Abstract
Pathogenic Helicobacter pylori strains can selectively activate epithelial mitogen-activated protein kinase (MAPK) signaling pathways linked with disease. We now demonstrate that H. pylori-induced hemolysis is strain specific and is mediated by phospholipases PldA1 and PldD. Inactivation of PldD inhibited activation of extracellular signal-regulated kinases 1 and 2 (ERK1/2), indicating that H. pylori hemolytic phospholipases also harbor MAPK-activating properties.
TEXT
Helicobacter pylori colonizes the stomachs of at least half of the world's population and is a strong risk factor for gastric adenocarcinoma (24, 26). H. pylori can activate epithelial signaling pathways, such as the mitogen-activated protein kinase (MAPK) extracellular signal-regulated kinases 1 and 2 (ERK1/2), that lower the threshold for carcinogenesis. The best-studied H. pylori virulence factors are the cag pathogenicity island (PAI) and VacA, the vacuolating cytotoxin. The cag PAI encodes components of a type IV secretion system (T4SS) that can inject bacterial effectors into eukaryotic cells (24, 26). The gene encoding VacA (vacA) is found in all strains of H. pylori, but not all strains secrete VacA, due to vacA allelic variation. However, most persons infected with strains that possess these virulence constituents do not develop disease, indicating that other microbial, host, and/or environmental factors may contribute to pathogenesis (for reviews, see references 5, 8, and 28). Dubois and Boren have recently reported that H. pylori not only resides within the gastric mucosa but is also closely associated with red blood cells (10), raising the possibility that this bacterium may exert effects on components of the hematopoietic system.
Synergistic hemolysis was initially described as the extent of hemolysis observed when Staphylococcus aureus and Streptococcus agalactiae were grown in close proximity on blood agar plates. Regions containing diffusing sphingomyelinase (SMase) from S. aureus and the pore-forming toxin of S. agalactiae were found to exhibit a higher degree of hemolysis than zones containing only one of these virulence factors (7), a conjoined phenotype commonly known as the CAMP (Christie-Atkins-Munch-Perkins) test. Synergistic hemolysis has subsequently been observed with diffusible factors from organisms as phylogenetically distant as dermatophytes (fungi) and Listeria ivanovii (31). Two recent reports have demonstrated that H. pylori is a component of a complex gastric microbial ecosystem, which likely promotes dynamic interspecies interactions (6, 16). Here, we examined the ability of H. pylori to induce synergistic hemolysis and demonstrate that a microbial constituent which possesses the ability to induce hemolysis can also activate MAPK signaling in gastric epithelial cells.
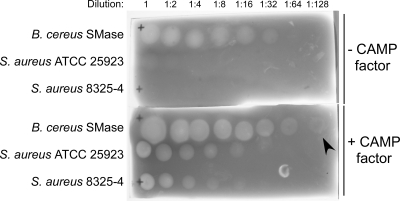
S. agalactiae and S. aureus were initially tested for the CAMP reaction on TSAII blood agar plates and reproducibly exhibited synergistic hemolysis (data not shown) as previously reported (4). Since SMases (17) as well as pore-forming toxins (20) have been described within H. pylori, we developed a protocol to test for H. pylori-induced synergistic hemolysis. Media containing tryptic soy agar with 5% sheep blood (TSBA), TSBA with CAMP factor (from S. agalactiae supernatant), or TSBA with SMase (from S. aureus filtered supernatant) were prepared, and an assay was performed using S. aureus supernatant and Bacillus cereus sphingomyelinase C (Sigma) as positive controls for synergistic hemolysis (Fig. 1).
Fig 1.
Synergistic hemolysis on blood agar medium. The sensitivity of synergistic hemolysis was estimated using medium containing CAMP factor (16 μg/ml) from S. agalactiae filter-sterilized culture supernatant. B. cereus SMase (top row) or S. aureus culture supernatants from strain ATCC 25923 (middle row) or 8325-4 (bottom row) were applied at 2-fold dilutions as indicated. The area of hemolysis on the top row in the lower slide (arrowhead) indicates that the limit of detection for this assay is 0.00165 units of sphingomyelinase activity.
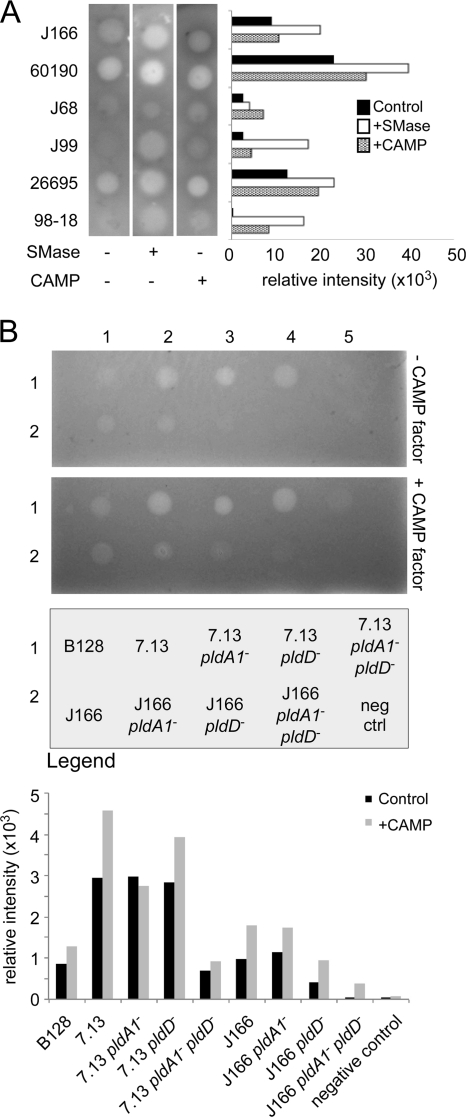
To investigate the ability of H. pylori to induce synergistic hemolysis, strains (n = 6, ∼107 CFU) were spotted on medium and incubated overnight. Hemolysis developed directly under areas of bacterial growth (Fig. 2A), consistent with previous observations that hemolytic factors are likely not secreted by H. pylori (3). H. pylori strains exhibited different levels of hemolysis on either control, CAMP-containing, or SMase-containing plates (Table 1; Fig. 2A). For example, strains J99 and 98-18 did not exhibit hemolysis when grown on TSBA plates alone; however, synergistic hemolysis developed on both SMase- and CAMP-containing plates, highlighting the utility of this approach for examining strain-specific hemolytic differences (Fig. 2A). The degree of hemolysis was significantly greater (P = 0.0067) when H. pylori strains were grown on CAMP-containing plates than on control plates. We also examined potential roles exerted by the cag PAI by use of cag PAI deletion mutations in strains 7.13 and 26695; however, no difference was found between wild-type strains and their corresponding mutants (data not shown).
Fig 2.
Variability of hemolysis among strains of H. pylori. (A) Hemolytic ability of H. pylori strains was determined by inoculating 3 to 4 μl (107 CFU) onto TSB agar containing 140 mM NaCl and crude preparations of either SMase or CAMP factor. Hemolysis under each condition was quantified as relative intensity using NIH ImageJ and is shown in the bar graph on the right. Data shown are from an experiment representative of several replicates. (B) Indicator plates estimating SMase activity in wild-type strains J166 and 7.13 and mutants in phospholipase genes in J166 and 7.13 backgrounds as well as the parental clinical strain B128 are shown. The lower plate contains CAMP factor. Row 1, H. pylori B128, 7.13, 7.13 pldA1 mutant, 7.13 pldD mutant, and 7.13 pldA1 pldD mutant; row 2, H. pylori J166, J166 pldA1 mutant, J166 pldD mutant, J166 pldA1 pldD mutant, and broth (negative control). Hemolysis under each condition was quantified as relative intensity using NIH ImageJ and is shown in the bar graph.
Table 1.
List of bacterial strains with a description of clinical outcome, cag status, and hemolytic ability
| Strain | Genotype/relevant characteristics and hemolytic ability (if applicable) | Reference(s) or source |
|---|---|---|
| H. pylori | ||
| J166 | Duodenal ulcer, cagA+ strain of H. pylori; hemolytic, synergistic hemolysis | 25 |
| 60190 | Lab derivative of ATCC 49503, cagA+; hemolytic, enhanced synergistic hemolysis on SMase plate relative to CAMP plate | |
| J68 | Duodenal ulcer, cagA mutant; minimally hemolytic, enhanced synergistic hemolysis on CAMP plate compared to SMase plate | |
| J99 | Duodenal ulcer, cagA+; sequenced strain; minimally hemolytic; synergistic hemolysis on both plates, maximum enhancement on SMase plate | 2 |
| 26695 | Gastritis, cagA+; sequenced strain; hemolytic, synergistic hemolysis | 1, 33 |
| 26695 Δcag | 26695 with the cag PAI deleted; hemolytic, synergistic hemolysis | 11 |
| 98-18 | Gastric ulcer, cagA+; minimally hemolytic, enhanced synergistic hemolysis on SMase plate compared to CAMP plate | 27 |
| J166 pldA1 | Mutant in pldA1; kanamycin resistant; hemolytic, synergistic hemolysis on CAMP plate | This work |
| J166 pldD | Mutant in pldD; chloramphenicol resistant; hemolytic, synergistic hemolysis on CAMP plate | This work |
| J166 pldA1 pldD | Double mutant in pldA1 and pldD; kanamycin and chloramphenicol resistant; nonhemolytic, no synergistic hemolysis on CAMP plate | This work |
| B128 | Gastric ulcer; cagA+; minimally hemolytic, synergistic hemolysis | 13 |
| 7.13 | Carcinogenic strain of H. pylori recovered after infection of a gerbil with strain B128; hemolytic, synergistic hemolysis on CAMP plate | 12 |
| 7.13 Δcag | Mutant of 7.13 lacking the cag PAI; hemolytic, enhanced synergistic hemolysis on CAMP plate relative to SMase plate | Lab strain |
| 7.13 pldA1 | Mutant in pldA1; kanamycin resistant; hemolytic, synergistic hemolysis on CAMP plate | This work |
| 7.13 pldD | Mutant in pldD; chloramphenicol resistant; hemolytic, synergistic hemolysis on CAMP plate | This work |
| 7.13 pldA1 pldD | Double mutant in pldA1 and pldD; kanamycin and chloramphenicol resistant; nonhemolytic, no synergistic hemolysis on CAMP plate | This work |
| E. coli | ||
| DH5α | E. coli K-12; F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsd R17(rK− mK−) phoA supE44 thi-1 gyrA96 relA1 mutant; strain used for cloning | Lab strain |
| BL21 | E. coli B; F−dcm ompT hsdS(rB− mB−) gal; Dam+ Dcm− | Stratagene |
| Other bacteria | ||
| Streptococcus agalactiae ATCC 12386 | Secretes CAMP factor into culture medium | American Type Culture Collection |
| Staphylococcus aureus subsp. aureus ATCC 25923 | Secretes sphingomyelinase D into culture medium, observed to cause hemolysis to a greater extent than strain 8325-4 (below) | American Type Culture Collection |
| Staphylococcus aureus 8325-4 | Secretes sphingomyelinase D into culture medium | 21 |
HP0499 encodes a membrane-bound phospholipase A (PldA1) in H. pylori (9). HPAG1_0184 was identified as a “phospholipase D-family protein” (http://cmr.jcvi.org/tigr-scripts/CMR/shared/GenePage.cgi?locus=NTL03HP0185), and HP0190 of strain 26695 is predicted to be 94% identical to HPAG1_0184 at the protein level. An analysis of HP0190 with NCBI's Conserved Domain Database (19) revealed phospholipase D (PldD) active-site motifs. To define potential roles of these genes in synergistic hemolysis, we inactivated HP0499 and HP0190 individually and in tandem (HP0499 HP0190 mutant) in strains 7.13 and J166 (see Tables S1 and S2 in the supplemental material) (25). Wild-type strains and isogenic mutants were then tested for hemolytic ability on TSBA plates with or without CAMP factor (Fig. 2B). Single phospholipase mutants of strain J166 or 7.13 were similar to wild type (Fig. 2B). Double mutants of both strains were nonhemolytic, indicating that both phospholipases mediate synergistic hemolysis. We also examined strain B128, the parental strain of 7.13, and found that this induced less hemolysis than did strain 7.13.
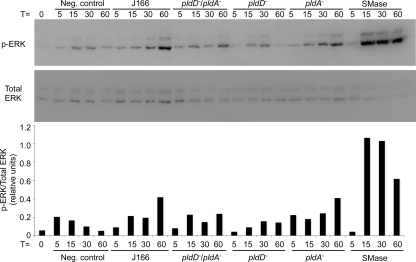
H. pylori can activate MAPK pathways in gastric epithelial cells in a strain-specific manner, and activation of ERK1/2 may play a role in pathogenesis (15). H. pylori encodes SMases that catalyze the conversion of host cell sphingomyelin to ceramide, which can activate MAPK (17, 34). We therefore determined if phospholipases HP0499 and HP0190 were required for activation of ERK1/2 signaling in gastric epithelial cells. AGS gastric epithelial cells were grown to confluence, then cultured in medium for 24 h, and then cocultured with H. pylori for specified times at a multiplicity of infection (MOI) of 100. H. pylori-infected and uninfected AGS cells were lysed in RIPA buffer (50 mM Tris, pH 7.2; 150 mM NaCl; 1% Triton X-100; 0.1% SDS), and protein concentrations were quantified by the bicinchoninic acid (BCA) assay (Pierce). Proteins (30 μg) were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Pall Corporation, Ann Arbor, MI). Levels of phosphorylated or total ERK1/2 were detected using an anti-phospho-ERK1/2 antibody (Promega) (1:5,000) or an anti-total-ERK1/2 antibody (Cell Signaling) (1:5,000). Primary antibodies were detected using horseradish peroxidase-conjugated secondary antibodies (Santa Cruz) and visualized by Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer) according to the manufacturer's instructions. Western blots were imaged and band intensities were quantified using the ChemiGenius Gel Bio Imaging System (Syngene) (22). Our previous studies determined that a bacterium/cell ratio of 100:1 and a time course ranging from 5 to 60 min were optimal to evaluate the ability of H. pylori wild-type and mutant strains to activate MAPK using AGS gastric epithelial cells (15); therefore, H. pylori or B. cereus SMase C (1 unit/ml) was added to gastric cells and ERK activation was detected 5 to 60 min later (Fig. 3). B. cereus SMase induced robust activation of ERK. Coculture of strain J166 with AGS cells led to gradual ERK activation over 60 min. Inactivation of pldA1 did not affect this activation; however, loss of pldD led to reduced ERK activation, peaking at a much lower level by 30 min (Fig. 3). The double pldD pldA mutant also activated ERK1/2 less potently than did the wild-type strain.
Fig 3.
Western blot assays to evaluate ERK1/2 activation in AGS cells. AGS cells were incubated with B. cereus SMase (1 unit/ml), wild-type H. pylori J166, or its isogenic single or double phospholipase mutants from 0 to 60 min. The blot was probed for phosphorylated ERK1/2 (top panel), stripped, and reprobed for total ERK1/2 (bottom panel). Densitometric analysis of phospho-ERK1/2 is shown below the Western blot.
In this study, we developed assays that incorporated cell-free culture supernatants containing CAMP factor or SMase into growth medium to permit testing for synergistic hemolysis in slow-growing fastidious bacteria. This allowed identification of cell-associated hemolytic factors, whereas previous studies detected only secreted hemolytic factors. We also demonstrate that both H. pylori phospholipases analyzed mediate synergistic hemolysis in combination with the S. agalactiae CAMP factor. PldA1 is highly conserved among H. pylori strains, but variation in pldA1 sequences exists among strains from different geographic locales (35). Our current experiments also indicate that H. pylori PldD can activate ERK1/2 in AGS cells and thus represent a novel function of H. pylori phospholipases.
Phospholipases are ubiquitous throughout the living world and have been implicated as virulence factors in several bacterial species such as Staphylococcus aureus (23), Pseudomonas aeruginosa (30), Yersinia pseudotuberculosis (14), Listeria monocytogenes (32), and Mycobacterium tuberculosis (29). The current work highlights an important issue regarding the effect of colonization by multiple bacterial species (as commensals, symbionts, or pathogens) on the host. The ability of H. pylori phospholipases to participate in synergistic hemolysis with a pore-forming toxin from S. agalactiae indicates that membrane-active factors of phylogenetically distant organisms can exhibit cooperative effects. Within the gastric niche, H. pylori is a resident of a complex microflora, containing >120 distinct phylotypes (6, 16). A recent report consistent with our findings (18) demonstrated that the severity of skin lesions in mice was exacerbated by the cooperative effect of the SMase of S. aureus and a CAMP factor from Propionibacterium acnes, an abundant commensal found on human skin. Neutralization of the CAMP factor and immunization against the SMase reduced the severity of lesions during coinfection of mice. Thus, complementary functions of membrane-damaging agents produced by unrelated microbial species may be a common theme in microbial biology.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants CA116087, CA77955, and DK58587 (to R.M.P.) and DK058404 (to the Vanderbilt Digestive Disease Research Center).
We thank Uma Krishna for technical assistance and Daniel O'Brien and Seth Ogden for intellectual contributions. Douglas Kernodle's gift of S. aureus strain 8325-4 is gratefully acknowledged.
Footnotes
Published ahead of print 28 December 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Akopyants NS, Eaton KA, Berg DE. 1995. Adaptive mutation and cocolonization during Helicobacter pylori infection of gnotobiotic piglets. Infect. Immun. 63:116–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alm RA, et al. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176–180 [DOI] [PubMed] [Google Scholar]
- 3. Ansorg R, Rein R, Spies A, von Recklinghausen G. 1993. Cell-associated haemolytic activity of Helicobacter pylori. Eur. J. Clin. Microbiol. Infect. Dis. 12:98–104 [DOI] [PubMed] [Google Scholar]
- 4. Beecher DJ, Wong AC. 2000. Cooperative, synergistic and antagonistic haemolytic interactions between haemolysin BL, phosphatidylcholine phospholipase C and sphingomyelinase from Bacillus cereus. Microbiology 146:3033–3039 [DOI] [PubMed] [Google Scholar]
- 5. Bergman M, Del Prete G, van Kooyk Y, Appelmelk B. 2006. Helicobacter pylori phase variation, immune modulation and gastric autoimmunity. Nat. Rev. Microbiol. 4:151–159 [DOI] [PubMed] [Google Scholar]
- 6. Bik EM, et al. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. U. S. A. 103:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Christie R, Atkins NE, Munch-Petersen E. 1944. A note on a lytic phenomenon shown by group B streptococci. Aust. J. Exp. Biol. Med. Sci. 22:197–200 [DOI] [PubMed] [Google Scholar]
- 8. Correa P, Houghton J. 2007. Carcinogenesis of Helicobacter pylori. Gastroenterology 133:659–672 [DOI] [PubMed] [Google Scholar]
- 9. Dorrell N, et al. 1999. Characterization of Helicobacter pylori PldA, a phospholipase with a role in colonization of the gastric mucosa. Gastroenterology 117:1098–1104 [DOI] [PubMed] [Google Scholar]
- 10. Dubois A, Boren T. 2007. Helicobacter pylori is invasive and it may be a facultative intracellular organism. Cell. Microbiol. 9:1108–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eaton KA, et al. 2001. Role of Helicobacter pylori cag region genes in colonization and gastritis in two animal models. Infect. Immun. 69:2902–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franco AT, et al. 2005. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 102:10646–10651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Israel DA, et al. 2001. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J. Clin. Invest. 107:611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karlyshev AV, et al. 2001. Application of high-density array-based signature-tagged mutagenesis to discover novel Yersinia virulence-associated genes. Infect. Immun. 69:7810–7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keates S, et al. 1999. Differential activation of mitogen-activated protein kinases in AGS gastric epithelial cells by cag+ and cag− Helicobacter pylori. J. Immunol. 163:5552–5559 [PubMed] [Google Scholar]
- 16. Li XX, et al. 2009. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One 4:e7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin YL, Liu JS, Chen KT, Chen CT, Chan EC. 1998. Identification of neutral and acidic sphingomyelinases in Helicobacter pylori. FEBS Lett. 423:249–253 [DOI] [PubMed] [Google Scholar]
- 18. Lo CW, Lai YK, Liu YT, Gallo RL, Huang CM. 2011. Staphylococcus aureus hijacks a skin commensal to intensify its virulence: immunization targeting beta-hemolysin and CAMP factor. J. Investig. Dermatol. 131:401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marchler-Bauer A, et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martino MC, et al. 2001. Helicobacter pylori pore-forming cytolysin orthologue TlyA possesses in vitro hemolytic activity and has a role in colonization of the gastric mucosa. Infect. Immun. 69:1697–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Novick R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155–166 [DOI] [PubMed] [Google Scholar]
- 22. O'Brien DP, et al. 2008. Regulation of the Helicobacter pylori cellular receptor decay-accelerating factor. J. Biol. Chem. 283:23922–23930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park PW, et al. 2004. Activation of syndecan-1 ectodomain shedding by Staphylococcus aureus alpha-toxin and beta-toxin. J. Biol. Chem. 279:251–258 [DOI] [PubMed] [Google Scholar]
- 24. Peek RM, Jr, Blaser MJ. 2002. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2:28–37 [DOI] [PubMed] [Google Scholar]
- 25. Peek RM, Jr, et al. 1999. Helicobacter pylori strain-specific genotypes and modulation of the gastric epithelial cell cycle. Cancer Res. 59:6124–6131 [PubMed] [Google Scholar]
- 26. Polk DB, Peek RM., Jr 2010. Helicobacter pylori: gastric cancer and beyond. Nat. Rev. Cancer 10:403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pride DT, Meinersmann RJ, Blaser MJ. 2001. Allelic variation within Helicobacter pylori babA and babB. Infect. Immun. 69:1160–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prinz C, Hafsi N, Voland P. 2003. Helicobacter pylori virulence factors and the host immune response: implications for therapeutic vaccination. Trends Microbiol. 11:134–138 [DOI] [PubMed] [Google Scholar]
- 29. Raynaud C, et al. 2002. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 45:203–217 [DOI] [PubMed] [Google Scholar]
- 30. Sato H, Frank DW. 2004. ExoU is a potent intracellular phospholipase. Mol. Microbiol. 53:1279–1290 [DOI] [PubMed] [Google Scholar]
- 31. Schaufuss P, Brasch J, Steller U. 2005. Dermatophytes can trigger cooperative (CAMP-like) haemolytic reactions. Br. J. Dermatol. 153:584–590 [DOI] [PubMed] [Google Scholar]
- 32. Schwarzer N, et al. 1998. Two distinct phospholipases C of Listeria monocytogenes induce ceramide generation, nuclear factor-kappa B activation, and E-selectin expression in human endothelial cells. J. Immunol. 161:3010–3018 [PubMed] [Google Scholar]
- 33. Tomb JF, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547 [DOI] [PubMed] [Google Scholar]
- 34. Tseng HJ, Chan CC, Chan EC. 2004. Sphingomyelinase of Helicobacter pylori-induced cytotoxicity in AGS gastric epithelial cells via activation of JNK kinase. Biochem. Biophys. Res. Commun. 314:513–518 [DOI] [PubMed] [Google Scholar]
- 35. Xerry J, Owen RJ. 2001. Conservation and microdiversity of the phospholipase A (pldA) gene of Helicobacter pylori infecting dyspeptics from different countries. FEMS Immunol. Med. Microbiol. 32:17–25 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.