanger dideoxy sequencing of either 16S (bacterial) or D2 LSU (fungal) rRNA genes was utilized to confirm the presence of amplifiable nucleic acids in the specificity panel.
Accuracy/method comparison and testing for specimen matrix inhibition.
PCR results were compared to viral tube and shell vial culture results. Prospectively collected specimens were processed in parallel for PCR and viral tube and shell vial cultures to allow for comparison, while PCR results were compared to the original viral tube and shell vial culture results for archived specimens.
Due to the low number of positive clinical specimens obtained by prospective and retrospective testing, the plasmid positive control was also spiked into negative clinical specimens at approximately 10 to 100 copies/μl to evaluate the sensitivity of the assay in these specimen sources and to look for potential matrix-mediated inhibition. Spiked specimens comprised 30 body fluid, 30 whole-blood, 30 plasma, 20 respiratory, 20 CSF, 20 ocular swab, 20 urine, and 20 stool specimens. Inhibition was also assessed by spiking extracted stool and ocular swab specimens with plasmid positive control at an approximate concentration of 10 to 100 copies/μl and testing by real-time PCR. Extraction and/or PCR inhibition was indicated by loss of positivity as measured by melting-curve analysis.
To test for assay inclusivity, HAdV types 1 to 52, including variant strain 14a linked to a pneumonia outbreak in 2007 (8), were tested by the HAdV assay. Virus strains were ordered from ATCC or obtained from the California Department of Health or the Centers for Disease Control and Prevention (CDC) (see Table S3 in the supplemental material).
Since we were unable to obtain virus isolates for HAdV types 53 to 57, the oligonucleotide target region of each type was constructed by TIB MolBiol (Berlin, Germany) by PCR followed by insertion into TOP 10 F′ cells via the pCR2.1 TOPO vector (Life Technologies Invitrogen, Carlsbad, CA). Plasmids were extracted and purified, followed by confirmation of the product by EcoRI restriction digest analysis. Positive clones were sequenced using BigDye Terminator v 1.1 cycle sequencing kit and analyzed on an ABI Prism 310 genetic analyzer (Life Technologies Applied Biosystems, Carlsbad, CA).
RESULTS
Analytical sensitivity and specificity.
The established LoD of the novel HAdV PCR assay using the positive control plasmid was determined to be approximately 1 copy of target/μl in TE buffer and 10 copies of target/μl in specimen matrices (upper and lower respiratory, CSF, ocular, whole-blood, stool, urine, body fluid, and plasma specimens). To supplement these data using virus strains with one or more mutations under the targeted primer and probe regions, studies were performed using representative quantitated whole-virus isolates. All representative virus strains except HAdV 12 demonstrated LoDs of 10−1 to 101 TCID50/ml (strains 1, 3, 4, 9, 16, 19, and 41) (Table 2). Human adenovirus type 12 contained 3 mutations within the targeted primer/probe regions (3 mutations in the nucleotide region targeted by the probes, and 0 mutations in the primer region) and also had the lowest melting temperature of all 57 HAdV types, indicating the weakest affinity between the probes and targeted sequence; the LoD for this type was 103 TCID50/ml.
The BLAST search of the primer, probe, and target sequences for the HAdV penton base gene did not yield any highly similar sequences. In addition, testing of nucleic acids from 112 bacteria, fungi, and viruses demonstrated no positive results with this assay (see Tables S1 and S2 in the supplemental material).
Accuracy/method comparison and testing for specimen matrix inhibition.
Of the 791 clinical specimens tested by novel HAdV PCR and culture (763 prospective and 28 archived specimens), 75 specimens were positive for HAdV by both viral culture and the novel PCR (7/221 respiratory nonswab, 28/288 swab [comprising 279 respiratory swab, 8 anal-genital swab, and 1 dermal swab specimen], 29/125 ocular swab, 6/72 stool, 3/56 fresh tissue, 2/5 urine, 0/9 body fluid, and 0/15 miscellaneous specimens). Clinical sensitivity of the novel HAdV PCR was 100% with no culture-positive, PCR-negative specimens. Seven specimens (1 bronchial, 1 throat swab, 1 liver biopsy, 1 ocular swab, 2 urine, 1 stool) were positive by the novel PCR and negative by culture, while 709 specimens were negative by both culture and PCR (clinical specificity 99%). Stools and urine specimens are considered difficult sources to culture, which may explain 3 of the discrepant results. One PCR-positive, culture-negative liver biopsy specimen was obtained from a patient in which HAdV was grown from a liver biopsy specimen obtained 2 days previously. The remaining 3 discrepant specimens had high crossing points (Cps; >35) indicating low levels of virus which may not have been present in sufficient quantities for detection by cell culture. The low levels prevented confirmatory sequencing of the amplicon.
Additional studies using analyte-negative clinical specimens spiked with the adenovirus-positive plasmid control at 10 to 100 copies/μl followed by extraction were positive in 30/30 blood specimens, 30/30 body fluid specimens, 20/20 CSF specimens, 20/20 ocular specimens, 30/30 plasma specimens, 20/20 respiratory specimens, 20/20 stool specimens, and 20/20 urine specimens. Spiking studies therefore demonstrated a sensitivity of 100% for HAdV in all specimen types.
By spiking clinical specimens within 1 log of the LoD prior to extraction as described above, we were able to demonstrate that PCR inhibitors were not present or were effectively removed during the extraction process in representative specimens. In addition, spiking of 100 stool and 72 ocular swab specimens demonstrated no amplification inhibition. These data were combined with historic data for other PCR tests performed using similar design, extraction platform, master mix components, and cycling conditions, which demonstrated inhibition rates of <1%; specifically, inhibition was seen in only 2 of 32,642 blood specimens (0.01%), 6 of 851 body fluid specimens (0.71%), 3 of 1,946 CSF specimens (0.15%), 0 of 18,245 plasma specimens (0%), 9 of 17,645 respiratory specimens (0.05%), 6 of 644 stool specimens (0.93%), and 2 of 270 urine specimens (0.74%).
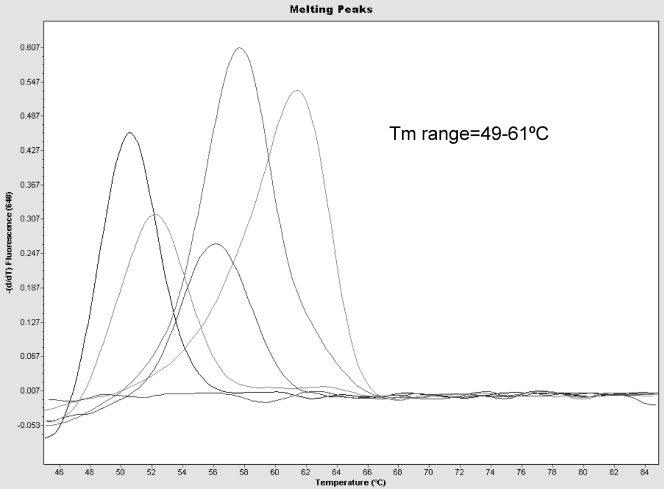
All 52 whole-virus strains and the 5 DNA constructs (HAdV types 53 to 57) tested positive by the novel HAdV PCR assay. The acceptable Tm peak range based on testing all confirmed HAdV types was noted to be 49°C to 62°C (Fig. 1). The presence of a Tm within this range that exceeded 0.02 fluorescence was used to determine assay positivity. Table 2 shows the Tms of the 57 HAdV types and amplified regions under the assay probes.
Fig 1.
Melt peak analysis of the results of the LC PCR assay. Most of the HAdV types will melt at 62°C, but the melting point can range from 49°C to 62°C depending on the type and number of mismatches under the probes.
DISCUSSION
We present a real-time PCR assay for qualitative detection of all 57 reported types of human adenovirus from multiple specimen types. To our knowledge, ours is the only assay currently described that will detect all 57 types, including recently described epidemic strains. The described assay may be used for qualitative detection from diverse specimen sources, including stool specimens, urine specimens, lower respiratory specimens, and specimens collected on a swab. Given the potential for multiple organ system involvement by this virus and the organ tropism of some HAdV types, it is desirable that a PCR assay be validated for multiple specimen types. For example, HAdV is a known cause of pharyngoconjunctival fever, and respiratory and ocular specimens are commonly submitted for HAdV PCR. HAdV is also a common cause of gastrointestinal disease, and detection of HAdV DNA in stool may precede viremia (20, 21). Finally, detection of HAdV DNA by PCR in serum may also predict disseminated infection in severe disease (6). Quantitative detection may be useful in some situations (21, 19), and additional studies may be performed to determine the suitability of the currently described assay for monitoring HAdV type viral load over time.
There are currently three FDA approved tests for qualitative detection of HAdVs. However, these are approved for use with nasopharyngeal swabs only in patients with signs and symptoms of respiratory disease. The first assay to receive FDA approval was the Luminex xTAG respiratory viral panel (RVP; Luminex Corporation, Toronto, Canada) for simultaneous detection of multiple respiratory viruses from a nasopharyngeal swab. This assay is an “open” PCR system in that amplicon is first generated and then must be manipulated via the addition of proprietary beads before detection via the Luminex instrument. The original format of this assay can amplify and detect viruses in 96 specimens from a previously extracted specimen in an 8-h shift. The newly FDA-approved version (xTAG RVP Fast) is still an open system but offers some decreased contamination risk and takes approximately 5 h from amplification to detection and reporting. However, some investigators have reported decreased sensitivity for detection of some viruses by the new faster method (24).
Like the xTAG RVP, the FilmArray respiratory panel (RP; Idaho Technology, Inc., Salt Lake City, UT) is a multiplex assay for qualitative detection of multiple respiratory viruses including HAdV. It consists of a self-contained pouch system that contains all of the reagents needed for nucleic acid extraction and amplification. While it provides sensitivity equal to or greater than that of the xTAG RVP (26), it can test only a single patient specimen in 1 h. According to the package insert, testing of 17 HAdV types and in silico analysis indicate that the FilmArray can detect HAdV types 1 to 53 (12).
Finally, the Gen-Probe Prodesse ProAdeno+ is a singleplex assay for qualitative detection of HAdV DNA from extracted nasopharyngeal swab specimens. The package insert (7) states that it can detect HAdV serotypes 1 to 51; additional types were not tested.
In addition to the 2 FDA-approved tests, several qualitative and quantitative PCR tests for HAdV types have been described (3–5, 9–11, 15, 22, 25, 28, 34). The vast majority require nested, multiplex, or multiple singleplex PCRs for the detection of multiple virus types. Furthermore, it is unknown if any of these assays are capable of detecting the newly described HAdV types 53 to 57, responsible for human outbreaks in Japan (13, 16, 18), China (33), France (27), and the United States (14).
We describe an assay that uses a single set of primers and probes for detection of all reported HAdV types and allows sensitive and specific detection of HAdVs from a wide range of clinical specimens. Analytical sensitivity is excellent, with similar limits of detection determined for all HAdV strains tested, except HAdV 12, which had the lowest Tm of all 57 HAdV types, indicating the weakest bond between probes and the target sequence. It is therefore not surprising that the LoD was 2 dilutions lower for this type. Fortunately, HAdV 12 is rarely described in human infection.
The straightforward specimen processing protocols, including automated extraction, make this assay well suited for routine diagnostic applications. In addition, the closed-system, real-time PCR design with single primer and probe set facilitates implementation as a laboratory-developed test.
Melting temperature analysis is the detection phase of the assay and is performed at the end of the analysis after amplification has occurred. The production of a distinct Tm occurs only if the 33-base primer set has successfully amplified the targeted region and the 36-base probe set has annealed to produce a detectable fluorescent signal. The specificity of the assay is supported by the results of the BLAST search and the specificity studies which included a wide range of pathogens, commensal organisms, and other respiratory viruses. Polymorphisms in the region targeted by the FRET probes results in a shift in Tm peak, so that the Tms of known HAdV types range from 49°C to 62°C (Table 2; Fig. 1). While not a specific goal of this investigation, further work could be performed to determine if Tm differences could separate HAdV genotypes into groups. This may be useful for epidemiological purposes.
In summary, we present a highly sensitive and specific qualitative real-time PCR assay using a single set of primers/probes for detection of all 57 known HAdV types from a wide variety of clinical specimens commonly submitted for viral culture.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Schnurr from the Viral and Rickettsial Disease Lab, California Department of Health, for providing the HAdV type 51 strain used in this study. We also thank Ute Boetcher and Eduardo Thuroff from TIB MolBiol for the generation of DNA constructs containing the targeted region of HAdV types 53 to 57.
S.P.B., L.M.S., M.J.E., T.F.S., and Mayo Clinic have received royalties of less than the federal threshold for significant financial interest from the licensing of this technology.
Footnotes
Published ahead of print 14 December 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Aoki K, et al. 2008. Epidemic keratoconjunctivitis due to the novel hexon-chimeric-intermediate 22,37/H8 human adenovirus. J. Clin. Microbiol. 46:3259–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen EC, et al. 2011. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog. 7:e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Claas EC, et al. 2005. Internally controlled real-time PCR monitoring of adenovirus DNA load in serum or plasma of transplant recipients. J. Clin. Microbiol. 43:1738–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Damen M, et al. 2008. Real-time PCR with an internal control for detection of all known human adenovirus serotypes. J. Clin. Microbiol. 46:3997–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ebner K, Suda M, Watzinger F, Lion T. 2005. Molecular detection and quantitative analysis of the entire spectrum of human adenoviruses by a two-reaction real-time PCR assay. J. Clin. Microbiol. 43:3049–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Echavarria M, et al. 2001. Prediction of severe disseminated adenovirus infection by serum PCR. Lancet 358:384–385 [DOI] [PubMed] [Google Scholar]
- 7. Gen-Probe 2010. Prodesse ProAdeno+ instructions for use for detection of human adenovirus. Gen-Probe Prodesse Inc., Waukesha, WI [Google Scholar]
- 8. Gray GC, Chorazy ML. 2009. Human adenovirus 14a: a new epidemic threat. J. Infect. Dis. 199:1413–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gu Z, Belzer SW, Gibson CS, Bankowski MJ, Hayden RT. 2003. Multiplexed, real-time PCR for quantitative detection of human adenovirus. J. Clin. Microbiol. 41:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He JW, Jiang S. 2005. Quantification of enterococci and human adenoviruses in environmental samples by real-time PCR. Appl. Environ. Microbiol. 71:2250–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heim A, Ebnet C, Harste G, Pring-Akerblom P. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 70:228–239 [DOI] [PubMed] [Google Scholar]
- 12. Idaho Technology Inc 2011. The FilmArray respiratory panel instruction manual. Idaho Technology Inc., Salt Lake City, UT [Google Scholar]
- 13. Ishiko H, et al. 2008. Novel human adenovirus causing nosocomial epidemic keratoconjunctivitis. J. Clin. Microbiol. 46:2002–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones MS, II, et al. 2007. New adenovirus species found in a patient presenting with gastroenteritis. J. Virol. 81:5978–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jothikumar N, et al. 2005. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 71:3131–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaneko H, et al. 2011. Recombinant analysis of intermediate human adenovirus type 53 in Japan by complete genome sequence. J. Gen. Virol. 92:1251–1259 [DOI] [PubMed] [Google Scholar]
- 17. Kaneko H, et al. 2011. Complete genome analysis of a novel intertypic recombinant human adenovirus causing epidemic keratoconjunctivitis in Japan. J. Clin. Microbiol. 49:484–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaneko H, et al. 2011. Epidemiological and virological features of epidemic keratoconjunctivitis due to new human adenovirus type 54 in Japan. Br. J. Ophthalmol. 95:32–36 [DOI] [PubMed] [Google Scholar]
- 19. Leen AM, Rooney CM. 2005. Adenovirus as an emerging pathogen in immunocompromised patients. Br. J. Haematol. 128:135–144 [DOI] [PubMed] [Google Scholar]
- 20. Lion T, et al. 2003. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood 102:1114–1120 [DOI] [PubMed] [Google Scholar]
- 21. Lion T, et al. 2010. Monitoring of adenovirus load in stool by real-time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia 24:706–714 [DOI] [PubMed] [Google Scholar]
- 22. Logan C, O'Leary JJ, O'Sullivan N. 2006. Real-time reverse transcription-PCR for detection of rotavirus and adenovirus as causative agents of acute viral gastroenteritis in children. J. Clin. Microbiol. 44:3189–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mandell GL, Bennett JE, Dolin R. 2009. Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 7th ed Churchill Livingstone/Elsevier, Philadelphia, PA [Google Scholar]
- 24. Pabbaraju K, Wong S, Tokaryk KL, Fonseca K, Drews SJ. 2011. Comparison of the Luminex xTAG respiratory viral panel with xTAG respiratory viral panel fast for diagnosis of respiratory virus infections. J. Clin. Microbiol. 49:1738–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pehler-Harrington K, Khanna M, Waters CR, Henrickson KJ. 2004. Rapid detection and identification of human adenovirus species by adenoplex, a multiplex PCR-enzyme hybridization assay. J. Clin. Microbiol. 42:4072–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rand KH, Rampersaud H, Houck HJ. 2011. Comparison of two multiplex methods for detection of respiratory viruses: FilmArray RP and xTAG RVP. J. Clin. Microbiol. 49:2449–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinson CM, et al. 2011. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology 409:141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sarantis H, Johnson G, Brown M, Petric M, Tellier R. 2004. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J. Clin. Microbiol. 42:3963–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seto D, Chodosh J, Brister JR, Jones MS. 2011. Using the whole genome sequence to characterize and name human adenoviruses. J. Virol. 85:5701–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shetty AK, et al. 2003. Comparison of conventional viral cultures with direct fluorescent antibody stains for diagnosis of community-acquired respiratory virus infections in hospitalized children. Pediatr. Infect. Dis. J. 22:789–794 [DOI] [PubMed] [Google Scholar]
- 31. Stroparo E, et al. 2010. Adenovirus respiratory infection: significant increase in diagnosis using PCR comparing with antigen detection and culture methods. Rev. Inst. Med. Trop. Sao Paulo 52:317–321 [DOI] [PubMed] [Google Scholar]
- 32. Walsh MP, et al. 2009. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 4:e5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walsh MP, et al. 2010. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J. Clin. Microbiol. 48:991–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watzinger F, et al. 2004. Real-time quantitative PCR assays for detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. J. Clin. Microbiol. 42:5189–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.