Abstract
Staphylococcus aureus is a common causative agent of bovine mastitis in dairy herds. The emergence of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals as well as the community is a significant and costly public health concern. S. aureus-related bovine mastitis is a common reason for therapeutic and/or prophylactic use of antibiotics on dairy farms. In this study, herd prevalence of S. aureus, including MRSA, was estimated from bulk tank milk (BTM) from Minnesota farms. A total of 150 pooled BTM samples from 50 farms, collected over 3 seasons (spring, summer, and fall of 2009), were assessed. Herd prevalence of methicillin-susceptible S. aureus (MSSA) was 84%, while MRSA herd prevalence was 4%. A total of 93 MSSA isolates and 2 MRSA isolates were recovered from 150 BTM samples. Antibiotic susceptibility testing of S. aureus isolates showed pansusceptibility in 54 isolates, resistance to a single antibiotic class in 21 isolates, resistance to two antibiotic classes in 13 isolates, and resistance to ≥3 antibiotics classes and thus multidrug resistance in 5 isolates. The two MRSA isolates displayed resistance to β-lactams, cephalosporins, and lincosamides and were multiresistant. Staphylococcal protein A gene (spa) typing identified spa types t529 and t034 most frequently among methicillin-susceptible isolates, while t121 was observed in MRSA isolates. Seven isolates, including the two MRSA isolates, produced staphylococcal enterotoxins B, C, D, and E on overnight culture. MRSA isolates were further genotyped using multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE). Of the 2 MRSA isolates, one had a composite genotype profile of MLST ST 5-PFGE USA100-unknown spa type, which has been reported among hospital-associated MRSA isolates, while the second isolate carried the MLST ST 8-PFGE USA300-spa type t121 genotype, commonly identified among community-associated MRSA isolates. These results suggest that MRSA genotypes associated with hospitals and community can be isolated from milk at very low rates.
INTRODUCTION
The emergence of antibiotic-resistant microorganisms in farm animal environments poses a potential public health concern. Staphylococcus aureus is an important opportunistic pathogen both in humans and in dairy cattle. It is also a common cause of mastitis in dairy cows (59, 64), a primary reason for antibiotic use on farms. However, the proportion of contagious pathogens (such as S. aureus and Streptococcus agalactiae) has decreased relative to that of environmental pathogens in the recent past (from 1994 to 2001) (40). The use of antimicrobial agents on dairy farms as well as in other food animal production systems is a major concern in the emergence of resistant zoonotic bacterial pathogens (50). Although different antibiotic classes of drugs are used in animal health management and in human medicine, the selection of resistance to one drug class may lead to cross-resistance to another (50). Antibiotics on dairy operations are used to treat highly prevalent infections, such as subclinical mastitis, and as a preventive measure during dry cow therapy. Monitoring the emergence of resistant pathogens in animal reservoirs is important particularly for those with zoonotic potential.
Methicillin-resistant Staphylococcus aureus (MRSA) has emerged as a major cause of health care-associated (HA) and community-associated (CA) infections (33). In addition, livestock-associated (LA) MRSA genotypically classified under clonal complex 398 (CC398) has been detected among pigs and swine farmers in the Netherlands and other countries (15, 16, 31) and is known to cause infections in humans and animals (14, 37, 65). MRSA isolates are frequently multidrug resistant (MDR), which can result in higher costs, longer treatment times, and higher rates of hospitalization and comorbidities (10, 47). MRSA was first reported from bovine sources in 1975 (17), although it has occasionally been reported since then (21, 25, 30, 61). The presence of MRSA in bovine milk and dairy environments poses potential risk to farm workers, veterinarians (30, 36), and farm animals that are exposed to contaminated cattle.
Limited data about the prevalence and genetic spread of MRSA in dairy environments in the United States are available. Previous studies have reported zero to low occurrence of MRSA among S. aureus isolates from bovine milk—0.6% of 846 S. aureus isolates in Michigan (20), 1.8% of 2,132 S. aureus isolates in Wisconsin (39), and 0% of 357 S. aureus isolates in North Carolina and Virginia (2). In the recent past, very low prevalence rates of MRSA in either individual cow milk or bulk tank milk (BTM) have been reported internationally. In Hungary, although a prevalence of 70% was reported for S. aureus on farms (14 of 20 farms), no MRSA isolates were detected from BTM (49). Similarly, in Korea, a prevalence of 5.6% was reported for S. aureus from milk samples from individual cow udders (835 of 14,688 samples). However, only 2.8% of these isolates were identified as MRSA (42).
In comparison, for S. aureus or methicillin-susceptible Staphylococcus aureus (MSSA), a prevalence rate of 38%, i.e., 38 S. aureus isolates from 101 raw milk samples (collected from bulk tank, milk cans, and cheese vats), has been reported in Vermont; farm prevalence of S. aureus as reported in the same study was 67% (14 of 21 farms) (11, 29, 48). Similarly, 39 S. aureus isolates were reported from 126 bulk tanks in Pennsylvania (31% prevalence) (11, 29, 48), while 256 S. aureus isolates were reported from 732 individual cow milk samples in Louisiana (35% prevalence) (11, 29, 48). It is important to monitor the occurrence and distribution of MSSA and MRSA isolates in dairy herds to detect the emergence of new clonal types, similar to the emergence of LA MRSA CC398 (31).
S. aureus is also the third most reported cause of food-borne diseases in the world (46, 67). Growth of S. aureus in foods leads to the production of staphylococcal enterotoxins (SEs), a cause of food poisoning (1). Contaminated milk and milk products have frequently been implicated in staphylococcal food poisoning (13). SEs are heat resistant and hence may be present even when S. aureus is not viable (49). Presence of staphylococcal enterotoxin genes and production of SEs by S. aureus of bovine origin have been reported in numerous studies. Eleven of 94 S. aureus isolates (11.7%) in BTM from Argentina showed production of enterotoxins (45). One hundred nine of 291 S. aureus isolates (37.5%) from milk from mastitic cows, BTM, and cheese from Brazil showed production of one or more enterotoxins (3). Nineteen of 102 S. aureus isolates (18.6%) recovered from BTM and milk filters from national milk-producing herds in Ireland showed the presence of one or more enterotoxin genes (44). Conversely, a higher percentage of isolates, 46.9% of 130, associated with subclinical bovine mastitis in Turkey showed the presence of one or more enterotoxin genes (23). The aim of the present study was to characterize and document the prevalence and distribution of MSSA and MRSA in bulk tank milk samples. We also investigated enterotoxin production by isolates. Both MSSA and MRSA isolates were phenotypically and genotypically characterized.
MATERIALS AND METHODS
Farm sampling.
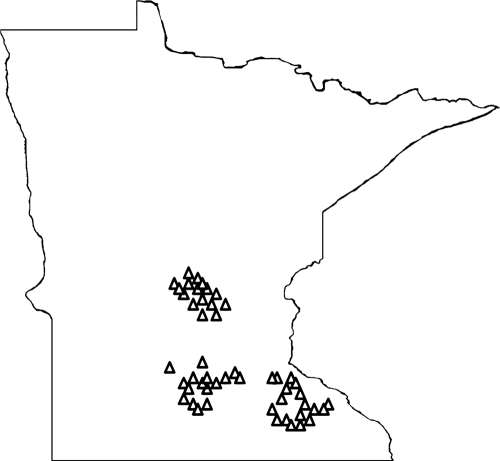
Fifty dairy herds in Minnesota (MN) were sampled for this study (Fig. 1). Of approximately 4,700 dairy herds in MN, about 2,500 are enrolled with the MN Dairy Herd Improvement Association (DHIA). Herds for the study were selected from among the DHIA herds according to their location in the three major dairy regions in MN, the southeast region (n = 18), the south central region (n = 16), and the north central region (n = 16). Herds were also categorized as small (40 to 99 cattle) (n = 15), medium (100 to 299) (n = 19), or large (≥300) (n = 16), according to the number of milking cows. Criteria for inclusion in the study included location of farms in the three main dairy regions and less than 2 h away from the Twin Cities. Participation was voluntary for the farms which met the criteria. Fifty herds were randomly selected from this pool to equally represent the three dairy regions as well as the three herd size categories.
Fig 1.
Spatial distribution of participating dairy herds in Minnesota (n = 50). The 50 study farms are represented on the physical map of Minnesota. These farms were selected to represent the three major dairy regions in Minnesota, south central, southeast, and north central, as well as to have equal distributions of small (40 to 99 cattle), medium (100 to 299 cattle), and large (>300 cattle) farms.
Bulk tank milk (BTM) samples were collected, in duplicate, on three consecutive days in spring, summer, and fall of 2009. Samples for spring were collected in the months of April and May, while samples for summer were collected in July and August and samples for fall were collected in October and November. An additional questionnaire documenting farm management practices was administered by university personnel. Bulk tank milk samples were frozen on the farm (−20°C) and then shipped on ice to the University of Minnesota, Laboratory of Udder Health. Samples collected from three consecutive days were pooled for a single herd from a single season for a total of 150 pooled BTM samples (50 each from spring, summer, and fall) for S. aureus assessment.
Isolation of MSSA.
S. aureus was cultured from 1 ml of pooled milk by direct plating onto factor medium (University of Minnesota, St. Paul) using a spreader and incubated at 37°C for 24 to 48 h. S. aureus colonies were identified on the basis of colony morphology and confirmed using a Sure-Vue Color Staph ID confirmatory test (Fisher Healthcare, TX). Isolates were transferred to Luria-Bertani (LB) broth (BD, MD), and DNA was extracted using a QIAamp DNA minikit (Qiagen, MD) per manufacturer's instructions. All isolates were also tested for growth on MRSA Select plates (Bio-Rad Laboratories, WA) to determine the methicillin susceptibility status of isolates.
Isolation of MRSA.
MRSA was isolated from a duplicate set of 150 pooled BTM samples. Isolation involved a two-step enrichment procedure, with the inoculation of pooled milk into Mueller-Hinton broth (Teknova, Inc., CA) supplemented with 6.5% NaCl (24 h at 37°C) followed by phenol red mannitol broth supplemented with 4 mg/liter oxacillin (24 h at 37°C). The samples were then streaked on MRSA Select medium, a selective medium for detection of MRSA, for 24 h at 37°C. If growth was detected, colonies were transferred to CNA plates (BBL; BD, MD) for 24 h at 37°C to check for beta-hemolysis. Isolates were further confirmed as S. aureus by a coagulase test using coagulase plasma with EDTA (BBL; BD, MD). Thus, selectively enriched isolates that grew on MRSA Select plates and showed classic positive beta-hemolysis in CNA medium as well as coagulase positivity were considered suspect MRSA. These isolates were then transferred to LB broth, and DNA was extracted using a QIAamp DNA minikit as mentioned above.
Genotypic and phenotypic characterization of MSSA.
All MSSA isolates were confirmed as S. aureus by species-specific 16S rRNA gene PCR using the primers 16s1 (5′ CAG CTC GTG TCG TGA GAT GT 3′) and 16s2 (5′ AAT CAT TTG TCC CAC CTT CG 3′) (7). 16S rRNA gene amplicons which represented a fragment of the 16S rRNA gene were sequenced using standard Sanger sequencing at the Biomedical Genomics Center (BMGC), University of Minnesota, using a primer concentration of 3.2 pmol and approximately 20 ng of amplified DNA as a template (http://www.bmgc.umn.edu/facilities/sequencing/services/sequencing/home.html). Sequences obtained were compared to a reference S. aureus 16S rRNA gene sequence (NCBI accession no. AM980864) using the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov), and confirmation to the species level was carried out if sequences showed 98 to 100% similarity to the reference S. aureus gene.
Staphylococcal protein A gene, or spa, typing was carried out for all S. aureus isolates as described previously (55). spa types were obtained using the sequence comparisons against egenomics (http://www.egenomics.com) and ridom (http://www.spaserver.ridom.de) databases.
MSSA isolates were screened for their antimicrobial agent susceptibility patterns using the Kirby Bauer disk diffusion method with commercially available antibiotic disks (BBL, Sparks, MD). Table 1 lists antibiotic disks used and their concentrations. Interpretations of results as susceptible or resistant were done according to the Clinical and Laboratory Standards Institute M31-A3 guidelines (9).
Table 1.
Antibiotics used in the Kirby Bauer disk diffusion method and concentrations of antibiotics used
| Antimicrobial agent | Concn (μg) of antibiotic in disk |
|---|---|
| Amoxicillin-ampicillin | 10 |
| Ceftiofur | 30 |
| Cephalothin | 30 |
| Cloxacillin | 1 |
| Erythromycin | 15 |
| Novobiocin | 30 |
| Penicillin | 10 |
| Penicillin-novobiocin | 10/30a |
| Pirlimycin | 2 |
| Streptomycin | 300 |
| Tetracycline | 30 |
Penicillin, 10 μg; novobiocin, 30 μg.
Genotypic and phenotypic characterization of MRSA.
All suspect MRSA isolates were confirmed as S. aureus by 16S rRNA gene sequencing as mentioned above for MSSA. Detection of the mecA gene, encoding the modified penicillin binding protein PBP 2a, was performed on confirmed S. aureus isolates. A mecA conventional PCR was performed using a primer set targeting an upstream region of mecA, i.e., mecA1 (5′ GTA GAA ATG ACT GAA CGT CCG ATA A 3′) and mecA2 (5′ CCA ATT CCA CAT TG TTC GGT CTA A 3′). Amplicons were sequenced and compared for sequence similarity to a reference gene (NCBI accession no. EU790490) using BLAST. The detection was repeated for confirmation at the Minnesota Department of Health (MDH) using real-time PCR with a second set of established primers as specified by Killgore et al. (32). The mecA detection using the primers from Killgore et al. was also repeated in our laboratory using conventional PCR for all suspect MRSA isolates. The use of two different primer sets (the Killgore et al. primer set and the primer set designed by our lab), which target the upstream and downstream ends of the mecA gene, identified possible differences in mediation of methicillin resistance by this gene. Further, staphylococcal cassette chromosome mec element (SCCmec) typing for the suspected MRSA isolates was carried out as outlined by Zhang et al. (66).
Genotypic characterization involved spa typing for all suspect MRSA isolates as mentioned above. All suspect MRSA isolates were further characterized by multilocus sequence typing (MLST) as described by Enright et al. (19). Sequence types were assigned using an established MLST database (http://www.mlst.net). These isolates were also characterized using pulsed-field gel electrophoresis (PFGE) using standard CDC protocols at the MDH (41).
All suspect MRSA isolates were subjected to antibiotic sensitivity testing using the broth microdilution method (compared to the Kirby Bauer method used for MSSA strains) to obtain precise concentrations of antibiotics at which the methicillin-resistant strains would be growth inhibited. Two different Sensititre antimicrobial agent panels were used: one at the University of Minnesota, Veterinary Diagnostic Laboratory (VDL), which compared against the veterinary CLSI reference M31-A3 (9), and the other at the Minnesota Department of Health (MDH), which compared against the human CLSI reference M100-S21 (Tables 2 and 3 list the antimicrobial agents tested by these respective methods). These two different panels were used to test sensitivities to antibiotics in human as well as animal medicine, as MRSA in animals may pose an animal as well as a public health concern. Both antimicrobial agent panels tested for oxacillin resistance as an indicator of methicillin resistance.
Table 2.
Broth microdilution panel used at the University of Minnesota, Veterinary Diagnostic Laboratory (VDL), showing MICs of the two MRSA isolates
| Antimicrobial agent | MIC in μg/ml (profile)a for: |
|
|---|---|---|
| Sp12 | Sp19 | |
| Amikacin | 16.00 (S) | ≤4.00 (S) |
| Amoxicillin-clavulanic acid | 32.00 (R) | ≤4.00 (R) |
| Ampicillin | >16.00 (R) | >16.00 (R) |
| Cefazolin | >16.00 (R) | ≤4.00 (R) |
| Cefovecin | >8.00 (NI) | >8.00 (NI) |
| Cefoxitin | >16.00 (R) | 16.00 (R) |
| Cefpodoxime | >16.00 (R) | >16.00 (R) |
| Ceftiofur | >4.00 (R) | >4.00 (R) |
| Chloramphenicol | 8.00 (S) | 8.00 (S) |
| Clindamycin | ≤0.50 (R) | ≤0.50 (R) |
| Doxycycline | ≤2.00 (S) | ≤2.00 (S) |
| Enrofloxacin | >2.00 (R) | ≤0.25 (S) |
| Erythromycin | >4.00 (R) | >4.00 (R) |
| Gentamicin | ≤1.00 (S) | ≤1.00 (S) |
| Imipenem | 4.00 (R) | ≤1.00 (R) |
| Marbofloxacin | >2.00 (R) | ≤0.25 (S) |
| Oxacillin + 2% NaCl | >4.00 (R) | >4.00 (R) |
| Penicillin | >8.00 (R) | >8.00 (R) |
| Rifampin | ≤1.00 (NI) | ≤1.00 (NI) |
| Ticarcillin | 64.00 (R) | 32.00 (R) |
| Ticarcillin-clavulanic cid | 32.00 (R) | ≤8.00 (R) |
| Trimethoprim-sulfamethoxazole | ≤0.50 (S) | ≤0.50 (S) |
S, susceptible; R, resistant; NI, not interpretable (no interpretations from CLSI).
Table 3.
Broth microdilution panel used at the Minnesota Department of Health (MDH), showing MICs of the two MRSA isolates
| Antimicrobial agent | MIC in μg/ml (profile)a for: |
|
|---|---|---|
| Sp12 | Sp19 | |
| Clindamycin | ≤0.25 (S) | ≤0.25 (S) |
| Trimethoprim-sulfamethoxazole | ≤1.00 (S) | ≤1.00 (S) |
| Daptomycin | ≤0.50 (S) | ≤0.50 (S) |
| Quinupristin-dalfopristin | ≤0.25 (S) | ≤0.25 (S) |
| Mupirocin | <1.00 (U) | <1.00 (U) |
| Cefoxitinb | >16.00 (U) | >16.00 (U) |
| Tetracycline | ≤1.00 (S) | ≤1.00 (S) |
| Linezolid | 2.00 (S) | 2.00 (S) |
| Minocycline | ≤0.50 (S) | ≤0.50 (S) |
| Oxacillin | >32.00 (R) | 32.00 (R) |
| Gentamicin | ≤2.00 (S) | ≤2.00 (S) |
| Vancomycin | 1.00 (S) | 1.00 (S) |
| Levofloxacin | >8.00 (R) | >8.00 (R) |
| Doxycycline | ≤1.00 (S) | ≤1.00 (S) |
| Erythromycin | >8.00 (R) | ≤0.12 (S) |
| Rifampin | ≤0.25 (S) | ≤0.25 (S) |
Interpretations of susceptible, intermediate, and resistant profiles were taken from CLSI M100-S21. Sp12 was found positive for inducible clindamycin resistance, while Sp19 was found negative. Inducible clindamycin resistance testing was performed only on erythromycin-resistant and clindamycin-sensitive or -intermediate isolates.
Results for cefoxitin are noted as undefined (U). A cefoxitin MIC of ≤4 correlates with oxacillin susceptibility, and a cefoxitin MIC of ≥8 or correlates with oxacillin resistance.
PCR amplification of genes encoding PVL toxin and TSST in suspect MRSA isolates.
The genomic DNA was extracted from the suspect MRSA isolates using a QIAamp DNA minikit as mentioned above and was used to screen for the toxic shock syndrome toxin (TSST) gene and the Panton-Valentine leukocidin (PVL) toxin gene. PCR for the TSST (62) and PVL toxin (38) genes was performed as described previously.
Detection of staphylococcal enterotoxin A, B, C, D, and E production in suspect MRSA isolates.
The suspected MRSA isolates were grown in LB medium overnight at 37°C. The supernatant of the cultures was collected by centrifugation at 3,500 × g for 5 min at 15°C. Sterile filtration of the supernatant was carried out using Millipore Steriflip membranes (0.22 μm) to avoid transfer of microorganisms to the assay. The commercially available Ridascreen SET A, B, C, D, E (R-Biopharm GmbH, Germany) sandwich immunoassay was used for the detection of staphylococcal enterotoxin (SE) in the supernatants of culture fluids. Protocol was followed as per manufacturer instructions for the enzyme immunoassay. Optical density at 450 nm (OD450) results were interpreted using cutoff values derived by adding 0.15 to the average negative-control OD450. Samples with OD450 less than the cutoff value were considered negative, while those with OD450 equal to or greater than the cutoff were considered positive for the respective toxin.
RESULTS
MSSA isolate prevalence and characterization.
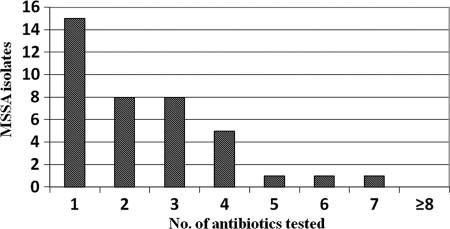
Ninety-three MSSA isolates were detected from the 150 bulk tank milk (BTM) samples, a 62% prevalence in BTM. Herd level prevalence of MSSA was 84% (42 of 50 farms, S. aureus detection in at least one season in 2009 was considered positive). Of these 93 S. aureus isolates recovered from BTM samples using conventional isolation methods, none showed growth on selective medium for MRSA, the MRSA Select plates, indicating that they were all MSSA isolates. All isolates were confirmed as S. aureus in that they carried a 98 to 100% sequence similarity of 16S rRNA genes to the reference gene. Of the 93 MSSA isolates detected, 54 were pansusceptible by the disk diffusion method, 21 were resistant to one class of antibiotic, 13 were resistant to 2 classes of antibiotic, and 5 were resistant to three or more classes of antibiotic and thus multiresistant (Fig. 2). Of the 93 isolates from 42 farms, 29% were resistant to cloxacillin, 17% to ampicillin, 16% to penicillin and tetracycline each, 7% to erythromycin, 5% to pirlimycin, 4% to novobiocin, 3% to streptomycin, and 1% to ceftiofur and cephalothin each, as determined by the Kirby Bauer disk diffusion method.
Fig 2.
Frequency distribution of antimicrobial agent resistance patterns of MSSA. Shown on the x axis are the numbers of antibiotics to which the isolates were resistant. Of the 93 MSSA isolates, 54 were pansusceptible to all antibiotics, 15 were resistant to at least one antibiotic, and the remaining 24 were resistant to 2 or more antibiotics, as tested by the Kirby Bauer method.
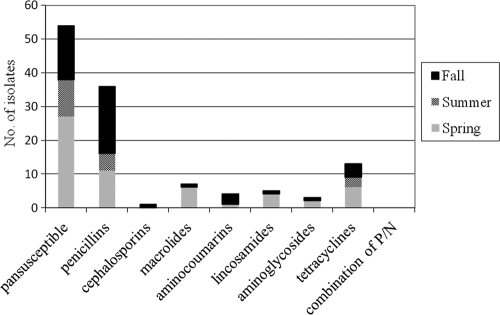
MSSA isolates were classified into the following spa types: t529 (n = 25), t034 (n = 10), t359 (n = 2), t189 (n = 2), t203 (n = 2), t044 (n = 1), t084 (n = 1), t1166 (n = 1), t267 (n = 4), t2734 (n = 1), t337 (n = 1), t4173 (n = 1), and t521 (n = 1). Among the 15 MDR isolates, t034 (n = 5) and t529 (n = 4) spa types dominated. MSSA isolates showed the most resistance to beta-lactam antibiotics, followed by tetracyclines, across all 3 seasons (Fig. 3).
Fig 3.
Distribution of antimicrobial resistance to different classes of antibiotics among the 93 MSSA isolates across the 3 seasons in 2009. Antimicrobial agent resistance was determined for all MSSA isolates by the Kirby Bauer method with individual antibiotic disks. Proportions of MSSA isolates varied among the three seasons, as follows: summer, n = 16; spring, n = 41; and fall, n = 36. P/N, penicillin-novobiocin.
MRSA isolate prevalence and characterization.
Both MRSA isolates were also confirmed as S. aureus by a 98 to 100% sequence similarity of 16S rRNA genes to the reference gene. Any S. aureus isolate was characterized as MRSA if it showed the presence of the mecA gene and displayed phenotypic resistance to oxacillin at a MIC of ≥4 μg/μl. Eight isolates were identified as suspect MRSA, as they amplified an upstream region of mecA and their sequences aligned (98 to 100% nucleotide identities) with S. aureus mecA gene sequences in the NCBI databases. However, the results of the real-time PCR assay from the Minnesota Department of Health (MDH) using an established primer set showed that only 2 of the 8 suspect MRSA isolates contained the mecA gene segment. Repetition of the MDH mecA gene PCR in our lab using conventional PCR produced consistent results. Sequence alignment of primers used in our lab and those of Killgore et al. (32) showed that the primers amplified different regions of the open reading frame (ORF) of the mecA gene (regions separated by 500 bp). Further, the Sensititre results for the 8 suspected MRSA isolates validated that the two isolates which were identified as containing the mecA gene by the MDH assay were MRSA because they were resistant to oxacillin, with MICs of >32 μg/ml. Another isolate (Fa25) showed a low MIC to oxacillin on only one of the panels but did not carry the mecA gene in the MDH assay and hence was not considered MRSA. The remaining 5 isolates were all sensitive to oxacillin, confirming them as false positives.
Two isolates (1.3%) from 150 BTM samples were confirmed as MRSA after enrichment and screening for the presence of the mecA gene. The prevalence of MRSA in BTM is 1.33%, while the herd prevalence of MRSA is 4% (2 of 50).
The two MRSA isolates each showed resistance to oxacillin, at MICs of >32 μg/μl. The MRSA isolate, Sp19, showed resistance to β-lactams, lincosamides, macrolides, and narrow-spectrum, expanded-spectrum, and broad-spectrum cephalosporins, while the other MRSA isolate, Sp12, showed resistance to fluoroquinolones, carboxypenicillins, and oxazolidinones in addition to the antibiotic classes to which Sp19 was resistant, as shown in Tables 2 and 3.
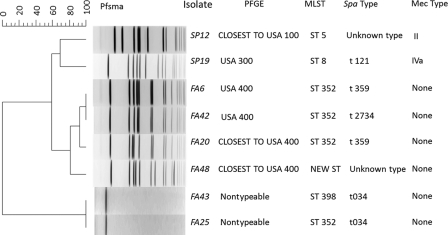
Subtyping using MLST, PFGE, SCCmec typing, and spa typing was performed for the 2 MRSA isolates and the 6 suspect isolates (Fig. 4). One MRSA isolate, Sp12, shows a composite profile of unknown spa type-PFGE type close to USA100-ST 5-mec type II, while the second isolate, Sp19, shows a composite profile of spa type t121-PFGE type USA300-ST 8-mec type IVa. Other PFGE types among the suspect MRSA isolates include USA400 (n = 2), closest to USA400 (n = 2), and nontypeable (n = 2). MLST types found in the false positives include ST 352 (n = 4), ST 398 (n = 1), and unassigned ST. No mec types could be obtained for any of the false-positive isolates. spa type t121 was the only spa type identified among the 2 MRSA isolates, while types t034 (n = 2), t359 (n = 2), t2734 (n = 1), and unknown type (n = 1) were detected in the suspect MRSA isolates.
Fig 4.
PFGE patterns of MRSA and staphylococcal enterotoxin-producing MSSA isolates, compared with subtypes obtained by MLST, spa typing, and SCCmec typing. The composite genotypes obtained by the combination of PFGE pulsotype with MLST sequence type, spa type, and SCCmec type provide important information about the epidemiologies of the MRSA and MSSA isolates.
Presence of TSST and PVL toxin genes and SE production in suspect MRSA isolates.
The 8 suspect MRSA isolates were analyzed further for their toxin production profiles and the presence of TSST and PVL toxin genes. Although only 2 of the 8 isolates were confirmed as MRSA by detection of the mecA gene and phenotypic expression of methicillin resistance, toxin data from the 6 suspect isolates are presented to depict the toxigenic potential of non-methicillin-resistant strains circulating in milk. MRSA isolate Sp12 produced SE B, SE C, and SE D, while isolate Sp19 produced SE C, SE D, and SE E (Table 4). Among the 8 S. aureus isolates analyzed, the order of SE production (produced by most number of isolates) is as follows: SE C > SE D > SE B and SE E. None of the 8 isolates showed the presence of the TSST gene, while the presence of the PVL toxin gene was detected in one isolate alone, the Sp19 MRSA isolate.
Table 4.
Staphylococcal enterotoxins produced and toxin genes detected for MRSA and MSSA isolates
| Isolatea | Organism | Toxin production analyzed for SE: |
Toxin gene detected by PCR | ||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | |||
| Sp12 | MRSA | − | + | + | + | − | − |
| Sp19 | MRSA | − | − | + | + | + | PVL |
| Fa6 | MSSA | − | − | + | − | − | − |
| Fa20 | MSSA | − | − | + | + | − | − |
| Fa25 | MSSA | − | − | + | − | − | − |
| Fa42 | MSSA | − | − | + | + | − | − |
| Fa43 | MSSA | − | − | − | − | − | − |
| Fa48 | MSSA | − | + | + | + | + | − |
Only Sp12 and Sp19 are MRSA isolates; the rest were suspect MRSA but were confirmed as MSSA isolates.
DISCUSSION
MRSA has increasingly been recognized in farm animal populations in recent years. Some studies have investigated MRSA prevalence in pig populations in North America (31, 57); however, limited information with respect to the genetic characterization of MRSA in cattle or in bulk tank milk in the United States is available.
The 50 farms sampled in our study are representative of 2.3% of all dairy herds enrolled with the MN DHIA (50 of 2,159 farms). The mean milk production in terms of rolling herd averages (lb milk/cow/year) for the 50 farms (21,586 lb) was reflective of the mean rolling herd averages of the farms enrolled with the MN DHIA (21,329 lb).
Our results suggest that there is a low prevalence of MRSA in MN dairy herds. This is in accordance with the NAHMS Dairy 2007 study (including samples from MN), which found no MRSA in bulk tank milk samples (63). Our methods were more robust and used selective enrichment for oxacillin-resistant S. aureus prior to characterization, which may explain why some MRSA isolates were detected in the samples we collected. Also to be noted is the fact that 150 pooled BTM samples from three consecutive days were analyzed, which could represent anywhere between 450 and 300 separate milk loads (assuming milk was emptied every day and every other day, respectively). This sampling protocol in addition to the selective double enrichment protocol for MRSA may account for success in detecting MRSA and the higher rates of prevalence of MSSA isolates than were found with previous studies. Consistent with our findings, other countries have reported various rates of prevalence of MRSA in bovine milk: 1.4% in Switzerland (25), 1.5% in Japan (24), 2.4% in Korea (42), 0 to 7.4% in Belgium (61), and 5.1 to 16.7% in southwest Germany (58). Recently, a study of S. aureus from mastitic cattle in India reported a high prevalence of MRSA, i.e., 13% of S. aureus isolates (14 of 107 samples) (34). Such high prevalence has not been reported in studies from the United States or Europe, with the exception of southwest Germany (58). However, only 10 of 14 isolates classified as MRSA by the authors showed the presence of the mecA gene by PCR.
Although 6 isolates were detected on MRSA Select plates, they did not amplify the mecA gene by MDH primers or show resistance to oxacillin, with the exception of one isolate, which showed a low MIC on one of the two panels. They did amplify a segment of the mecA gene, suggesting a possible truncation in the open reading frame. It is likely that these isolates showed poor expression of the mecA gene or overproduction of beta-lactamase, as they remained susceptible to oxacillin plus 2% NaCl but resistant to penicillin (26, 42). Another factor that may explain this discrepancy could be that phenotypic expression of resistance varies with growth condition (pH, osmolarity of medium, etc.) (8, 35). On the other hand, higher MICs of oxacillin resistance (on the order of 16 μg/ml) have been observed upon induction (i.e., culturing in the presence of oxacillin) in MRSA isolates which previously exhibited low MICs of 1 to 2 μg/ml. Hence, false positives could also arise due to limitations in the microbiological detection methods (56). Because of this, all suspect MRSA isolates from animal environments should also be subjected to broth microdilution-based MIC detection and confirmation for the presence of mecA by PCR (32).
Also of interest is the recent finding in S. aureus isolates from bulk milk as well as milk from mastitic cows in England of a divergent mecA gene which is not detectable by routinely used PCR primers and which showed 70% amino acid similarity to currently identified mecA (22). This divergent mecA was associated primarily with spa type t843 and CC130 and has also been identified in human MRSA isolates from Scotland, England, Denmark, and Ireland (22, 56). The discovery of novel mecA homologs suggests the possibility of the presence of such a divergent homolog in the 6 suspect MRSA isolates, which were confirmed as mecA negative by established primers described by Killgore et al. (32) but showed the presence of mecA fragments amplified by primers used in our lab. Also to be noted is the growth of these six S. aureus isolates in a selective medium for identification of MRSA, the MRSA Select plates, indicating that mechanisms of resistance to methicillin may be different among some animal strains.
In a recent study of MRSA isolated from milk samples from mastitic cows, 100% of isolates were resistant to erythromycin, clindamycin, chloramphenicol, and gentamicin (60). In another study in Switzerland, two MRSA isolates from 142 milk samples from mastitic cows were resistant to ampicillin, ceftiofur, clindamycin, erythromycin, oxacillin, penicillin, and tetracycline (25), while Vanderhaeghen et al. (61) reported resistance to tetracycline, with frequent resistance to macrolides, lincosamides, and aminoglycosides, in MRSA from milk from mastitic cows. Resistance profiles of MRSA isolates from the previous two studies closely resemble the profiles of both MSSA and MRSA isolates in our current study. Further studies are needed to analyze the relationship between patterns of antibiotic use on farms and the emergence of resistance to specific drugs. Few studies have analyzed the statistical relationship between antibiotic use and emergence of antibiotic-resistant pathogens on dairy farms. Poll and Ruegg (51) reported a heterogeneous relationship of S. aureus MICs with drug usage on farms and concluded that a clear dose-response interaction was not observed. Further, they observed no significant associations between level of antibiotic exposure (in organic versus conventional farms) and MICs of mastitis pathogens (51). Erskine et al. also detected no indication of increased resistance of mastitis isolates (including S. aureus) to commonly used antibiotics and instead reported an increase in proportion of isolates susceptible to ampicillin, penicillin, and erythromycin over a 7-year period (2, 20, 39). Rajala-Schultz et al. analyzed the susceptibility patterns of mastitis pathogens isolated at calving from first-lactation and older cows and reported no significant difference in the proportions of resistant isolates between first-lactation and older cows (53). A study conducted by Roesch et al. in Switzerland compared the rates of occurrence of resistant mastitis pathogens between organic and conventional dairy farms and reported no significant differences between the two (54).
The most prevalent spa type among MRSA isolates from bovine sources is t011, as is corroborated by reports from Germany, Belgium, and Switzerland (21, 25, 58, 61). MRSA isolates from bovine mastitis samples in Germany and Belgium (21, 61) also included other spa types—t034, t2576, and t567. Limited data on spa type distribution are available for MSSA isolates from bovine sources. Of the spa types obtained in our study, two of the MSSA isolates belonged to the type found in Germany (21), t034, while one MRSA isolate belonged to type t121, which is common throughout France, Belgium, Germany, Switzerland, and the United States and is associated with MLST ST 8 (http://www.spaserver.ridom.de).
Isolate Sp12 carried a composite genotype of ST5-close to USA100-mec type II, which is a genotype commonly reported among hospital-associated MRSA isolates, while isolate Sp19 carried a composite profile of ST8-USA300-mec type IVa, reported in community-associated MRSA lineages with the smaller SCCmec cassette IV (41). In contrast, MRSA isolates from milk from mastitic cows in Switzerland and Belgium belonged to MLST ST 398, which is a LA MRSA sequence type (21, 25, 41). Bovine MRSA isolates from Germany were also untypeable by SmaI PFGE (21, 25). Such untypeable ST 398 isolates have frequently been detected from MRSA isolates from pigs in the United States as well as in other countries (15, 57). Two of the suspect MRSA isolates which were later confirmed susceptible to methicillin show spa type, MLST, and PFGE composite profiles similar to those seen in LA MRSA genotypes, indicating that such genotypes are circulating in dairy farms but remain susceptible to methicillin.
While the prevalence of MRSA is low in MN dairies and S. aureus is eliminated by pasteurization, there is an unproved but potential risk of transmission of MRSA to humans in situations where raw milk is utilized either directly for consumption or in raw milk cheese production, especially young ripened cheeses. At present, raw milk sales are legal in MN if the consumer purchases directly from the dairy herd. Dairy producers also commonly consume raw milk produced on their farms. Two surveys conducted in 1999 and 2001 by Jayarao et al. (27, 28) reported that 60% of dairy producers in eastern South Dakota and western Minnesota consumed raw milk. In our current study, 36% of farms reported consumption of unpasteurized raw milk produced on farm.
Staphylococcal enterotoxins are capable of retaining immunological and biological activity following pasteurization. When the concentration of an enterotoxigenic strain of S. aureus exceeds 105 CFU/ml or CFU/g, the strain is capable of producing toxin sufficient to cause intoxication symptoms (5). However, one of the limitations of this study is the unavailability of colony counts from isolated MSSA and MRSA. In the case of MRSA at least, CFU/ml is not representative of actual numbers of the organism present in BTM, as the organism has been isolated after a double enrichment and selection protocol. Of the 8 isolates analyzed for toxin production, 7 showed production of at least one enterotoxin, which could be explained based on the relatively high percentage of classical SE-producing strains from bovine subclinical mastitis (4, 6, 12, 52). As previously observed in literature, none of the S. aureus isolates from milk produced SE E (6, 43, 52, 68), but we found one SE E-producing MSSA isolate in this study.
In summary, two MDR MRSA isolates capable of producing multiple staphylococcal enterotoxins and carrying genotypes commonly isolated in both health care and community settings were found in two MN dairy herds. Different states of interaction can exist between S. aureus and its host: infection, carriage, colonization, and contamination (18). Healthy humans can act as carriers if persistently or intermittently colonized by MRSA (colonization is a major risk factor) and can play a role in its spread among animals, the dairy environment, and the community. Transmission of MRSA between humans and animals, in particular, between cows and humans, has been reported previously (30).
The presence of MRSA isolates in bulk tank milk may present a potential public health risk, and MRSA may spread between animals if the numbers of isolates in dairy environments were to increase. Further, resistance to multiple antibiotics is a more realistic risk to animal and public health if found to cause invasive diseases other than MRSA. Therefore, careful monitoring of the resistance status of S. aureus in dairy environments is needed, as Staphylococcus aureus transmission is dynamic and involves humans, animals, and likely the farm production environment. Future studies are also needed to help identify critical areas that allow for contamination and spread within the farm environment.
ACKNOWLEDGMENTS
This study was supported by a grant from the University of Minnesota, College of Veterinary Medicine, Agricultural Experiment Station.
We thank the 50 dairy farms for their participation in the study. We also acknowledge the assistance of Andriy Vatalin, Habner Bastos, and Christiano Gontijo in the collection of field milk samples.
Footnotes
Published ahead of print 14 December 2011
REFERENCES
- 1. Akineden O, et al. 2001. Toxin genes and other characteristics of Staphylococcus aureus isolates from milk of cows with mastitis. Clin. Diagn. Lab. Immunol. 8:959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson KL, Lyman RL, Bodeis-Jones SM, White DG. 2006. Genetic diversity and antimicrobial susceptibility profiles among mastitis-causing Staphylococcus aureus isolated from bovine milk samples. Am. J. Vet. Res. 67:1185–1191 [DOI] [PubMed] [Google Scholar]
- 3. Arcuri EF, et al. 2010. Toxigenic status of Staphylococcus aureus isolated from bovine raw milk and Minas frescal cheese in Brazil. J. Food Prot. 73:2225–2231 [DOI] [PubMed] [Google Scholar]
- 4. Balaban N, Rasooly A. 2000. Staphylococcal enterotoxins. Int. J. Food Microbiol. 61:1–10 [DOI] [PubMed] [Google Scholar]
- 5. Bergdoll MS. 1997. Toxic shock syndrome. Toxins 1:6–21 [Google Scholar]
- 6. Boynukara B, Gulhan T, Alisarli M, Gurturk K, Solmaz H. 2008. Classical enterotoxigenic characteristics of Staphylococcus aureus strains isolated from bovine subclinical mastitis in Van, Turkey. Int. J. Food Microbiol. 125:209–211 [DOI] [PubMed] [Google Scholar]
- 7. Chakravorty S, Helb D, Burday M, Connell N, Alland D. 2007. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69:330–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chambers HF. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. American Society for Microbiology, Washington, DC: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. CLSI 2006. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A3. CLSI, Wayne, PA [Google Scholar]
- 10. Cosgrove SE. 2006. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clini. Infect. Dis. 42:S82–S89 [DOI] [PubMed] [Google Scholar]
- 11. D'Amico DJ, Donnelly CW. 2010. Microbiological quality of raw milk used for small-scale artisan cheese production in Vermont: effect of farm characteristics and practices. J. Dairy Sci. 93:134–147 [DOI] [PubMed] [Google Scholar]
- 12. da Silva ER, do Carmo LS, da Silva N. 2005. Detection of the enterotoxins A, B, and C genes in Staphylococcus aureus from goat and bovine mastitis in Brazilian dairy herds. Vet. Microbiol. 106:103–107 [DOI] [PubMed] [Google Scholar]
- 13. De Buyser ML, Dufour B, Maire M, Lafarge V. 2001. Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. Int. J. Food Microbiol. 67:1–17 [DOI] [PubMed] [Google Scholar]
- 14. Declercq P, Petre D, Gordts B, Voss A. 2008. Complicated community-acquired soft tissue infection by MRSA from porcine origin. Infection 36:590–592 [DOI] [PubMed] [Google Scholar]
- 15. de Neeling AJ, et al. 2007. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet. Microbiol. 122:366–372 [DOI] [PubMed] [Google Scholar]
- 16. Denis O, et al. 2009. Methicillin-resistant Staphylococcus aureus ST398 in swine farm personnel, Belgium. Emerg. Infect. Dis. 15:1098–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Devriese LA, Hommez J. 1975. Epidemiology of methicillin-resistant Staphylococcus aureus in dairy herds. Res. Vet. Sci. 19:23–27 [PubMed] [Google Scholar]
- 18. EFSA Panel on Biological Hazards 2009. Assessment of the public health significance of meticillin resistant Staphylococcus aureus (MRSA) in animals and food. EFSA J. 933:1–3 [Google Scholar]
- 19. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Erskine RJ, Walker RD, Bolin CA, Bartlett PC, White DG. 2002. Trends in antibacterial susceptibility of mastitis pathogens during a seven-year period. J. Dairy Sci. 85:1111–1118 [DOI] [PubMed] [Google Scholar]
- 21. Fessler A, et al. 2010. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65:619–625 [DOI] [PubMed] [Google Scholar]
- 22. Garcia-Alvarez L, et al. 2011. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 11:595–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gunaydin B, Aslantas O, Demir C. 2011. Detection of superantigenic toxin genes in Staphylococcus aureus strains from subclinical bovine mastitis. Trop. Anim. Health Prod. 43:1633–1637 [DOI] [PubMed] [Google Scholar]
- 24. Hata E, et al. 2010. Genetic variation among Staphylococcus aureus strains from bovine milk and their relevance to methicillin-resistant isolates from humans. J. Clin. Microbiol. 48:2130–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huber H, Koller S, Giezendanner N, Stephan R, Zweifel C. 2009. Prevalence and characteristics of meticillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland. Euro Surveill. 15:1–4 [PubMed] [Google Scholar]
- 26. Hulya Turutoglu MH, Dilek Ozturk Murat Yildirim SS. 15 August 2009. Methicillin and aminoglycoside resistancein Staphylococcus aureus isolates from bovine mastitis and sequence analysis of their mecA genes. Vet. Res. Commun. 33:945–956 [DOI] [PubMed] [Google Scholar]
- 27. Jayarao BM, Cassel EK. 1999. Mastitis prevention and milk hygiene practices adopted by dairy practitioners. Large Anim. Pract. 20:6–14 [Google Scholar]
- 28. Jayarao BM, Henning DR. 2001. Prevalence of foodborne pathogens in bulk tank milk. J. Dairy Sci. 84:2157–2162 [DOI] [PubMed] [Google Scholar]
- 29. Jayarao BM, Pillai SR, Sawant AA, Wolfgang DR, Hegde NV. 2004. Guidelines for monitoring bulk tank milk somatic cell and bacterial counts. J. Dairy Sci. 87:3561–3573 [DOI] [PubMed] [Google Scholar]
- 30. Juhász-Kaszanyitzky E, et al. 2007. MRSA transmission between cows and humans. Emerg. Infect. Dis. 13:630–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khanna T, Friendship R, Dewey C, Weese JS. 2008. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet. Microbiol. 128:298–303 [DOI] [PubMed] [Google Scholar]
- 32. Killgore GE, Holloway B, Tenover FC. 2000. A 5′ nuclease PCR (TaqMan) high-throughput assay for detection of the mecA gene in staphylococci. J. Clin. Microbiol. 38:2516–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klein E, Smith DL, Laxminarayan R. 2007. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg. Infect. Dis. 13:1840–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kumar R, Yadav BR, Singh RS. 2011. Antibiotic resistance and pathogenicity factors in Staphylococcus aureus isolated from mastitic Sahiwal cattle. J. Biosci. 36:175–188 [DOI] [PubMed] [Google Scholar]
- 35. Kumar R, Yadav BR, Singh RS. 2010. Genetic determinants of antibiotic resistance in Staphylococcus aureus isolates from milk of mastitic crossbred cattle. Curr. Microbiol. 60:379–386 [DOI] [PubMed] [Google Scholar]
- 36. Lee JH. 2003. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 69:6489–6494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lewis HC, et al. 2008. Pigs as source of methicillin-resistant Staphylococcus aureus CC398 infections in humans, Denmark. Emerg. Infect. Dis. 14:1383–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lina G, et al. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128–1132 [DOI] [PubMed] [Google Scholar]
- 39. Makovec JA, Ruegg PL. 2003. Antimicrobial resistance of bacteria isolated from dairy cow milk samples submitted for bacterial culture: 8,905 samples (1994–2001). J. Am. Vet. Med. Assoc. 222:1582–1589 [DOI] [PubMed] [Google Scholar]
- 40. Makovec JA, Ruegg PL. 2003. Results of milk samples submitted for microbiological examination in Wisconsin from 1994 to 2001. J. Dairy Sci. 86:3466–3472 [DOI] [PubMed] [Google Scholar]
- 41. McDougal LK, et al. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moon JS, et al. 2007. Phenotypic and genetic antibiogram of methicillin-resistant staphylococci isolated from bovine mastitis in Korea. J. Dairy Sci. 90:1176–1185 [DOI] [PubMed] [Google Scholar]
- 43. Morandi S, Brasca M, Lodi R, Cremonesi P, Castiglioni B. 2007. Detection of classical enterotoxins and identification of enterotoxin genes in Staphylococcus aureus from milk and dairy products. Vet. Microbiol. 124:66–72 [DOI] [PubMed] [Google Scholar]
- 44. Murphy BP, O'Mahony E, Buckley JF, O'Brien S, Fanning S. 2010. Characterization of Staphylococcus aureus isolated from dairy animals in Ireland. Zoonoses Public Health 57:249–257 [DOI] [PubMed] [Google Scholar]
- 45. Neder VE, Canavesio VR, Calvinho LF. 2011. Presence of enterotoxigenic Staphylococcus aureus in bulk tank milk from Argentine dairy farms. Rev. Argent. Microbiol. 43:104–106 [DOI] [PubMed] [Google Scholar]
- 46. Normanno G, et al. 2005. Coagulase-positive staphylococci and Staphylococcus aureus in food products marketed in Italy. Int. J. Food Microbiol. 98:73–79 [DOI] [PubMed] [Google Scholar]
- 47. Noskin GA, et al. 2005. The burden of Staphylococcus aureus infections on hospitals in the United States: an analysis of the 2000 and 2001 Nationwide Inpatient Sample Database. Arch. Intern. Med. 165:1756–1761 [DOI] [PubMed] [Google Scholar]
- 48. Owens WE, Watts JL. 1988. Antimicrobial susceptibility and beta-lactamase testing of staphylococci isolated from dairy herds. J. Dairy Sci. 71:1934–1939 [DOI] [PubMed] [Google Scholar]
- 49. Peles F, et al. 2007. Characterization of Staphylococcus aureus strains isolated from bovine milk in Hungary. Int. J. Food Microbiol. 118:186–193 [DOI] [PubMed] [Google Scholar]
- 50. Piddock LJV. 1996. Does the use of antimicrobial agents in veterinary medicine and animal husbandry select antibiotic resistant bacteria that infect man and compromise antimicrobial chemotherapy? J. Antimicrob. Chemother. 38:1–3 [DOI] [PubMed] [Google Scholar]
- 51. Pol M, Ruegg PL. 2007. Relationship between antimicrobial drug usage and antimicrobial susceptibility of gram-positive mastitis pathogens. J. Dairy Sci. 90:262–273 [DOI] [PubMed] [Google Scholar]
- 52. Rahimi E, Safai HG. 2010. Detection of classical enterotoxins of Staphylococcus aureus strains isolated from bovine subclinical mastitis in Isfahan, Iran. Vet. Microbiol. 141:393–394 [DOI] [PubMed] [Google Scholar]
- 53. Rajala-Schultz PJ, Smith KL, Hogan JS, Love BC. 2004. Antimicrobial susceptibility of mastitis pathogens from first lactation and older cows. Vet. Microbiol. 102:33–42 [DOI] [PubMed] [Google Scholar]
- 54. Roesch M, et al. 2006. Comparison of antibiotic resistance of udder pathogens in dairy cows kept on organic and on conventional farms. J. Dairy Sci. 89:989–997 [DOI] [PubMed] [Google Scholar]
- 55. Shopsin B, et al. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shore AC, et al. 2011. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 55:3765–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith TC, et al. 2009. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One 4:e4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Spohr M, et al. 2011. Methicillin-resistant Staphylococcus aureus (MRSA) in three dairy herds in southwest Germany. Zoonoses Public Health 58:252–261 [DOI] [PubMed] [Google Scholar]
- 59. Trinidad P, Nickerson SC, Alley TK. 1990. Prevalence of intramammary infection and teat canal colonization in unbred and primigravid dairy heifers. J. Dairy Sci. 73:107–114 [DOI] [PubMed] [Google Scholar]
- 60. Turkyilmaz S, Tekbiyik S, Oryasin E, Bozdogan B. 2010. Molecular epidemiology and antimicrobial resistance mechanisms of methicillin-resistant Staphylococcus aureus isolated from bovine milk. Zoonoses Public Health 57:197–203 [DOI] [PubMed] [Google Scholar]
- 61. Vanderhaeghen W, et al. 2010. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet. Microbiol. 144:166–171 [DOI] [PubMed] [Google Scholar]
- 62. Varshney AK, et al. 2009. Diverse enterotoxin gene profiles among clonal complexes of Staphylococcus aureus isolates from the Bronx, New York. Appl. Environ. Microbiol. 75:6839–6849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Virgin JE, Van Slyke TM, Lombard JE, Zadoks RN. 2009. Short communication: methicillin-resistant Staphylococcus aureus detection in US bulk tank milk. J. Dairy Sci. 92:4988–4991 [DOI] [PubMed] [Google Scholar]
- 64. Waage S, et al. 1999. Bacteria associated with clinical mastitis in dairy heifers. J. Dairy Sci. 82:712–719 [DOI] [PubMed] [Google Scholar]
- 65. Witte W, Strommenger B, Stanek C, Cuny C. 2007. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, central Europe. Emerg. Infect. Dis. 13:255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang S, Iandolo JJ, Stewart GC. 1998. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 168:227–233 [DOI] [PubMed] [Google Scholar]
- 68. Zouharova M, Rysanek D. 2008. Multiplex PCR and RPLA identification of Staphylococcus aureus enterotoxigenic strains from bulk tank milk. Zoonoses Public Health 55:313–319 [DOI] [PubMed] [Google Scholar]