Abstract
Real-time PCR methodology can be applied to rapidly and accurately detect influenza viruses. During times of surge testing or enhanced pandemic surveillance, public health laboratories (PHLs) may experience overwhelming demand for testing, even while the prevalence of positive specimens remains low. To improve laboratory capacity and testing efficiency during surges, we evaluated whether nasopharyngeal (NP)/throat swab specimens can be pooled and tested for the presence of the 2009 H1N1 influenza virus without a reduction in sensitivity. Pools of 10 specimens were extracted and concentrated upon elution on the MagNA Pure LC instrument, and real-time PCR was performed on the Applied Biosystems 7500 Fast platform, using the CDC swine influenza virus real-time RT-PCR detection panel (rRT-PCR swine flu panel). Specimens in positive pools were singly re-extracted and retested by PCR to identify individual positive samples. Initial studies showed that spiking a pool of nine negative specimens (100 μl each) or 900 μl of virus transport medium with 100 μl of a positive clinical specimen caused no loss of sensitivity by rRT-PCR testing. Pools containing either multiple positive specimens or specimens positive for other respiratory viruses also showed no negative effect on crossing threshold (CT) values. To test the robustness of the pooling protocol, a panel of 50 blinded samples was sent to three PHLs and tested in five pools of 10. All PHLs correctly identified the positive specimens. This study demonstrates the feasibility of using a pooling strategy to increase capacity and conserve resources during surge testing and periods of enhanced influenza surveillance when the prevalence is low.
INTRODUCTION
Reverse transcription–real-time PCR (rRT-PCR) is an important diagnostic tool for detection of influenza virus in public health laboratories. The Centers for Disease Control and Prevention (CDC) human influenza virus rRT-PCR detection and characterization panel (rRT-PCR flu panel) was cleared by the U.S. Food and Drug Administration (FDA) for diagnosing human influenza virus infections and H5N1 viruses (6) in September 2008. Likewise, the CDC real-time RT-PCR protocol for detection and characterization of swine influenza virus (rRT-PCR swine flu panel) (24) was authorized by the FDA under an emergency use authorization (EUA) to diagnose 2009 H1N1 influenza virus infections (7) in April 2009 and subsequently cleared by the FDA in June 2010. A combination of the seasonal flu panel with the swine flu panel was cleared by the FDA in September 2011 (8). The use of either protocol allows for rapid detection of influenza viruses from clinical specimens. However, during an influenza outbreak or pandemic, test workloads in public health laboratories may rapidly outstrip testing capacity, and reagent shortages may occur. The recent outbreak of a novel influenza A/H1N1 virus (2009 H1N1) (1, 2) is a clear example. At the start of the pandemic, there was a high volume of specimens that required testing but a low prevalence of the 2009 H1N1 strain. One possible way to increase testing capacity and conserve reagents is to pool specimens prior to RNA extraction, concentrate the RNA, test the pools, and subsequently retest single specimens from positive pools to identify the positive specimens.
Pooling specimens to increase efficiency of testing and cost effectiveness is not unprecedented. Testing specimens in pools has been used to detect infections such as human immunodeficiency virus (HIV) and the hepatitis B and C viruses (5, 18). Blood banks worldwide are able to screen millions of blood donations by implementing the minipool nucleic acid amplification technology (NAT) testing method to detect transfusion-transmissible viruses (9, 14, 18, 20, 21, 23). The size of the minipools differs from country to country, but each sample in a positive pool is retested individually. Pooling of specimens has also been evaluated for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae as a measure to reduce cost and labor while maintaining accuracy (4, 15, 16, 19, 22).
In this study, we investigated the potential of pooling nasopharyngeal (NP)/throat swab specimens for testing using the CDC rRT-PCR swine flu panel on the Applied Biosystems (ABI) 7500 Fast platform and the MagNA Pure LC instrument as the extraction method. The investigation consisted of two major parts, a proof-of-concept component and a small multisite trial study. This is the first publication describing a pooling method for detection of a respiratory virus. Public health laboratories currently using the CDC rRT-PCR flu panel or rRT-PCR swine flu panel could easily implement a pooling protocol to increase testing capacity and conserve resources during low influenza prevalence without additional purchase of costly equipment or reagents or dramatic modification of their workflow.
MATERIALS AND METHODS
Specimen type and processing.
The human seasonal influenza virus A/H1N1 strain A/South Dakota/06/2007 was provided by the CDC as a representative influenza virus A strain. Clinical NP/throat swab specimens used in this investigation were submitted to the Wisconsin State Laboratory of Hygiene (WSLH) by Wisconsin hospitals and clinics. The clinical NP/throat swab specimens were transported in various virus transport media (VTM), refrigerated, and processed within 72 h of collection. Volumes of specimen samples in excess of that required for initial testing were frozen and stored at −80°C. Aliquots of these frozen specimens were used in this study.
Clinical NP/throat swab specimens used in this study were previously tested by PCR for the presence of the 2009 H1N1 influenza virus. “Negative” specimens in this study were negative for seasonal influenza viruses (A/H1, A/H3, and B) and 15 additional respiratory viruses (respiratory syncytial virus types A and B; parainfluenza virus types 1, 2, 3, 4a, and 4b; human metapneumovirus; adenovirus subgroups B, C, and E; coronaviruses OC43, NL63, and 229E; and human rhinovirus). Detection of these 15 respiratory viruses was performed using a real-time PCR protocol designed to detect multiple targets in a single assay (17).
Nucleic acid extraction.
Extraction of nucleic acid from samples was performed using either the MagNA Pure LC total nucleic acid isolation kit (TNA kit) or the MagNA Pure LC total nucleic acid isolation kit—large volume (large-volume TNA kit) on the MagNA Pure LC instrument (Roche Diagnostics, Indianapolis, IN). Extractions using both kits were performed according to the manufacturer's instructions. For use with the large-volume TNA kit, 1-ml volumes were extracted and eluted into 100 μl of elution buffer, and for the TNA kit, 100-μl volumes were extracted and eluted into 100 μl of elution buffer.
Reverse transcription and DNA amplification.
Detection of the 2009 H1N1 virus and seasonal influenza viruses A and B was performed using the CDC rRT-PCR swine flu panel (24) and the CDC rRT-PCR flu panel, respectively. The CDC rRT-PCR swine flu panel included primers and TaqMan probes for seasonal influenza viruses (InfA), universal swine influenza virus A (swInfA), the swine influenza virus H1N1 (swH1) subtype, and RNase P (RP). The InfA primer and probe set was designed to detect all influenza A viruses, that for swInfA to detect all swine influenza A viruses, and that for swH1 to specifically detect the 2009 H1N1 virus. The RP primer and probe are designed to detect the human RNase P gene, and they serve as an internal positive control for human nucleic acid. PCR was performed according to the CDC protocol (24) using the SuperScript III platinum one-step quantitative RT-PCR kit (Invitrogen Corporation, Carlsbad, CA) on the Applied Biosystems (ABI) 7500 Fast real-time PCR system (Applied Biosystems, Inc., Foster City, CA).
Comparison of large-volume TNA kit to TNA kit.
For the experiment comparing the large-volume TNA kit to the TNA kit, 15 clinical specimens that previously tested positive for the 2009 H1N1 influenza virus by PCR were selected. Five demonstrated crossing threshold (CT) values in the range of 30 to 35, five had CT values of 24 to 29, and five had CT values of 15 to 20. One hundred microliters of clinical specimen was mixed with 900 μl VTM, and nucleic acid was extracted using the large-volume TNA kit. Another 100 μl of the same clinical specimen was also extracted for nucleic acid using the TNA kit. Extracts from both kits were eluted in 100 μl of elution buffer; thus the 1 ml of input material for the large-volume TNA kit was concentrated to the same final volume of 100 μl as the input material for the TNA kit. PCR was performed in triplicate for each extracted specimen.
Pools with CDC reference strain.
For pooling of the CDC influenza virus reference strain with negative clinical specimens, two dilutions of the reference strain, 50% egg infectious doses (EID50) 4.2 and 2.2, were each spiked into six pools of unique negative clinical specimens. The reference strain was also spiked into VTM, as a control in the experiment. Each pool was extracted once, and PCR was performed using the CDC rRT-PCR flu panel protocol on each extract in triplicate.
Specimen pools.
Ten 100-μl aliquots of various combinations of the influenza A virus reference strain, influenza A virus-positive clinical specimens, negative clinical specimens, and clinical specimens positive for a variety of respiratory viruses were used to create 1-ml pools. Each pool was extracted once using the large-volume TNA kit, and PCR was performed.
Multisite study.
Three state public health laboratories participated in this study. Each site was sent three sets of 50 blinded specimens. They were instructed to create five pools of 10 specified clinical specimens from each set of 50 specimens. Each pool was extracted using the large-volume TNA kit, and PCR was performed with the required controls according to the CDC rRT-PCR swine flu panel (24). Specimens from pools positive for the swInfA and swH1 markers were individually re-extracted and retested by PCR. Extraction of the individual specimens was performed using the large-volume TNA kit. One hundred microliters of the specimen was diluted in 900 μl of VTM and eluted in 100 μl of elution buffer. The laboratories were asked to test 50 specimens in pools of 10 on different days by different technologists if possible. All results were reported on a standardized worksheet provided for the study, and data were sent to the WSLH for analysis.
Data analysis.
PCR results were interpreted as recommended in the CDC rRT-PCR swine flu panel protocol (24). A pool, dilution, or sample was considered positive if the CT values of two out of the three replicates were less than 37.
RESULTS
Proof-of-concept study.
The extraction performance of the MagNA Pure large-volume TNA kit was compared to that of the MagNA Pure TNA kit (Roche Diagnostics, Indianapolis, IN) to ensure that the extraction efficiency was not sacrificed with a higher input volume (Materials and Methods). The average CT values obtained with testing specimens extracted with the MagNA Pure TNA kit and the large-volume TNA kit for each set of extracts were within one or two CT values (Table 1). This result suggested that the pooled specimens did not exceed the capacity of the extraction method.
Table 1.
Comparison of influenza A virus M gene target crossing threshold values resulting after extraction with the MagNA Pure TNA kit and large-volume TNA kit using influenza A virus-positive specimens representing various CT values
| Sample ID | Mean InfAa CT value with: |
|
|---|---|---|
| TNA kit | Large-volume TNA kit | |
| F | 28 | 28 |
| G | 27 | 25 |
| H | 27 | 28 |
| I | 28 | 28 |
| J | 25 | 25 |
| K | 30 | 29 |
| L | 33 | 33 |
| M | 32 | 32 |
| N | 30 | 33 |
| O | 30 | 31 |
InfA, influenza A virus matrix (M) gene target for detecting any influenza A virus.
The CDC seasonal A/H1N1 reference strain was also used to test for potential inhibitory effects of cellular material from pooling multiple clinical specimens. Instead of VTM, the influenza virus reference strain was mixed with negative clinical specimens (see Materials and Methods). The PCR results showed that samples of the 104.2 EID50 reference strain in VTM spiked into pools of negative clinical specimens were within one CT value of each other (data not shown). The reference strain with 102.2 EID50 showed a slightly wider difference, within 2 to 3 CT values, but the discrepancy was inside the standard deviation of the replicates (data not shown). These results demonstrated that concentration of specimen materials via pooling did not lead to inhibition of PCR.
Clinical specimens previously established as positive for the 2009 H1N1 influenza virus were chosen to determine whether they were detectable when mixed with negative specimens. Pools of 10 clinical specimens were examined, each pool containing one positive and nine negative specimens. Positive specimens with a different range of CT values were chosen, and PCR was performed in triplicate for determination of reproducibility. The mean CT value of the pooled specimen was compared to the mean CT value of the positive specimen diluted in VTM. The results showed differences of one to two CT values for the InfA, swInfA, and swH1 markers, demonstrating the effectiveness of pooling 10 or fewer specimens (Table 2).
Table 2.
Comparison of individual influenza A virus-positive results to results when influenza A virus-positive specimens were pooled with negative specimensa
| Specimen |
CT value forb: |
|||
|---|---|---|---|---|
| InfA | swInfA | swH1 | RP | |
| CS1 | 20 ± 1 | 21 ± 2 | 25 ± 1 | 25 ± 1 |
| Spiked pools | 20 ± 1 | 21 ± 2 | 24 ± 1 | 22 ± 1 |
| CS2 | 20 ± 1 | 20 ± 3 | 24 ± 1 | 24 ± 1 |
| Spiked pools | 20 ± 1 | 21 ± 2 | 24 ± 1 | 22 ± 2 |
| CS3 | 26 ± 1 | 26 ± 1 | 29 ± 0 | 26 ± 1 |
| Spiked pools | 26 ± 1 | 26 ± 2 | 28 ± 1 | 24 ± 2 |
Nine negative clinical specimens and one specimen positive for the 2009 H1N1 virus were combined to create pools of 10 clinical specimens. Three unique clinical specimens (CS1, CS2, and CS3) positive for the 2009 H1N1 virus were each spiked into five pools of negative specimens. The positive specimens were diluted in VTM for use as controls. The crossing threshold values of CS1, CS2, and CS3 diluted in VTM were compared to the PCR results of pools containing the corresponding positive specimens (spiked pools). Each pool and specimen in VTM was extracted once, and PCR was performed in triplicates. The values are the means ± standard deviations of the triplicates.
InfA, influenza A virus matrix (M) gene target for detecting any influenza A virus; swInfA, influenza A virus nucleoprotein (NP) gene from swine for detecting swine influenza virus in the genetic lineage of 2009 pandemic influenza A (H1N1); swH1, highly conserved region of hemagglutinin (HA) gene for detecting the 2009 pandemic influenza A virus (H1N1), subtype H1; RP, human ribonucleoprotein (RP) for detecting human RNase P RNA, used with human clinical specimens to indicate that adequate isolation of nucleic acid resulted from extraction of the clinical specimen.
To test the possibility that higher concentrations of viral nucleic acid in a pool have an inhibitory effect on PCR, pools containing multiple positive specimens were tested and were shown to have no effect on the detection of a positive pool. Nine pools of 10 specimens each containing two, five, seven, and eight positive specimens were tested. The required number of negative specimens was mixed with positive specimens to create the pools of 10. Using the InfA, swInfA, and swH1 primer and probe sets, these pools generated CT values similar to those of the specimen in the pool with the lowest CT value (Table 3). Higher concentrations of viral nucleic acid from the presence of seasonal influenza viruses and other respiratory viruses in a pool also did not inhibit detection of the 2009 H1N1 influenza virus strain by PCR. The results for a representative pool are shown in Table 4. As expected, a pool containing seasonal influenza A/H1 and A/H3 viruses and the 2009 H1N1 virus generated a lower CT value for the InfA target (Table 4). The results with the swInfA and swH1 targets for these pools were within one or two CT values of those of the individual positive specimens. Additionally, for pools that contained seasonal influenza B virus, the CT values for the influenza B virus target were unaffected (data not shown). In a pool lacking the 2009 H1N1 virus but containing a seasonal influenza A virus, the InfA target was positive and the 2009 H1N1 subtype-specific targets (swInfA and swH1) were negative, as expected (data not shown). Pools comprised of other respiratory viruses but lacking any influenza viruses yielded negative PCR results for all influenza virus targets (data not shown), indicating no effect of the presence of specimens containing other viruses on the specificity of the PCR.
Table 3.
Representative PCR results of a pool containing eight specimens positive for the 2009 H1N1 influenza virus and two negative specimens
| Specimen |
CT value fora: |
||
|---|---|---|---|
| InfA | swInfA | swH1 | |
| 1 | 21 | 25 | 28 |
| 2 | 21 | 25 | 27 |
| 3 | 21 | 24 | 27 |
| 4 | 17 | 19 | 22 |
| 5 | 22 | 25 | 28 |
| 6 | 20 | 23 | 26 |
| 7 | 23 | 25 | 28 |
| 8 | 26 | 31 | 32 |
| 9 | Neg | Neg | Neg |
| 10 | Neg | Neg | Neg |
| Poolb | 16 ± 0.6 | 18 ± 0 | 21 ± 0.6 |
InfA, influenza A virus matrix (M) gene target for detecting any influenza A virus; swInfA, influenza A virus nucleoprotein (NP) gene from swine for detecting swine influenza virus in the genetic lineage of 2009 pandemic influenza A (H1N1); swH1, highly conserved region of hemagglutinin (HA) gene for detecting the 2009 pandemic influenza A virus (H1N1), subtype H1; Neg, negative.
Mean CT values of pool with standard deviations of the triplicate PCR results.
Table 4.
Results for pooling specimens positive for influenza with specimens positive for other respiratory viruses
| PCR markera |
CT value for: |
|
|---|---|---|
| 2009 A/H1N1 specimen | Poolb | |
| InfA | 25 | 18 ± 0 |
| swInfA | 26 | 26 ± 0 |
| swH1 | 28 | 29 ± 1 |
| InfB | NA | 22 ± 0 |
InfA, influenza A virus matrix (M) gene target for detecting any influenza A virus; swInfA, influenza A virus nucleoprotein (NP) gene from swine for detecting swine influenza virus in the genetic lineage of 2009 pandemic influenza A (H1N1); swH1, highly conserved region of hemagglutinin (HA) gene for detecting the 2009 pandemic influenza A virus (H1N1), subtype H1; InfB, influenza B virus nonstructural protein (NS) for detecting any influenza B virus.
Representative PCR results (crossing threshold, CT) for pool of 10 specimens containing one that is positive for the 2009 A/H1N1 virus and others positive for other respiratory viruses. The pool contains one specimen positive for 2009 A/H1N1, one for human coronavirus OC43, one for human metapneumovirus, one for rhinovirus, one for seasonal influenza B virus, one for seasonal influenza A/H3 virus, three for seasonal influenza A/H1 viruses, and one negative specimen. PCR was performed in triplicate for the pool sample. Data are the mean CT values of the pool with the standard deviations of the triplicate PCR results. NA, not applicable.
Multisite study.
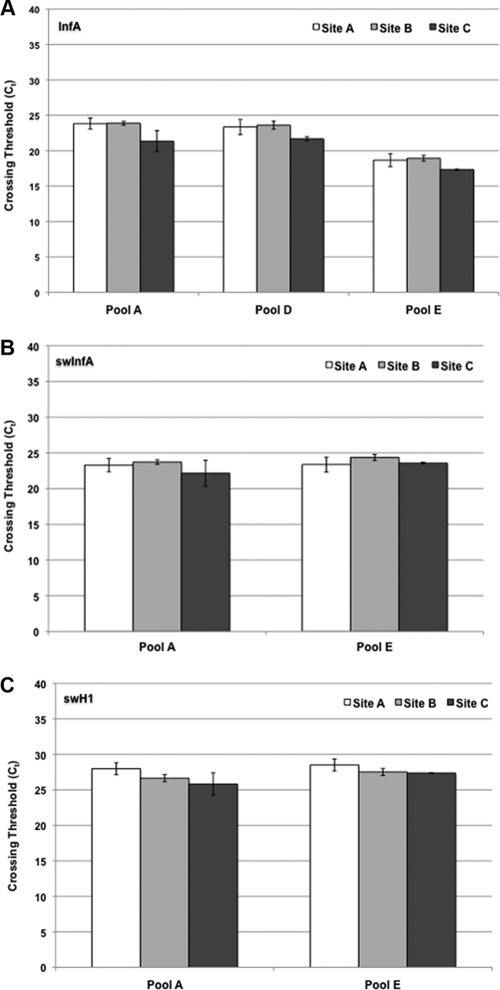
To test the feasibility of use and reproducibility of the pooling influenza virus protocol, a panel containing 50 blinded specimens with instructions for creating five pools of 10 samples each was sent to three state public health laboratories for testing. All three sites correctly identified the negative and positive pools from the testing panel. Pools A and E were positive for the 2009 H1N1 (swine) influenza virus markers, and pool D was positive for just the influenza A virus marker as it contained seasonal influenza viruses A/H1 and A/H3 (Fig. 1). Pools B and C contained specimens negative for influenza viruses. The average CT values of the pools were similar among the three sites for the three 2009 H1N1 influenza virus markers (InfA, swInfA, and swH1) (Fig. 1).
Fig 1.
PCR results of pools from multisite study. Three state public health laboratories tested five blinded pools of 10 specimens for the 2009 H1N1 influenza virus by PCR using markers for influenza A virus (InfA) (A), swine influenza A virus (swInfA) (B), and swine influenza virus H1 subtype (swH1) (C) from the CDC rRT-PCR protocol for detection and characterization of swine influenza virus. (A) Pools B and C were negative for influenza A virus. (B and C) Pools B, C, and D were negative for the swine influenza A virus and swine influenza virus H1 markers. All five pools were positive for RP. Each site tested the five pools three times on three different days. The error bars denote standard deviations of PCR results performed on three separate days.
Specimens from pools positive for the 2009 H1N1 influenza virus were individually extracted, and PCR was performed using the InfA, swInfA, swH1, and RP markers. All three sites correctly identified the negative specimens from positive pools (data not shown). Among the 50 specimens from the test panel, three were positive for the 2009 H1N1 influenza virus. All three sites successfully identified the three positive specimens. The CT values of the individual specimens with the InfA, swInfA, swH1, and RP markers were similar among the sites (Table 5 and data not shown). Pool E contained seasonal influenza viruses A/H1 and A/H3 and the 2009 H1N1. The three sites were also able to distinguish between specimens positive for seasonal influenza viruses and the 2009 H1N1 virus (data not shown).
Table 5.
Results for positive specimens from multisite studya
| Sample | Site |
CT value for: |
|||
|---|---|---|---|---|---|
| InfA | swInfA | swH1 | RP | ||
| 9 | A | 24 ± 1 | 25 ± 1 | 28 ± 0 | 26 ± 1 |
| B | 27 ± 2 | 27 ± 4 | 28 ± 0 | 27 ± 1 | |
| C | 24 ± 0 | 24 ± 0 | 28 ± 0 | 25 ± 0 | |
| 10 | A | 24 ± 2 | 26 ± 2 | 28 ± 0 | 25 ± 1 |
| B | 25 ± 2 | 27 ± 3 | 29 ± 1 | 27 ± 1 | |
| C | 24 ± 0 | 25 ± 0 | 32 ± 0 | 24 ± 0 | |
| 44 | A | 24 ± 1 | 25 ± 1 | 28 ± 1 | 25 ± 1 |
| B | 26 ± 2 | 25 ± 2 | 28 ± 0 | 25 ± 1 | |
| C | 23 ± 0 | 24 ± 0 | 28 ± 0 | 23 ± 1 | |
Specimens from pools that tested positive for the 2009 H1N1 influenza virus were re-extracted and retested by PCR individually. The average PCR results of samples 9, 10, and 44 reported by sites A, B, and C are represented above. Samples 9 and 10 are from pool A, and sample 44 is from pool E. Samples 1 to 8 from pool A were negative for influenza A virus (InfA), swine influenza A virus (swInfA), and swine influenza virus H1 (swH1) and were positive for RNase P (RP). Samples 41 and 45 from pool E were positive for influenza A, but negative for the swInfA and swH1 markers. InfA, influenza A virus matrix (M) gene target for detecting any influenza A virus; swInfA, influenza A virus nucleoprotein (NP) gene from swine for detecting swine influenza virus in the genetic lineage of 2009 pandemic influenza A virus (H1N1); swH1, highly conserved region of hemagglutinin (HA) gene for detecting the 2009 pandemic influenza A virus (H1N1), subtype H1; RP, human ribonucleoprotein (RP) for detecting human RNase P RNA, used with human clinical specimens to indicate that adequate isolation of nucleic acid resulted from extraction of the clinical specimen. The values are the means and standard deviations of the PCR results from three separate days.
DISCUSSION
In this study, we presented data demonstrating that a strategy of pooling specimens can be used to test for the presence of influenza virus using the CDC rRT-PCR swine flu panel (24). Our data showed that the increase in cellular material, including nucleic acid, due to pooling of multiple specimens did not affect detection of the 2009 H1N1 influenza virus by rRT-PCR (Table 1). Additionally, there was no loss of sensitivity or dilution of a positive specimen in a pool since the extraction of each pool was concentrated at the elution step. As expected, the pooling of multiple specimens positive for the 2009 A/H1N1 virus with higher concentrations of viral nucleic acid did not have an inhibitory effect on PCR, as demonstrated by the CT values of pools being similar to those of the specimen with the lowest CT value (Table 3). Furthermore, the presence of other respiratory viruses, such as adenovirus and parainfluenza virus, or multiple positive influenza virus specimens did not impede PCR amplification of the 2009 H1N1 influenza virus (Table 4). Finally, the results reported by three state public health laboratories testing a blinded pooling panel showed both inter- and intralaboratory reproducibility (Fig. 1 and Table 5). These results suggest that NP/throat swab specimens could be pooled, extracted, and tested for influenza virus by PCR with levels of detection and reproducibility similar to those obtained by processing each specimen individually.
The MagNA Pure TNA kit is one of the extraction methods validated for the CDC rRT-PCR swine flu panel (24). To optimize pooling of up to 10 specimens requires a larger input volume than that recommended for the MagNA Pure TNA kit (Roche Diagnostics, Indianapolis, IN). The large-volume TNA kit (Roche Diagnostics, Indianapolis, IN) allowed for processing of sample volumes up to 1 ml on the MagNA Pure LC instrument. Ten specimens were pooled, extracted, and concentrated upon elution using this kit. The sensitivity of the large-volume TNA kit was similar to that of the small-volume MagNA Pure TNA kit (Table 1). Detection of a specimen with a CT value in the high 20s was not compromised when mixed with four or nine negative specimens (Table 1 and data not shown). This demonstrated that viral nucleic acid is not diluted in pools of negative specimens, since the final elution volume of the large-volume TNA kit is 100 μl, the same as the elution volume of the small-volume MagNA Pure TNA kit recommended by the CDC rRT-PCR flu panel protocol. Hence, essentially all nucleic acid from the large-volume TNA kit is captured and not diluted. Others have also evaluated the MagNA Pure LC large-volume TNA kit and shown acceptable sensitivity when used in conjunction with real-time PCR for detection of other viruses, such as hepatitis B and C viruses (3, 11, 12, 13).
The pooling strategy proposed by this study does not necessitate large structural or workflow changes for laboratories currently performing the CDC rRT-PCR flu panel or the rRT-PCR swine flu panel using the MagNA Pure LC or other instruments recommended for these two protocols. Three state public health laboratories successfully tested a blinded influenza virus pooling panel without extensive supplementary training or the purchase of additional equipment. Laboratories, however, are required to develop and validate an algorithm and workflow scheme for tracking specimens in pools.
For laboratories lacking automated extraction instrumentation, manual extraction methods, such as the QIAamp viral RNA minikit (Qiagen, Valencia, CA), one of the manual extraction methods validated for the CDC rRT-PCR flu panel, can be used to extract and concentrate pools of specimens. However, laboratories should take into consideration that manual extraction methods can be more labor intensive than automated systems. Additional studies need to be performed in order to determine whether manual extraction methods will provide similar results using a pooling protocol and be useful in surge situations.
The decision to implement a pooling protocol should take into consideration the current positivity rate. The benefit of pooling is nullified if every pool yields a positive result, which may occur during periods of high positivity rate, and thus demands subsequent testing to reassess every specimen in the pool individually. One strategy to circumvent this is to alter the size of the pool to account for the prevalence of the situation. For instance, if the positivity rate is near 10%, pools of five specimens may prove more practical than 10 specimens. Pools of one positive and four negative specimens showed similar results in terms of CT values to pooling one positive and nine negative specimens (data not shown).
This investigation did not address the potential cost effectiveness of pooling influenza virus specimens. However, pooling schemes have been shown to increase testing capacity and lower the cost per test for detection of viral infections such as Chlamydia trachomatis, HIV, and hepatitis C (4, 10, 19, 22, 23). These studies also optimized the pooling strategy such that sensitivity was not diminished due to dilution of positive specimens in pools of increasing size. Laboratories should be aware, however, that a negative pool result would not distinguish between a true negative and an indeterminate/inconclusive result due to poor specimen collection or handling. Theoretically, our pooling design, if applied properly, can increase testing capacity and conserve reagents during times of pandemic surveillance, surge testing, clearing of backlogs, and in limited-resource settings. In addition, this study may serve as a model for pooling specimens for diagnosis of other infectious diseases.
In summary, we have demonstrated that NP/throat swab specimens can be pooled and tested for the presence of influenza virus without sacrificing sensitivity. The pooling scheme can increase testing capacity if used in combination with an automated extraction method, such as the MagNA Pure LC instrument, and the CDC real-time PCR assay for influenza virus.
ACKNOWLEDGMENTS
We acknowledge Stephen Lindstrom (CDC) and Roy Johnson (Battelle) for their intellectual contributions to this study and Denny Russell (Washington State Public Health Laboratory), Sandy Smole (Massachusetts State Public Health Laboratory), and T. J. Whyte (Wisconsin State Laboratory of Hygiene) for participating in the clinical trial. We also acknowledge the Association of Public Health Laboratories (Tricia Aden) for assistance in organizing the clinical trial and publication of the manuscript.
This research was supported in part by an appointment to the Emerging Infectious Diseases Fellowship Program (Tam T. Van was an Emerging Infectious Diseases Research Fellow) administered by the Association of Public Health Laboratories (APHL) and funded by the Centers for Disease Control and Prevention (CDC) cooperative agreement number U38 HM000012.
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the CDC.
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Centers for Disease Control and Prevention 2009. Swine influenza A (H1N1) infection in two children—Southern California, March–April 2009. MMWR Morb. Mortal. Wkly. Rep. 58:400–402 [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention 2010. Update: influenza activity—United States, August 30, 2009–January 9, 2010. MMWR Morb. Mortal. Wkly. Rep. 59:38–43 [PubMed] [Google Scholar]
- 3. Cook L, Ng K-W, Bagabag A, Corey L, Jerome KR. 2004. Use of the MagNA Pure LC automated nucleic acid extraction system followed by real-time reverse transcription-PCR for ultrasensitive quantitation of hepatitis C virus RNA. J. Clin. Microbiol. 42:4130–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Currie MJ, McNiven M, Yee T, Schiemer U, Bowden FJ. 2004. Pooling of clinical specimens prior to testing for Chlamydia trachomatis by PCR is accurate and cost saving. J. Clin. Microbiol. 42:4866–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Emmanuel JC, Bassett MT, Smith HJ, Jacobs JA. 1988. Pooling of sera for human immunodeficiency virus (HIV) testing: an economical method for use in developing countries. J. Clin. Pathol. 41:582–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Food and Drug Administration 30 September 2008. FDA clears new CDC test to detect human influenza. U.S. Food and Drug Administration, Washington, DC: http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm123591.htm [Google Scholar]
- 7. Food and Drug Administration 18 December 2009. Letter of authorization: CDC swine influenza virus real-time RT-PCR detection panel. U.S. Food and Drug Administration, Washington, DC: http://www.fda.gov/MedicalDevices/Safety/EmergencySituations/ucm195385.htm [Google Scholar]
- 8. Food and Drug Administration 23 August 2011, posting date CDC human influenza virus real-time RT-PCR diagnostic panel. 510(K) number k111507. U.S. Food and Drug Administration, Washington, DC: http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfivd/index.cfm?db=pmn&id=K111507 [Google Scholar]
- 9. Gallarda JL, Dragon E. 2000. Blood screening by nucleic acid amplification technology: current issues, future challenges. Mol. Diagn. 5:11–22 [DOI] [PubMed] [Google Scholar]
- 10. Garcia Z, et al. 1996. Evaluation of a pooling method for routine anti-HCV screening of blood donors to lower the cost burden on blood banks in countries under development. J. Med. Virol. 49:218–222 [DOI] [PubMed] [Google Scholar]
- 11. Germer JJ, et al. 2003. Evaluation of the MagNA Pure LC instrument for extraction of hepatitis C virus RNA for the COBAS AMPLICOR hepatitis C virus test, version 2.0. J. Clin. Microbiol. 41:3503–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Germer JJ, Harmsen WS, Mandrekar JN, Mitchell PS, Yao JDC. 2005. Evaluation of the COBAS TaqMan HCV test with automated sample processing using the MagNA Pure LC instrument. J. Clin. Microbiol. 43:293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gintowt AA, Germer JJ, Mitchell PS, Yao JDC. 2005. Evaluation of the MagNA Pure LC used with the TRUGENE HBV genotyping kit. J. Clin. Virol. 34:155–157 [DOI] [PubMed] [Google Scholar]
- 14. Hourfar MK, et al. 2008. Experience of German Red Cross blood donor services with nucleic acid testing: results of screening more than 30 million blood donations for human immunodeficiency virus-1, hepatitis C virus, and hepatitis B virus. Transfusion 48:1558–1566 [DOI] [PubMed] [Google Scholar]
- 15. Lindan C, et al. 2005. Utility of pooled urine specimens for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in men attending public sexually transmitted infection clinics in Mumbai, India, by PCR. J. Clin. Microbiol. 43:1674–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lisby G, Scheibel J, Abrahamsson L-O, Christensen ES, Paloheimo S. 1994. Detection of Chlamydia trachomatis in individual and pooled endocervical and urethral scrapes by a commercially available polymerase chain reaction. APMIS 102:797–800 [PubMed] [Google Scholar]
- 17. Marshall DJ, et al. 2007. Evaluation of a multiplexed PCR assay for detection of respiratory viral pathogens in a public health laboratory setting. J. Clin. Microbiol. 45:3875–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mine H, et al. 2003. High throughput screening of 16 million serologically negative blood donors for hepatitis B virus, hepatitis C virus and human immunodeficiency virus type-1 by nucleic acid amplification testing with specific and sensitive multiplex reagent in Japan. J. Virol. Methods 112:145–151 [DOI] [PubMed] [Google Scholar]
- 19. Morre SA, et al. 2001. Pooling cervical swabs for detection of Chlamydia trachomatis by PCR: sensitivity, dilution, inhibition, and cost-saving aspects. J. Clin. Microbiol. 39:2375–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ohnuma H, et al. 2001. The first large-scale nucleic acid amplification testing (NAT) of donated blood using multiplex reagent for simultaneous detection of HBV, HCV, and HIV-1 and significance of NAT for HBV. Microbiol. Immunol. 45:667–672 [DOI] [PubMed] [Google Scholar]
- 21. Roth WK, et al. 2002. Yield of HCV and HIV-1 NAT after screening of 3.6 million blood donations in central Europe. Transfusion 42:862–868 [DOI] [PubMed] [Google Scholar]
- 22. Rours GIJG, et al. 2005. Use of pooled urine samples and automated DNA isolation to achieve improved sensitivity and cost-effectiveness of large-scale testing for Chlamydia trachomatis in pregnant women. J. Clin. Microbiol. 43:4684–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shang G, et al. 2009. Two HBV DNA+/HBsAg- blood donors identified by HBV NAT in Shenzhen, China. Transfus. Apher. Sci. 41:3–7 [DOI] [PubMed] [Google Scholar]
- 24. World Health Organization 2009. CDC protocol of realtime RTPCR for influenza A(H1N1). World Health Organization, Geneva, Switzerland: http://www.who.int/csr/resources/publications/swineflu/realtimeptpcr/en/index.html [Google Scholar]