Abstract
The chitin synthase structural gene WdCHS5 was isolated from the black fungal pathogen of humans Wangiella dermatitidis. Sequence analysis revealed that the gene has a myosin motor-like-encoding region at its 5′ end and a chitin synthase (class V)-encoding region at its 3′ end. Northern blotting showed that WdCHS5 is expressed at high levels under conditions of stress. Analysis of the 5′ upstream region of WdCHS5 fused to a reporter gene indicated that one or more of the potential regulatory elements present may have contributed to the high expression levels. Disruption of WdCHS5 produced mutants that grow normally at 25°C but have severe growth and cellular abnormalities at 37°C. Osmotic stabilizers, such as sorbitol and sucrose, rescued the wild-type phenotype, which indicated that the loss of WdChs5p causes cell wall integrity defects. Animal survival tests with a mouse model of acute infection showed that all wdchs5Δ mutants are less virulent than the parental strain. Reintroduction of the WdCHS5 gene into the wdchs5Δ mutants abolished the temperature-sensitive phenotype and reestablished their virulence. We conclude that the product of WdCHS5 is required for the sustained growth of W. dermatitidis at 37°C and is of critical importance to its virulence.
Wangiella (Exophiala) dermatitidis is a polymorphic, dematiaceous (melanized) fungal pathogen of humans which exists predominantly as a yeast form in vitro but can be easily manipulated to undergo morphological transitions to produce isotropically enlarged yeast, multicellular forms and various types of hyphae (10, 11, 17, 44, 45, 54). In vivo, this fungus also produces various morphological forms, such as budding yeast, pseudohyphae, true hyphae, isotropically enlarged yeast cells, and sclerotic bodies (18, 21, 31). Although considered a paradigm for phaeohyphomycosis, because of its increasing detection in cutaneous, subcutaneous, and central nervous system infections (18, 21, 22), it is better known as a model for the more than 100 other dematiaceous agents of mycoses (11, 44, 45). The well-defined polymorphic features and well-documented cell wall structure of W. dermatitidis, together with its increasing molecular tractability, make it also an excellent system for studies of fungal cell wall biosynthesis, as well as fungal cell wall-related virulence factors (27, 44). To date, both melanin and chitin have been shown to have relevance to the full virulence of this fungus (12, 19, 44, 50)
Chitin, the β(1,4)-linked homopolymer of N-acetylglucosamine, is an essential structural component of fungal cell walls and plays an important role in fungal morphogenesis (8, 35, 37). Chitin synthases (UDP-N-acetyl-d-glucosamine:chitin 4-β-N-acetylglucosamine transferase; EC 2.4.1.16), which are membrane-bound proteins, are responsible for the synthesis and deposition of this chitin (28, 30, 35). The presence of distinct classes of chitin synthases was initially indicated by the identification and analysis of multiple chitin synthase genes in various fungi, which have more recently been distributed between two families (3, 4, 28, 32, 35, 37). Extensive study of the chitin synthases of Saccharomyces cerevisiae (ScChsp) has documented that a specific function can be assigned to each of its three chitin synthases (8, 33). Briefly, ScChs1p (class I) is responsible for the synthesis of chitin after cell separation and counterbalances any chitinase activity: it is thus considered to be a repair enzyme (5, 6). ScChs2p (class II) is responsible for chitin deposition in the primary septum (40, 41). Finally, ScChs3p (class IV) is responsible for chitin deposition in the ring and lateral cell wall and contributes to the synthesis of most of the cell wall chitin during vegetative growth (36, 40).
Unlike the situation in S. cerevisiae, there is no general rule for defining the specific function of each chitin synthase among other fungi. To the contrary, it has been suggested that chitin synthases of the same class may play different roles in different fungi (28). Furthermore, the complicated life cycles of filamentous fungi may require them to have more chitin synthases than yeast, and isozymes of as many as six classes have been identified in some aspergilli (4, 7, 13, 24, 42). By use of gene disruption technologies, many of these chitin synthases have been proven to contribute directly to cell wall integrity and cell morphogenesis (28). In pathogenic fungi, some have also been shown to contribute directly or indirectly to virulence (28, 50).
In W. dermatitidis, chitin is found throughout the cell wall in hyphae and isotropic forms, but the primary site of chitin localization in yeast cells is in septal regions (9, 15, 23). Initially, four different chitin synthase structural genes (WdCHS) were identified in this fungus; each of these encodes a member of a different chitin synthase (WdChsp) class, as follows: WdCHS2, class I; WdCHS1, class II; WdCHS3, class III; WdCHS4, class IV (44). Characterization of mutants with each of these four WdCHS genes disrupted singly or in all possible double-mutant combinations revealed that no single WdChsp is essential for the viability of W. dermatitidis at 25°C (44). However, double mutants with both WdCHS1 and WdCHS2 disrupted grow poorly at 25°C, are incapable of growth at 37°C, and are avirulent (44, 48), whereas mutants with both WdCHS2 and WdCHS3 disrupted have reduced virulence but grow normally at both 25 and 37°C (50). In this report, we characterize a fifth chitin synthase structural gene (WdCHS5) in W. dermatitidis; this gene encodes a class V chitin synthase with a myosin motor-like domain at its N-terminal region. We also report that disruption of this single gene produced mutants that were normal at 25°C but could not sustain growth at 37°C and were less virulent. Our discovery of WdCHS5 broadens the range of this kind of gene to a polymorphic fungus, which we think will eventually allow a more definitive functional characterization of a class V isozyme with relevance to human pathogenicity.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The W. dermatitidis strains used in this work are listed in Table 1. The wild-type W. dermatitidis strain 8656 (ATCC 34100 [E. dermatitidis CBS525.76]) and the temperature-sensitive mutants Mc3 (wdcdc2; ATCC 38716) and Hf1 have been described in detail previously (10, 49, 54). Routine propagation of these strains was carried out in the rich medium YPD (2% peptone, 1% yeast extract, 2% dextrose) as reported previously (48, 56). The synthetic defined (SD) medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.2% ammonium nitrate, 0.1% asparagine, 1% glucose) and the media for inducing development of the sclerotic or hyphal morphologies were prepared as described previously (49). Methods for transformation by electroporation of intact yeast cells have also been described previously (48, 56). Drug selection plates for isolating W. dermatitidis transformants were made by adding agar (1.5%, wt/vol) and hygromycin B (HmB; Sigma, St. Louis, Mo.) to YPD for selection for resistance conferred by the hygromycin phosphotransferase (hph) gene at a final concentration of 50 μg/ml or by adding chlorimuron ethyl (provided by J. Sweigard, Dupont Co., Wilmington, Del.) to SD for detection of resistance conferred by the sulfonyl urea resistance (sur) gene at a final concentration of 20 μg/ml. Escherichia coli XL-1 Blue (Stratagene, La Jolla, Calif.), which was used for subcloning and plasmid preparation, was grown in Luria-Bertani medium supplemented with ampicillin (100 μg/ml) or chloramphenicol (25 μg/ml). Growth rates of the wild type and the wdchs5Δ mutants were determined by three different methods, using cultures inoculated with 106 cells/ml in 50 ml of YPD. Samples from cultures grown at 25 and 37°C were collected at 4- or 8-h intervals and subjected to spectrophotometric, hemacytometric, and plate viable counting procedures. Three independent measurements were performed, and the average at each time was used for the growth plots.
TABLE 1.
Strains used in this study
| Strain or mutationa | Parent strain | Genotype or properties | Reference or source |
|---|---|---|---|
| Wd8656 | Wild type | ATCC 34100 | |
| Mc3 | Wd8656 | wdcdc2; grows isotropically at 37°C | ATCC 38716 |
| Hf1 | Wd8656 | Temperature-sensitive mutant; grows as hyphae at 37°C | 49 |
| wdchs5Δ11 | Wd8656 | wdchs5::hph, WdCHS5 replacement | This work |
| wdchs5Δ236 | Wd8656 | wdchs5::hph, myosin-motor domain Δb | This work |
| wdchs5Δ316 | Wd8656 | wdchs5::hph, chitin synthase domain Δb | This work |
| wdchs5Δ11-1 | wdchs5Δ11 | wdchs5::hph-WdCHS5-sur | This work |
| wdchsΔ236-1 | wdchs5Δ236 | wdchs5::hph-WdCHS5-sur | This work |
| wdchsΔ316-1 | wdchs5Δ316 | wdchs5::hph-WdCHS5-sur | This work |
All mutants used in this study were derived from the wild-type parental strain 8656.
Δ indicates site of WdCHS5 disruption.
Nucleic acid manipulations and plasmids.
W. dermatitidis genomic DNA was isolated either as described previously (48, 56) by spheroplasting with Zymolyase 20T (ICB Biomedicals, Inc., Aurora, Ohio) followed by detergent lysis or by using a glass bead method adapted from reference 2. Briefly, the latter method involved pelleting and washing cells with distilled, deionized water by centrifugation, adding breaking buffer (2% Triton X-100, 1% sodium dodecyl sulfate, 100 mM NaCl, 10 mM Tris-Cl [pH 8.0], 1 mM EDTA), glass beads (diameter, 400 to 520 μm; Thomas Scientific, Swedesboro, N.J.), and phenol-chloroform, and then vortexing vigorously using a Multi-Tube Vortexer (VWR International, West Chester, Pa.) for 3 min prior to the addition of Tris-EDTA (pH 8.0). After brief vortexing and centrifugation, the genomic DNA was precipitated using 100% ethanol, then washed with 75% ethanol and treated with RNase (Roche Applied Science, Indianapolis, Ind.). Methods for the isolation of total RNA and for Southern and Northern blot analyses were as described previously (48, 56). DNA fragments used for probes were labeled with [α-32P]dCTP by using the DECA prime II DNA labeling kit (Ambion, Austin, Tex.). PCR amplification conditions and primers chs51, chs53, chs5F, and chs5R, used for amplifying the WdCHS5 fragment, have been described previously (19, 47). Primers used for reverse transcription-PCR (RT-PCR) were designed as follows: pATG, 5′-ATGGCCACTCGAGGGAACGTC; pIntron1, 5′-GCTTTGGAAGCAGTTGGGTCG; pTGA, 5′-TCACAGTTGCCCAGACAAAAT; pIntron2, 5′-CTTCCCACTGGCTCTGTCTAT. Primers used to amplify 5′ upstream sequences were as follows: psmaI, 5′-TGTCCCGGGGGTGAACTTCAATGGC (the SmaI site is underlined); p1.2, 5′-TCAGGGCCCATCAGAAGGAGCGGTA; p1.0, 5′-TCAGGGCCCAACTTGACCTCGACTT; p0.88, 5′-TCAGGGCCCAGAGGTAGGTTGGAAT; p0.68, 5′-TCAGGGCCCTATTCTAGAGGGTCTA; and p0.2, 5′-TCAGGGCCCTTGATTACGACTTGA (the ApaI sites are underlined). Methods for the cDNA synthesis and subsequent PCR and RT-PCR have been described previously (47). Methods used for construction of the cosmid library and the ZAPII cDNA library have also been described previously (12, 49). The partial genomic library was constructed by using genomic DNA completely digested with EcoRI. Following electrophoresis, the resulting 2.5- to 3.5-kb fragments were excised from the gel and ligated into the pBS-KS(+) vector, which was cut by EcoRI and dephosphorylated with calf intestine alkaline phosphatase (Promega, Madison, Wis.). The ligation product was then transformed into XL-l Blue competent cells, and the resulting subgenomic library of about 6,000 independent clones was screened by colony hybridization using the PCR product generated by primers chs51 and chs53 as a probe.
The WdCHS5 integrative gene disruption plasmids, pHB0320 and pHB0510, were constructed by cloning 1.8-kb SacI and 1.2-kb SalI fragments from the 5′ and 3′ ends of WdCHS5 into corresponding sites of vectors pCB1636 and pCB1004 (50), respectively. After the resulting vectors were linearized with BclI or BstEII, respectively, to target integration into genomic sites of WdCHS5, they were used to transform W. dermatitidis competent cells by electroporation as described previously (48, 54). To construct the one-step replacement disruption vector pHB0280, the 1-kb EcoRI-BamHI fragment of the 3′ end was cloned into the EcoRI and BamHI sites, and the 0.5-kb KpnI-PstI fragment (blunt ended by filling in the PstI site with Klenow enzyme) of the 5′ end of WdCHS5 was cloned into the KpnI and ApaI sites (blunt ended by filling in the ApaI site with Klenow enzyme), of vector pCB1636, which positioned the hph gene to the middle of the construct. The 4.5-kb fragment obtained by digestion of pHB0280 with KpnI and BamHI was then used for transformation. The complementation vector pHB2080, which contained the full-length WdCHS5 gene, was constructed by cloning a 3-kb EcoRI fragment of the 3′ end of WdCHS5 into the EcoRI site of plasmid pHB0051, which contained the 5′ end of WdCHS5. The combined full-length WdCHS5 was then released with BssHII digestion, filled in with Klenow enzyme, and subsequently blunt-end ligated into the SmaI site of pCB1551 (provided by J. Sweigard), which has a sur gene marker for selection. The resulting circular plasmid was then used for complementation of the wdchs5Δ mutants directly. Putative randomly reconstituted strains were selected by prescreening for temperature-insensitive revertants, which were subsequently confirmed to contain WdCHS5 by Southern blot analysis. All the plasmids for analysis of the 5′ upstream sequence of WdCHS5 were derived from pYEX303-gal (53). Briefly, the 5′ upstream sequences of different lengths before the ATG start codon were amplified with primer psmalI (which has a SmaI restriction enzyme site) from one end and primers p1.2, p1.0, p0.88, p0.68, and p0.2 (which have ApaI restriction enzyme sites) from the other end, respectively. All the PCR products were subjected to SmaI digestion first and then to partial ApaI digestion. The corresponding 1.2-, 1.0-, 0.88-, 0.68-, and 0.2-kb fragments, and the 0.45-kb fragment obtained from complete digestion of the 1.2-kb PCR product with SmaI and ApaI, were used to replace the glaA promoter in plasmid pYEX303-gal to generate pHB8040, pHB9000, pHB9010, pHB9020, pHB9030, and pHB8050, respectively. Prior to transformation, the plasmids were linearized with FseI. The WdPKS1 fragment incorporated into these plasmids allowed strains with site-specific integrations among HmB-resistant transformants to be identified as white colonies (53).
Photomicroscopy.
Light photomicroscopy of W. dermatitidis wild-type and mutant cells was performed by using an Olympus BX-60 microscope. The procedures for staining cell wall chitin with Calcofluor (Sigma) or staining nuclei with DAPI (4′,6′-diamidino-2-phenylindole; Accurate Chemical, Westbury, N.Y.) have been described previously (54). Scanning electron microscopy was carried out as described previously (48), except that after fixation with cacodylate-buffered 2.5% glutaraldehyde, cells were not attached to coverslips pretreated with polylysine.
Chitin synthase activity, chitin content, and β-galactosidase and alkaline phosphatase assays.
Chitin synthase activities, chitin contents, and β-galactosidase activities were measured by methods described previously (48, 49, 56). The relative activities of alkaline phosphatase were determined by the modified method of Nombela and coworkers (26). All assays were carried out at least three times. Differences among groups were evaluated for statistical significance by the parametric one-way analysis of variance using the Newman-Keuls method for paired data. Statistical analyses were performed with the PRISM software package (version 2.0; GraphPad Software, Inc., San Diego, Calif.). Probability values of <0.05 were considered significant.
DNA sequence analysis.
WdCHS5 was sequenced by the Institute of Cellular and Molecular Biology of the University of Texas at Austin. The deduced amino acid sequence of WdCHS5 was determined, and sequences were compared, by using the BLAST software system from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.rlm.nih.gov/BLAST) and the ClustalW program from the European Molecular Biology Laboratory (EMBL) (http://www.ebi.ac.uk/clustalw). Promoter sequence analysis was performed by using MacInspect software and the TRANSFAC program (version 4.0; BCM Search Launcher).
Virulence tests and tissue burden analysis.
Tests for virulence in an immunocompetent mouse model system were performed as described previously (12, 48). Survival fractions in the virulence tests were calculated by the Kaplan-Meier method, and survival curves were tested for significant difference (P < 0.01) by the Mantel-Haenszel test using GraphPad Prism software (version 3.0 for Windows). Probability values of <0.05 were considered significant. Tissue burden analysis involved injecting the wdchs5Δ11 mutant into mice and sacrificing them at days 1, 3, 5, and 10 postinfection to determine the fungal burdens of the brain, kidney, liver, and spleen.
Nucleotide sequence accession number.
The nucleotide sequence of WdCHS5 was assigned GenBank accession number AF469116.
RESULTS
The WdCHS5 gene encodes a class V chitin synthase (WdChs5p) with a myosin motor-like domain
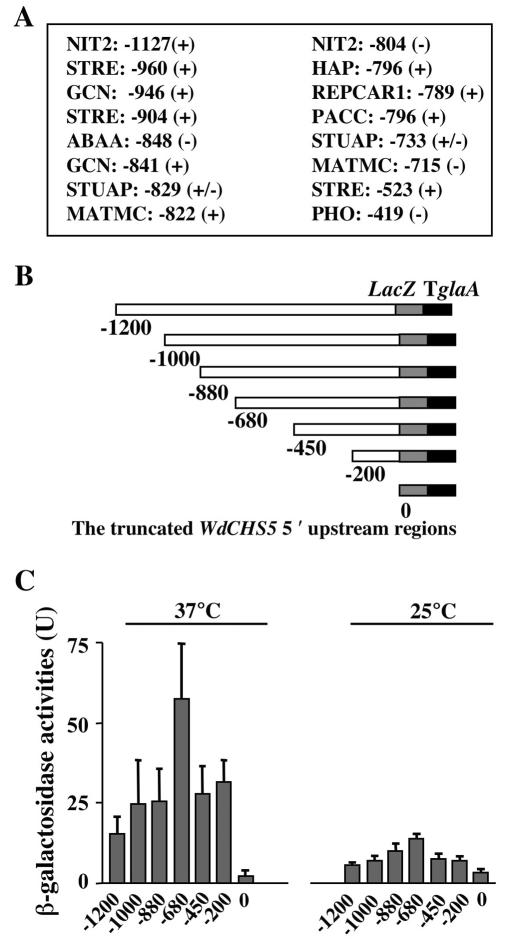
A 362-bp PCR product was amplified by using the degenerate primers chs51 and chs53 (19). Sequence analysis of the PCR product showed that it encoded a peptide with high homology to other class V chitin synthases. To isolate the whole gene, a cosmid library and a 3-kb EcoRI subgenomic library were screened by colony hybridization using the WdCHS5 PCR product as a probe. Clones from each were then isolated, shown to contain the WdCHS5 gene, and subjected to restriction mapping (19) and sequence analysis. The nucleotide sequence of the cloned WdCHS5 gene contained a single open reading frame of 5,655 bp interrupted by two introns of 53 and 57 bp at its 5′ and 3′ ends, respectively (data not shown). The WdCHS5 gene encoded a putative protein (WdChs5p) of 1,885 amino acids that had six putative transmembrane helices, a calculated mass of 208.9 kDa, and a pI of 7.76. Both introns had a consensus splice site that began with GT and ended with AG and were subsequently confirmed by RT-PCR (Fig. 1) and by sequence analysis of the RT-PCR products (data not shown). Comparisons by BLAST and ClustalW analysis (NCBI and EMBL, respectively) of the deduced protein sequence of WdChs5p with those of other class V chitin synthases having myosin motor-like domains indicated that it had 67, 66, 65, 65, 63, and 41% identity to CsmA of Aspergillus nidulans (13), Chs2 of Blumeria graminis (55), ChsA of Glomerella graminicola (1), ChsV of Fusarium oxysporum (20), Csm1 of Magnaporthe grisea (34), and PbrChs5p of Paracoccidioides brasiliensis (sequence analysis available at http//www.ncbi.nlm.nih.gov), respectively. The affiliation of WdChs5p with the class V family of chitin synthases was further supported by the identification in its N-terminal domain (first 800 residues) of a myosin motor-like region, which to date is associated only with members of this isozyme class (28). Further support for a class V affiliation for WdChs5p was provided by the identification of a characteristic ATP- or GTP-binding site motif (P-loop; GESGAGKT), located in the myosin motor-like region from amino acid residues 99 to 106, and the presence of switch I (TASKAG) and switch II (DFPGF) motifs at residues 148 to 153 and 407 to 411, respectively. Two putative unconventional TATA boxes were also found at bp −605 and −648. Cloning of the 3′ end of the cDNA of the WdCHS5 gene by cDNA library screening and subsequent analysis revealed that the polyadenylation signal sequence began at a position 225 bp downstream of the stop codon.
FIG. 1.
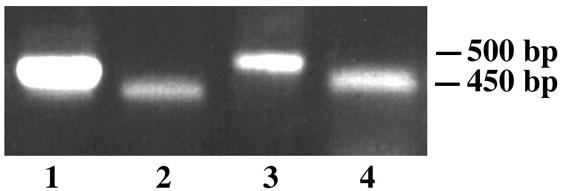
RT-PCR confirmation of the introns of WdCHS5. Primers pATG and pIntron1 were used for amplification of intron 1 (53 bp) from cDNA (lane 4) and genomic DNA (lane 3), whereas primers pTAG and pIntron2 were used for amplification of intron 2 (57 bp) from cDNA (lane 2) and genomic DNA (lane 1). The introns were also confirmed by direct sequencing of the RT-PCR products (data not shown).
Stress conditions result in increased cellular WdCHS5 mRNA levels.
Regulation of WdCHS5 expression was investigated by Northern blot analysis using a WdCHS5-specific PCR probe amplified with primers chs5F and chs5R. Total RNA for these assays was obtained from cells grown not only under conditions that included culture at elevated temperature (37°C) for the wild type and two temperature-sensitive morphological mutants (Mc3 and Hf1), in which WdCHS5 expression had been assayed previously by semiquantitative RT-PCR (47), but also under additional stress conditions that are known to initiate development of the sclerotic morphology (acidic conditions or Ca2+ limitation induced by increasing EGTA concentrations) or of hyphae (induced by nitrogen limitation) in the wild type. In response to all these conditions, WdCHS5 transcripts were detected at higher levels in stressed cells than in control cells (Fig. 2). Because a similar differential expression pattern was found previously for WdCHS3 (49), it was not surprising to find two identical regulatory elements, REPCAR1 and STUAP, and several similar regulatory elements, such as STRE, ABAA, and HAP, in the upstream regions of WdCHS3 (data not shown) and WdCHS5 (Fig. 3A), suggesting that these two genes may have similar mechanisms for regulating their transcription.
FIG. 2.
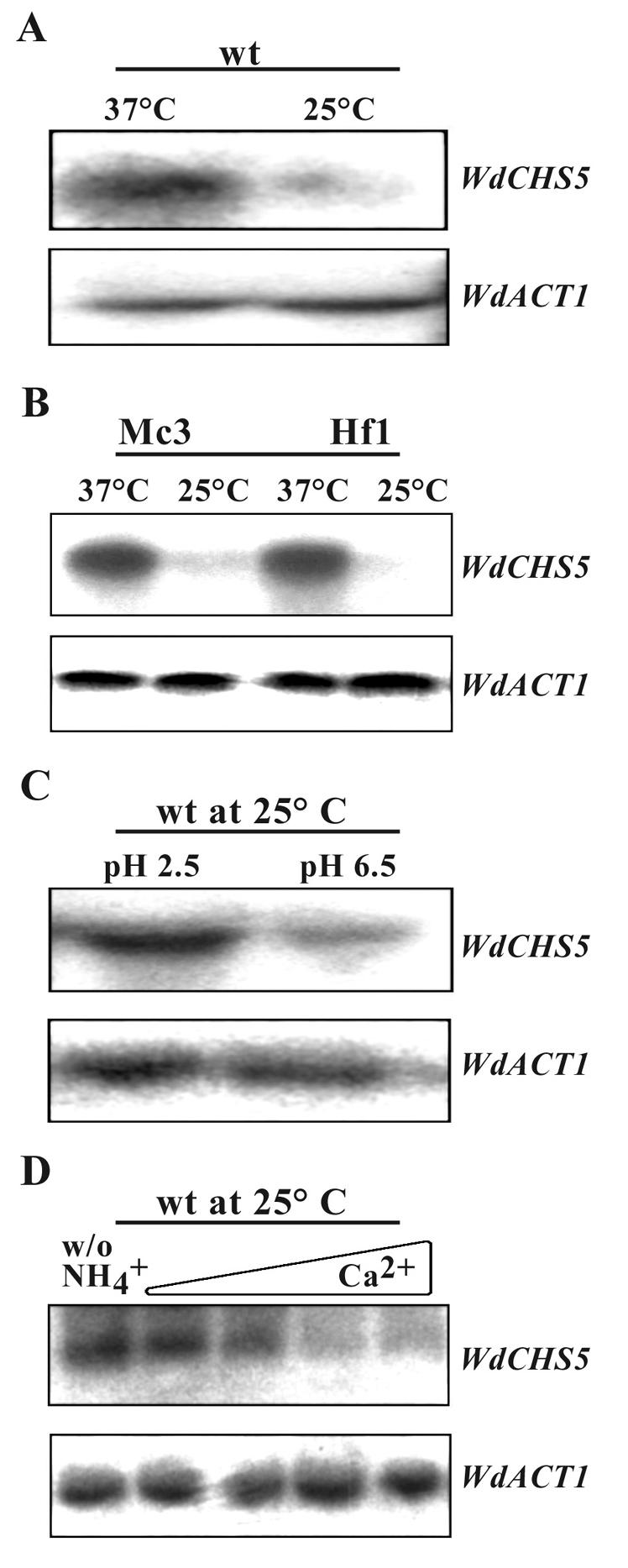
Northern blot analysis of WdCHS5 expression. Samples of total RNA were prepared from the wild-type strain (wt) grown in YPD at 37 or 25°C (A), the temperature-sensitive mutants Mc3 and Hf1 grown in YPD at 37 or 25°C (B), (C) the wild-type strain grown at 25°C in modified Czapek dextrose (MCD) broth at pH 2.5 or 6.5 (C), and the wild-type strain grown at 25°C in synthetic medium (SD) broth (pH 6.5) (D) without nitrogen (first lane) or with the addition of 20, 5, 0.5, or 0 mM EGTA (second to fifth lanes, respectively). After being probed with WdCHS5, the same membranes for each sample set (upper panels) were then stripped and probed with WdACT as controls to show the approximately equal amounts of RNA loading (lower panels).
FIG. 3.
Analysis of the 5′ URS of WdCHS5. (A) Putative binding sites for transcription factors. (B) Scheme of the upstream region of WdCHS5 and truncation sites of constructs used for the β-galactosidase expression studies. (C) Expression levels of WdCHS5::LacZ reporter fusions expressed in cells grown at 25 and 37°C.
The 5′ upstream sequence of WdCHS5 contains important regulatory regions.
A series of 5′ deletion fragments in the WdCHS5 upstream regulatory sequence (URS) was fused in frame with the LacZ gene (Fig. 3B) and then used to replace the original glaA promoter of plasmid pYEX303-gal, which also contains a WdPKS1 targeting sequence. The use of these constructs ensured that all alleles were integrated into the same nonessential WdPKS1 genomic locus, which is required for melanin biosynthesis (49, 53). After the constructs were linearized in the WdPKS1 sequence with FseI and transformed into yeast cells, transformants were selected for resistance to HmB, and mutants with a site-specific integration were identified by production of albino colonies. LacZ expression in cells grown at 25 and 37°C was quantitatively assessed by a β-galactosidase activity assay and by use of o-nitrophenyl-β-galactosidase as a chromogenic substrate. The resulting data (Fig. 3C) suggested two conclusions. First, all samples from cells grown at 37°C, except that from cells transformed with the plasmid control without any WdCHS5 URS, had significantly higher levels of β-galactosidase activity than those with corresponding constructs from cells grown at 25°C, a finding that was consistent with the Northern blot analysis results (Fig. 2). Second, the most dramatic changes in β-galactosidase activities were observed among samples from cells with constructs having truncations between bp −880 and −450, which is the region that includes most of the potential cis-acting elements identified by our sequence analysis. This finding indicated that at least one negative regulator binding sequence exists between bp −880 and −680 and that another regulatory binding site(s) is localized between bp −680 and −450.
Disruption of WdCHS5 produces mutants that are hyperpigmented and die at 37°C but are like the wild type at 25°C.
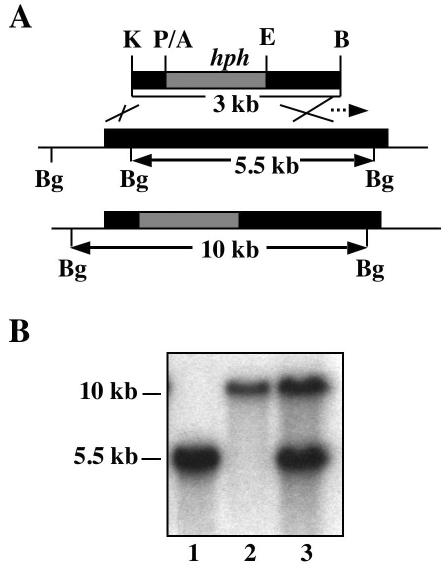
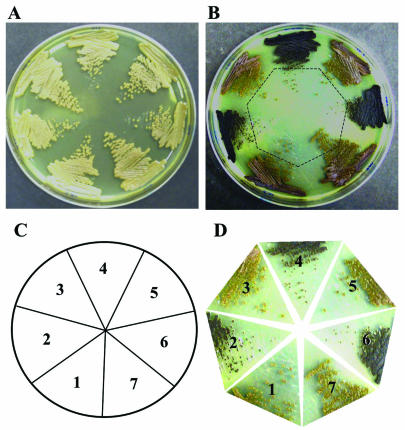
To begin to elucidate the functions of WdChs5p, as well as the possible function of each of its domains, three different disruption vectors were constructed. Two were then used for site-specific integrative gene disruptions that targeted the regions encoding the chitin synthase domain and the myosin motor-like domain, respectively (data not shown). The third was used for one-step replacement of the entire WdCHS5 gene (Fig. 4A). Southern blot analysis then identified numerous mutants (wdchs5Δ) of each type with site-specific gene disruptions that showed the expected band shifts for WdCHS5 (Fig. 4B; data shown only for the wdchs5Δ11 disruption mutant). All three types of wdchs5Δ mutants had identical phenotypes, which differed significantly from that of the wild type at 37°C but not at 25°C. For example, at 25°C, all wdchs5Δ disruption mutant strains grew normally in the manner of the wild-type strain on YPD agar medium (Fig. 5A). In contrast, at 37°C, the differences between the three types of wdchs5Δ disruption mutants and the wild type were very apparent on the agar medium by 72 h (Fig. 5B) and became even more obvious as time passed: the mutant growth became much darker, and isolated colonies were smaller than wild-type colonies (compare colony pigmentation in Fig. 5B; compare colony sizes in Fig. 5D, sector 1, with those in sectors 2, 4, and 6). Complementation of each type of wdchs5Δ disruption mutant with WdCHS5 returned each strain to its wild-type phenotype (compare colony pigmentation in Fig. 5B; compare colony sizes in Fig. 5D, sector 1, with those in sectors 3, 5, and 7). Darkening of wdchs5Δ mutants occurred similarly in YPD broth but was not apparent with the complemented strains grown identically (data not shown).
FIG. 4.
Disruption of WdCHS5 by one-step gene replacement. (A) Strategy for construction of the replacement vector. Abbreviations: K, KpnI; P, PstI; A, ApaI; E, EcoRI; B, BamHI; Bg, BglII. Broken arrow indicates the probe used for Southern blot analysis. (B) Southern blot analysis of the wild type (lane 1), a representative disruption mutant (wdchs5Δ11) (lane 2), and a representative complementation strain (wdchs5Δ11-1) (lane 3).
FIG. 5.
Colony characteristics of the wild-type strain, representative wdchs5Δ disruption mutants, and corresponding wdchs5Δ disruption mutants complemented with WdCHS5 after growth on YPD agar medium for 3 days. (A) Strains incubated at 25°C. (B) Strains incubated at 37°C. (C) Key to sectors: 1, wild type; 2, wdchs5Δ11; 3, wdchs5Δ11-1; 4, wdchs5Δ236; 5, wdchs5Δ236-1; 6, wdchs5Δ316; 7, wdchs5Δ316-1. (D) Enlarged images of the isolated colonies shown in each sector of panel B.
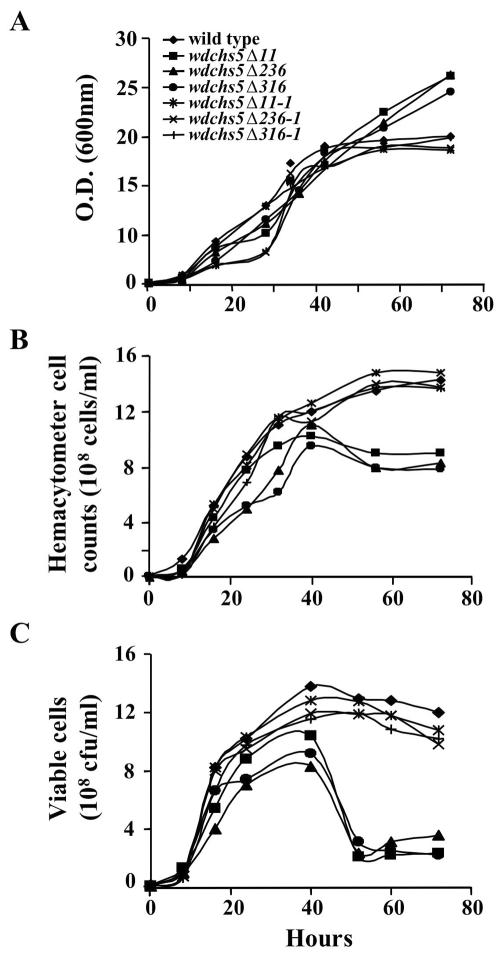
Subsequent quantitative growth studies of the wdchs5Δ mutants and the wild-type strain in YPD broth confirmed that disruption of WdCHS5 did not affect the kinetics of growth at 25°C when growth was measured by spectrophotometric, hemacytometric, and plate viable counting methods (data not shown). However, each of these methods provided somewhat different growth kinetic patterns when the mutants and the wild type were grown at 37°C (Fig. 6). Although subtle, the discrepancies detected were easily explained by correlating the microscopic and culture color change observations previously noted with data showing that death and lysis were not uncommon among cells grown at the higher temperature (see Fig. 7 and 8 below). For example, the optical density of the wdchs5Δ cultures at 600 nm continued to increase even after 40 h (Fig. 6A). However, this apparent increase was due to the mutant cells becoming much darker than the wild-type cells and not because there was an increase in the number of cells, as shown by both hemacytometry counting and viable counting procedures (Fig. 6B and C). More importantly, and as shown by the colony count data, the number of viable wdchs5Δ cells decreased rapidly after 40 h. Taken together, these results suggested that the wdchs5Δ mutants grew normally at 25°C but at 37°C lost viability during late-log or early-stationary phase. This strongly indicated that WdChs5p is important for sustained cell growth and cell maturation at 37°C. This hypothesis was strengthened by the demonstration that reintroduction of the wild-type WdCHS5 gene into the wdchs5Δ mutant backgrounds restored their wild-type growth kinetics (Fig. 6).
FIG. 6.
Comparisons of growth rate and viability of the wild-type strain, the wdchs5Δ11, wdchs5Δ236, and wdchs5Δ316 disruption mutants, and the wdchs5Δ11-1, wdchs5Δ236-1, and wdchs5Δ316-1 complementation strains grown in YPD liquid medium at 37°C and measured by optical density (A), hemacytometer counts (B), and viable counts (C). The initial inoculation level was 106 cells/ml.
FIG. 7.
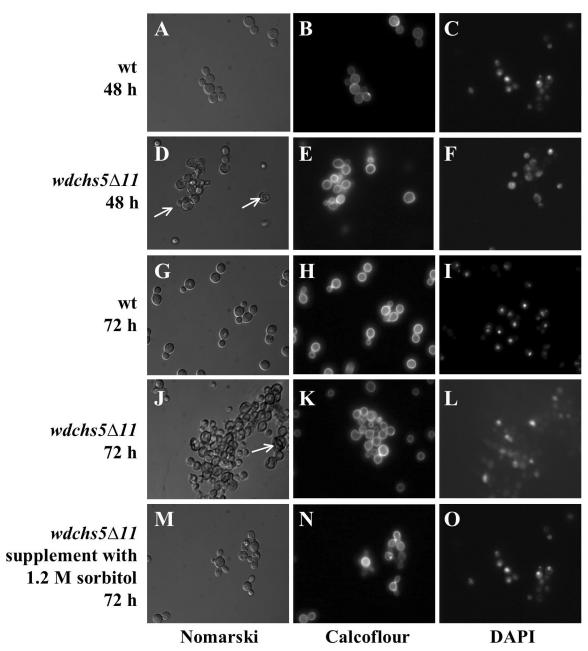
Cellular morphologies and staining properties of the wild-type strain and the wdchs5Δ11 disruption mutant. Wild-type (A, B, C, G, H, and I) and wdchs5Δ11 disruption mutant (D, E, F, J, K, L, M, N, and O) cells were grown at 37°C in YPD medium for 48 h (A, B, C, D, E, and F) or for 72 h without (G, H, I, J, K, and L) or with (M, N, and O) 1.2 M sorbitol, then fixed with 5% formaldehyde, stained with Calcofluor or DAPI, and viewed by Nomarski phase-contrast or fluorescence microscopy. All cells are shown at the same magnification. Arrows point to mutant cells that were swelled or enlarged and occasionally lost their nuclei.
FIG. 8.
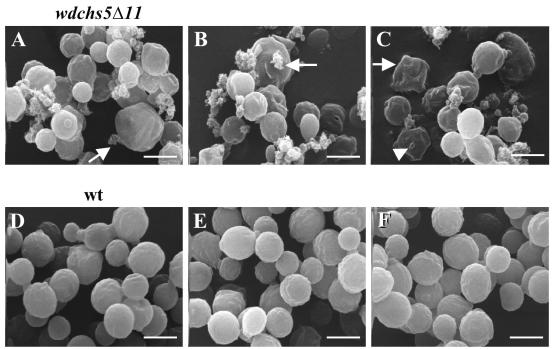
Scanning electron micrographs of the wdchs5Δ11 mutant (A, B, and C) and the wild type (wt) (D, E, and F) grown in YPD broth at 37°C for 72 h. Arrows in panels A through C point to enlarged or irregular mutant cells; arrowhead in panel C identifies a putative pore. Note also adhering materials in panels A through C. Bars, 5 μm.
The wdchs5Δ mutants have abnormal yeast morphology at 37°C.
Because the different wdchs5Δ mutants all had the same apparent phenotype, the wdchs5Δ11 mutant produced by the replacement disruption strategy was used for more-extensive microscopic investigations. Observations of this mutant showed that its phenotype could be distinguished from that of the wild type as early as 48 h of growth at 37°C in YPD liquid medium (Fig. 7A and D) and that by 72 h the aberrant cellular morphologies of the wdchs5Δ11 mutant had become considerably more obvious (Fig. 7G and J). By this time, the mutant cells had clearly begun to clump and form cell aggregations, whereas the wild-type cells did not. Also, individual cells of the mutant often had swelled, with some swelling to about double the size of normal cells as time increased. Eventually, most of these cells lost their smooth surfaces to the extent that they became obviously crinkled. Many of these cells also became flat or irregular and lost their cell wall integrity, and some even lysed. Calcofluor staining showed that the mutant cells often tended to have more cell wall chitin by 48 h (Fig. 7E), which was not as uniformly localized to septal regions as in the wild type (Fig. 7B). However, by 72 h the differences between the Calcofluor staining patterns of the two strains had become less apparent (Fig. 7H and K). Furthermore, DAPI staining showed that some of the mutant cells at 72 h had lost nuclei, indicating that they had probably leaked from the cytoplasm due to the cell wall damage (Fig. 7L). Interestingly, the aberrant phenotypes of the wdchs5Δ11 mutant could be reversed by adding osmotic stabilizers, such as 1 M sucrose (data not shown) or 1.2 M sorbitol, to either YPD broth (Fig. 7M, N, and O) or YPD agar medium (data not shown). Under these conditions the mutants did not swell or lyse, which suggested that the lysis and death of the mutant at 37°C was, in fact, due to cell wall damage and loss of cell wall integrity.
Scanning electron microscopy confirmed that wdchs5Δ11 cells had severe cellular defects when grown at 37°C for 72 h (Fig. 8A, B, and C) compared to wild-type cells (Fig. 8D, E, and F), with most mutant cells showing various irregular shapes; these cells often were enlarged and usually had more crinkled surfaces. In addition, adhering materials of unknown origin were associated with many wdchs5Δ11 cells; these possibly represented cellular products that had leaked through the pore-like structures seen on the surfaces of some cells. Support for our hypothesis that the adhering material was of cytoplasmic origin was provided by results from assays of alkaline phosphatase activity. These assays showed that by 72 h the relative alkaline phosphatase activity associated with the culture medium of wdchs5Δ11 cells was about six times higher (5.67 ± 0.626 [mean ± standard deviation for three independent measurements]) than that of wild-type cells (arbitrarily set at 1) and that this difference was eliminated by the addition of 1 M sorbitol (0.95 ± 0.545 versus 0.72 ± 0.545, respectively).
Disruption of WdCHS5 does not reduce overall chitin synthase activity but does induce an increase in chitin content at 37°C.
No significant differences were detected between the total chitin synthase activities of the wild-type strain and those of wdchs5Δ mutant strains at either 25 or 37°C when activities were evaluated under zymogenic or nonzymogenic assay conditions (data not shown). Interestingly, significantly more (P < 0.05) chitin (about 50%) was detected in the wdchs5Δ mutants than in the wild type, but only in cells cultured for at least 48 h at 37°C (data not shown). This paradox indicated that the cell wall damage caused by the disruption of WdCHS5 induced a compensatory pathway, which caused the other chitin synthases to be activated or stimulated to produce more chitin, as suggested previously for mutants with WdCHS4 disruptions (48). Nonetheless, this newly synthesized chitin was not able to rescue the damage caused by the loss of WdChs5p, because that damage was not repaired by the chitin produced by any of the other four WdChsp isozymes. As expected, complementation of the wdchs5Δ11 mutant with WdCHS5 (the wdchs5Δ11-1 strain) lowered the chitin content to that of the wild-type strain in cells cultured at 37°C for 48 h (data not shown).
The wdchs5Δ mutants have reduced virulence in mice.
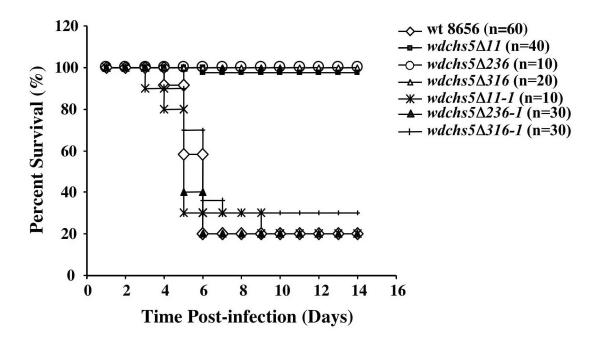
As might be expected of mutants with a temperature-lethal phenotype, all three types of wdchs5Δ mutants showed significant reductions in virulence compared to that of the wild-type strain when tested in an acute murine infection model (Fig. 9). Reintroduction of the WdCHS5 gene into each type of wdchs5Δ mutant background reconstituted virulence. To determine tissue burdens of the brain, kidney, and liver or spleen, mice injected with the wdchs5Δ11 mutant were sacrificed at days 1, 3, 5, and 10 postinfection. Starting on day 3, the viable counts began to decrease in all the organs examined, suggesting that the cells were being cleared from the different tissues (data not shown). Furthermore, the infection was totally cleared from the liver or spleen by day 5 and was 98% cleared from the brain and kidney by day 10. These results indicated that the mutant cells were incapable of sustained growth and survival under the elevated temperature associated with the mice.
FIG. 9.
Mouse survival analyses after injection with the wild type (wt 8656), wdchs5Δ disruption mutants, and wdchs5Δ mutants complemented with WdCHS5. One or more groups of 10 mice received injections of log-phase yeast cells of each strain. The injections contained 9 × 109 cells per mouse, and the mice were monitored for 14 days to determine the survival rate. When two or more groups were involved, the data presented are averages at each time point. Survival fractions were calculated by the Kaplan-Meier method, and survival curves were tested for significant difference (P < 0.01) by the Mantel-Haenszel test using GraphPad Prism software (version 3.00 for Windows).
DISCUSSION
This report documents that WdChs5p has relevance to the virulence of W. dermatitidis, because it is essential for sustained growth and viability at 37°C but not at 25°C. Cloning of the WdCHS5 gene and analysis of its deduced amino acid sequence showed that a myosin motor-like domain was fused to a chitin synthase domain, a condition only found among class V chitin synthases. Although the significance of this type of putative fusion gene remains unknown, it is becoming clear that genes encoding class V isozymes are probably not uncommon in filamentous fungi (28, 37). So far at least 10 similar chitin synthases with this unique structure have been identified. Furthermore, the class V chitin synthases are apparently present only in filamentous fungi and some dimorphic and polymorphic fungi that produce true hyphae (28, 38; this study). However, prior to the present investigation, only CsmA of A. nidulans and, to lesser extents, ChsA of G. graminicola and ChsV of F. oxysporum had received extensive study (1, 13, 16, 20, 46). Particularly, the studies of CsmA established for the first time the important role of this type of chitin synthase in the maintenance of hyphal wall integrity and polarized hyphal wall synthesis, and especially its importance in hyphal growth and morphogenesis under low osmotic conditions (16, 46). With G. graminicola it was found that ChsA is essential for conidial wall strength in media with high water potential and contributes to the strength of hyphal tips, whereas F. oxysporum mutants with defective ChsV have hyphae with abnormal swellings, which are eliminated by culture with osmotic stabilizers (1, 20). The results of our study similarly demonstrated that WdChs5p functions in the manner of CsmA, ChsA, and ChsV in maintaining cell wall integrity in W. dermatitidis, but only at 37°C.
Our Northern blot analysis provides support to the prior suggestion that WdCHS5 is a stress response gene (47). WdCHS5 mRNA was found at significantly higher levels in cells grown at 37°C (47; this study) and under a number of additional stress conditions known to promote morphology changes in W. dermatitidis, such as Ca2+ limitation, nitrogen starvation, and low pH. The importance of this gene to viability and virulence, and the interesting expression pattern of its transcription, prompted us to further characterize its promoter region. Our analyses suggested that a negative regulatory element(s) exists in the 5′ URS of WdCHS5. Studies have previously demonstrated that the CsmA gene of A. nidulans also displays different temporal patterns of expression and that its promoter region has several cis-acting elements similar to those found in the URS of WdCHS5, such as STRE, ABAA, and HAP (46). Similar findings have been made with WdCHS3 (37, 49; J.-H. Oh and P. J. Szaniszlo, unpublished data), indicating that W. dermatitidis has at least two chitin synthase genes that are differentially expressed in response to a variety of conditions. However, unlike the situation with disruption of WdCHS5, disruption of WdCHS3 does not produce a temperature-sensitive phenotype. Nonetheless, we speculate that these two genes share a similar regulatory mechanism or global regulation, although the exact mechanisms responsible for the increased mRNA levels of both WdCHS5 and WdCHS3 in stressed cells have not been determined. While it is still possible that posttranscriptional regulation is also contributing to the increased mRNA levels of both genes detected, we favor the hypothesis that the increased mRNA levels of WdCHS5 in the stressed cells resulted mainly from interactions between trans-acting factors and the cis-acting elements identified in its URS. Support for this possibility is provided by previous data from a semiquantitative RT-PCR study that detected comparable amounts of WdCHS5 mRNA in cells grown continuously at 37°C for 24 h and in cells grown at 25°C for 21 h and then shifted to 37°C and grown for an additional 3 h (47). Results from that study, at least for the response to the temperature shift, which are confirmed by the Northern blot analysis reported here, strongly argue that the increased level of WdCHS5 mRNA detected at 37°C was not due to the increased mRNA half-life, because the semiquantitative RT-PCR did not detect a larger amount of WdCHS5 mRNA in cells grown continuously at 37°C than in cells subjected to the temperature shift for just 3 h.
Studies of CsmA also suggest that posttranslational processing is necessary for activating the chitin synthase domain in A. nidulans, and they further gave rise to the notion that the myosin motor-like domain might contribute to CsmA localization. However, no evidence supports the latter hypothesis or suggests that the putative myosin motor associates with actin. Nonetheless, different myosins with different functions have been identified in cells, and some myosin tail domains contain a structural motif that may be used to direct the interaction of a given myosin with its cargo (25, 43). Thus, it remains tempting to suggest that the N-terminal domain of WdChs5p is, in fact, a myosin motor that is necessary at 37°C for the proper localization of the C-terminal chitin synthase domain and consequently the newly synthesized chitin by interaction with actin. Support for this idea is provided by results with S. cerevisiae, where Myo2p, a class V myosin, acts as a transport motor required for delivery of chitin synthase 3 to the growing buds (39). The recent finding that ChsZp of Aspergillus oryzae encodes a second chitin synthase with a myosin motor-like domain in that fungus, which is different enough from that of its ChsYp isozyme and those of other class V chitin synthases to suggest its inclusion in a new class (class VI), further indicates that the ultimate elucidation of the myosin motor-like domain's function is extremely important (7). Studies aimed at determining the mechanisms of WdChs5p localization and the possible interactions of its myosin motor-like domain with actin are in progress. Among the results of these studies was the preliminary finding that the wdchs5Δ11 mutant could not be complemented with just the chitin synthase domain of WdChs5p, or with only the lysine residue in the conserved P loop of Wdchs5p mutated to alanine (H. Liu and P. J. Szaniszlo, unpublished data).
Three different methods were used to disrupt WdCHS5. In each case, disruption of WdCHS5 resulted in mutants with temperature-sensitive phenotypes. At 25°C, the mutant yeast cells grew normally both in agar and in liquid medium, but at 37°C, dramatic changes in cell morphology were observed with prolonged incubation. This resulted in mutant cells that became hyperpigmented, were often swollen, and died in large numbers, frequently by lysis. These observations suggest that the loss of WdChs5p function results in yeast cell wall weakening at the elevated temperature, which in turn brings about the loss of cell viability. This scenario is consistent with the hypothesis that WdChs5p has an essential function at the temperature of infection. Reintroduction of the WdCHS5 gene successfully complemented the temperature-sensitive phenotype, confirming that that phenotype resulted solely from the loss of WdCHS5 itself. The fact that wdchs5Δ mutant cells are identical to wild-type cells at 25°C further suggests that either WdChs5p does not function at this temperature or one or more of the other chitin synthases compensate for the loss of WdChs5p at lower temperatures. In either case, it is obvious that none of the other four WdChsp isozymes can compensate for the loss of WdChs5p at the higher temperature. Under the scenario that WdChs5p does not have a function at 25°C, together with conclusions from our previous studies that none of the other four chitin synthases is essential for cell viability at 25°C, we hypothesized that the disruption of each other single WdCHS gene in the wdchs5Δ background would be without effect at 25°C. On the other hand, if one of the other WdChsp's compensated for the loss of function of WdChs5p at 25°C, then disruption of WdCHS5 in certain other wdchsΔ backgrounds should produce strains with major defects or incapable of growth at both 25 and 37°C. Investigations in progress, aimed at eliminating the latter possibility, have resulted in the production of all four types of double disruption mutants with either WdCHS1, WdCHS2, WdCHS3, or WdCHS4 disrupted together with WdCHS5 (unpublished data). Preliminary analyses of these double mutants indicate that when WdCHS5 is disrupted in a wdchsΔ background, the resulting yeast cells are similar to those of the wdchs5Δ single mutant at 37°C and either have a wild-type phenotype at 25°C or exhibit the very minor chaining abnormality characteristic of wdchs1Δ mutants or an even darker pigmentation than wdchs4Δ mutants, respectively, as reported previously for those single mutants (44, 48).
Cell wall weakening and abnormal cell shapes resulting from the disruption of a single chitin synthase gene have been documented in a number of different fungi, including Candida albicans (CaCHS1), A. nidulans (chsC, chsA, and CsmA), Aspergillus fumigatus (ChsE), Neurospora crassa (Chs1), G. graminicola (ChsA), and F. oxysporum (FoCHSV) (1, 14, 16, 20, 29, 42, 46, 52). Interestingly, the growth defects associated with these mutants resulted from the inactivation of chitin synthases of a number of classes. In addition, the defects were observed predominantly in older hyphal regions, and those defects could often be suppressed with an osmotic stabilizer. In W. dermatitidis, similarly, the weakening of the yeast cell wall and the loss of cell viability at 37°C caused by the disruption of WdCHS5 were observed only when mutant cells entered mature stages of growth. Thus, it appears that this temperature-sensitive phenotype is different from that of a wdchs1Δwdchs2Δ double mutant, which is not able to grow at all at 37°C (44, 48). This finding strongly suggests that WdChs5p, or the chitin synthesized by WdChs5p, is essential only for sustained or progressive yeast cell growth at elevated temperatures. We speculate that either WdChs1p or WdChs2p, or residual WdChs5p, is responsible for the initial cell growth at 37°C but that WdChs5p is then required for maintaining that growth at high temperatures. Possibly, mutant cells devoid of WdChs5p can grow for several generations at 37°C, but when cells become mature and older, especially while intracellular contents are increasing, their cell walls require extra strength to resist the rise in internal pressures. The observation that the temperature-sensitive phenotype can be rescued by supplementing cultures with osmotic stabilizers, such as sorbitol and sucrose, supported this idea and confirmed that the resulting cell lysis and death at 37°C are largely due to the loss of cell wall integrity. Under this scenario, the loss of virulence of wdchs5Δ mutants in our mouse model of acute infection would similarly be due to their inability to maintain cell wall integrity at temperatures of infection. This hypothesis is supported by necropsy data showing that the number of viable wdchs5Δ cells in mice decreased significantly with time: most or all of the infection was cleared from all the organs assayed by day 5 (data not shown). We suspect that the abnormal temperature-sensitive phenotype of the wdchs5Δ single mutants is a major factor contributing to this clearance and thus that the chitin contributed by WdChs5p is essential for the virulence of the wild-type strain.
To our knowledge, our data provide the first direct evidence that a chitin synthase can be a specific virulence factor in a conidiogenous fungal pathogen of humans. Previously, the only single chitin synthase disruption mutants unequivocally known to be less virulent were Chs3 (class IV) mutants of the dimorphic pathogenic yeast C. albicans, although Chs1 in that fungus can arguably also be considered essential for virulence because it is required for viability (28). Interestingly, inactivation of UmChs5p (class IV) in Usilago maydis and of ChsV in F. oxysporum produces mutants with reduced plant virulence (20, 51). Thus, among the filamentous fungi, drugs that target the production or function of chitin synthases of class IV or class V may be more efficacious than those that target members of the other chitin synthase classes.
Acknowledgments
We thank J. M. Mendenhall (Institute of Cellular and Molecular Biology, The University of Texas at Austin) for help with scanning electron microscopy.
This research was supported by a grant to P.J.S. from the National Institute of Allergy and Infectious Diseases (AI 33049).
REFERENCES
- 1.Amnuaykanjanasin, A., and L. Epstein. 2003. A class V chitin synthase gene, chsA is essential for conidial and hyphal wall strength in the fungus Colletotrichum graminicola (Glomerella graminicola). Fungal Genet. Biol. 38:272-285. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Current protocols in molecular biology, p. 13.11.1-13.11.4. John Wiley & Sons, New York, N.Y.
- 3.Beth-Din, A. B., C. A. Specht, P. W. Robbins, and O. Yarden. 1996. chs4, a class IV chitin synthase gene from Neurospora crassa. Mol. Gen. Genet. 250:214-222. [DOI] [PubMed] [Google Scholar]
- 4.Bowen, A. R., J. L. Chen-Wu, M. Momany, R. Young, P. J. Szaniszlo, and P. W. Robbins. 1992. Classification of fungal chitin synthases. Proc. Natl. Acad. Sci. USA 89:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulawa, C. E., M. Slater, E. Cabib, J. Au-Young, A. Sburlati, W. L. Adair, and P. W. Robbins. 1986. The Saccharomyces cerevisiae structural gene for chitin synthase is not required for chitin synthesis. Cell 46:213-215. [DOI] [PubMed] [Google Scholar]
- 6.Cabib, E., S. J. Silverman, and J. A. Shaw. 1992. Chitinase and chitin synthase 1: counterbalancing activities in cell separation of Saccharomyces cerevisiae. J. Gen. Microbiol. 138:97-102. [DOI] [PubMed] [Google Scholar]
- 7.Chagra, Y., K. Abe, K. Gomi, and T. Nakajima. 2002. chsZ, a gene for a novel class of chitin synthase from Aspergillus oryzae. Curr. Genet. 41:261-267. [DOI] [PubMed] [Google Scholar]
- 8.Cid, V. A., A. Duran, F. D. Rey, M. P. Synder, C. Nombela, and M. Sanchez. 1995. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol. Rev. 59:345-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper, C. R., Jr., J. L. Harris, C. W. Jacobs, and P. J. Szaniszlo. 1984. Effects of polyoxin on cellular development in Wangiella dermatitidis. Exp. Mycol. 89:349-363. [Google Scholar]
- 10.Cooper, C. R., Jr., and P. J. Szaniszlo. 1993. Evidence for two cell division cycle (CDC) genes that govern yeast bud emergence in the pathogenic fungus Wangiella dermatitidis. Infect. Immun. 61:2069-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.deHoog, G. S., K. Takeo, S. Yoshida, E. Gottlich, K. Nishimura, and M. Miyaji. 1994. Pleoanamorphic life cycle of Exophiala (Wangiella) dermatitidis. Antonie Leeuwenhoek 65:143-153. [DOI] [PubMed] [Google Scholar]
- 12.Feng, B., X. Wang, M. Hauser, S. Kaufmann, S. Jentsch, G. Haase, J. M. Becker, and P. J. Szaniszlo. 2001. Molecular cloning and characterization of WdPKS1, a gene involved in dihydroxynaphthalene melanin biosynthesis and virulence in Wangiella (Exophiala) dermatitidis. Infect. Immun. 69:1781-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujiwara, M., H. Horiuchi, A. Ohta, and M. Takagi. 1997. A novel fungal gene encoding chitin synthase with a myosin motor-like domain. Biochem. Biophys. Res. Commun. 236:75-78. [DOI] [PubMed] [Google Scholar]
- 14.Fujiwara, M., M. Ichinomiya, T. Motoyama, H. Horiuchi, A. Ohta, and M. Takagi. 2000. Evidence that the Aspergillus nidulans class I and class II chitin synthase genes, chsC and chsA, share critical roles in hyphal wall integrity and conidiophore development. J. Biochem. 127:359-366. [DOI] [PubMed] [Google Scholar]
- 15.Harris, J. L., and P. J. Szaniszlo. 1986. Localization of chitin in walls of Wangiella dermatitidis using colloidal gold-labeled chitinase. Mycologia 78:853-857. [Google Scholar]
- 16.Horiuchi, H., M. Jujiwara, S. Yamashita, A. Ohta, and M. Takagi. 1999. Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J. Bacteriol. 181:3721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karuppayil, S. M., and P. J. Szaniszlo. 1997. Importance of calcium to the regulation of polymorphism in Wangiella (Exophiala) dermatitidis. J. Vet. Med. Mycol. 35:379-388. [DOI] [PubMed] [Google Scholar]
- 18.Kwon-Chung, K. J., and J. E. Bennett. 1992. Medical mycology, p. 647-650. Lea and Febiger, Philadelphia, Pa.
- 19.Liu, H., Z. Wang, L. Zheng, M. Hauser, S. Kauffman, J. M. Becker, and P. J. Szaniszlo. 2001. Relevance of chitin and chitin synthases to virulence in Wangiella (Exophiala) dermatitidis, a model melanized pathogen of humans, p. 463-472. In R. A. A. Muzzarelli (ed.), Chitin enzymology 2001. Atec Edizioni, Grottammare, Italy.
- 20.Madrid, M. P., A. D. Pietro, and M. I. G. Roncero. 2003. Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defense compounds. Mol. Microbiol. 47:257-266. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto, T., T. Matsuda, M. R. McGinnis, and L. Ajello. 1993. Clinical and mycological spectra of Wangiella dermatitidis. Mycoses 36:145-155. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto, T., L. Ajello, T. Matsuda, P. J. Szaniszlo, and T. J. Walsh. 1994. Developments in hyalohyphomycosis and phaeohyphomycosis. J. Vet. Med. Mycol. 32(Suppl. 1):329-349. [DOI] [PubMed] [Google Scholar]
- 23.McIntosh, N. D. P. 1996. Yeast-to-hypha transition in Wangiella dermatitidis. M.A. thesis. The University of Texas at Austin, Austin.
- 24.Mellado, E., A. Aufauvre-Brown, C. A. Specht, P. W. Robbins, and D. W. Holden. 1995. A multigene family related to chitin synthase genes of yeast in the opportunistic pathogen Aspergillus fumigatus. Mol. Gen. Genet. 246:353-359. [DOI] [PubMed] [Google Scholar]
- 25.Mermall, V., P. L. Post, and M. S. Mooseker. 1998. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science 279:527-533. [DOI] [PubMed] [Google Scholar]
- 26.Molina, M., H. Martin, M. Sanchez, and C. Nombela. 1998. MAP kinase-mediated signal transduction pathways. Methods Microbiol. 26:3375-3393. [Google Scholar]
- 27.Montijn, R. C., P. V. Wolven, S. D. Hoog, and F. M. Klis. 1997. β-Glucosylated proteins in the cell wall of the black yeast Exophiala (Wangiella) dermatitidis. Microbiology 143:1673-1680. [DOI] [PubMed] [Google Scholar]
- 28.Munro, C. A., and N. A. R. Gow. 2001. Chitin synthesis in human pathogenic fungi. Med. Mycol. 39(Suppl. 1):41-53. [PubMed] [Google Scholar]
- 29.Munro, C. A., K. Winter, A. Buchan, K. Henry, J. M. Becker, A. J. P. Brown, C. E. Bulawa, and N. A. R. Gow. 2001. Chs1 of Candida albicans is an essential chitin synthase required for synthesis of the septum and for cell integrity. Mol. Microbiol. 39:1414-1426. [DOI] [PubMed] [Google Scholar]
- 30.Muzzarelli, R., C. Jeuniaux, and G. W. Gooday. 1986. Chitin in nature and technology. J. Gen. Microbiol. 99:1−11.
- 31.Nachman, S. A., O. Alpan, R. Malowitz, and E. D. Spitzer. 1996. Catheter-associated fungemia due to Wangiella (Exophiala) dermatitidis. J. Clin. Microbiol. 34:1011-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nino-Vega, G. A., C. A. Munro, G. San-Blas, G. W. Gooday, and N. A. R. Gow. 2000. Differential expression of chitin synthase genes during temperature-induced dimorphic transitions in Paracoccidioides brasiliensis. Med. Mycol. 38:31-39. [DOI] [PubMed] [Google Scholar]
- 33.Orlean, P. 1997. Biogenesis of yeast wall and surface components, p. 229-362. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 3. Cell cycle and biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Park, C., H. Horiuchi, C. W. Hwang, W. H. Yeh, A. Ohta, J. C. Ryu, and M. Takagi. 1999. Isolation of csm1 encoding a class V chitin synthase with a myosin motor-like domain from the rice blast fungus, Pyricularia oryzae. FEMS Microbiol. Lett. 170:131-139. [DOI] [PubMed] [Google Scholar]
- 35.Roncero, C. 2002. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 41:367-378. [DOI] [PubMed] [Google Scholar]
- 36.Roncero, C., M. H. Valdivieso, J. C. Ribas, and A. Duran. 1988. Effect of Calcofluor white on chitin synthases from Saccharomyces cerevisiae. J. Bacteriol. 170:1945-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz-Herrera, J., J. M. Gonzalez, and R. Ruiz-Medrano. 2001. Significance of multigenic control of chitin synthase in fungi. In R. A. A. Muzzarelli (ed.), Chitin enzymology 2001. Atec Edizioni, Grottammare, Italy.
- 38.San-Blas, G., G. Nino-Vega, and T. Iturriaga. 2002. Paracoccidioides brasiliensis and paracoccidioidomycosis: Molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med. Mycol. 40:225-242. [DOI] [PubMed] [Google Scholar]
- 39.Santos, B., and M. Snyder. 1997. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J. Cell Biol. 136:95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw, J. A., P. C. Mol, B. Bowers, S. J. Silverman, M. H. Valdivieso, A. Duran, and E. Cabib. 1991. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114:111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silverman, S. J., A. Sburlati, M. L. Slater, and E. Cabib. 1988. Chitin synthase 2 is essential for septum formation and cell division in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 85:4735-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Specht, C. A., Y. Liu, P. W. Robbins, C. E. Bulawa, N. Iartchouk, K. R. Winter, P. J. Riggle, J. C. Rhodes, C. L. Dodge, D. W. Culp, and P. T. Borgia. 1996. The chsD and chsE genes of Aspergillus nidulans and their role in chitin synthesis. Fungal Genet. Biol. 20:153-167. [DOI] [PubMed] [Google Scholar]
- 43.Steinberg, G. 2000. The cellular roles of molecular motors in fungi. Trends Microbiol. 8:162-168. [DOI] [PubMed] [Google Scholar]
- 44.Szaniszlo, P. J. 2002. Molecular genetic studies of the model dematiaceous pathogen Wangiella dermatitidis. Int. J. Med. Microbiol. 292:381-390. [DOI] [PubMed] [Google Scholar]
- 45.Szaniszlo, P. J., L. Mendoza, and S. M. Karuppayil. 1993. Clues about chromoblastomycosis and other dematiaceous pathogens based on Wangiella as a model, p. 241-255. In H. Vanden Bossche, F. C. Odds, and D. Kerrige (ed.), Dimorphic fungi in biology and medicine. Plenum Press, New York, N.Y.
- 46.Takashita, N., A. Ohta., and H. Horiuchi. 2002. csmA, a gene encoding a class V chitin synthase with a myosin motor-like domain of Aspergillus nidulans, is translated as a single polypeptide and regulated in response to osmotic conditions. Biochem. Biophys. Res. Commun. 298:103-109. [DOI] [PubMed] [Google Scholar]
- 47.Wang, Q., H. Liu, and P. J. Szaniszlo. 2002. Compensatory expression of five chitin synthase genes, a response to stress stimuli, in Wangiella (Exophiala) dermatitidis, a melanized fungal pathogen of humans. Microbiology 148:2811-2817. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Z., L. Zheng, M. Hauser, J. M. Becker, and P. J. Szaniszlo. 1999. WdChs4p, a homolog of chitin synthase 3 in Saccharomyces cerevisiae, alone cannot support growth of Wangiella (Exophiala) dermatitidis at the temperature of infection. Infect. Immun. 67:6619-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Z., and P. J. Szaniszlo. 2000. WdCHS3, a gene that encodes a class III chitin synthase in Wangiella (Exophiala) dermatitidis, is expressed differentially under stress conditions. J. Bacteriol. 182:874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Z., L. Zheng, H. Liu, Q. Wang, M. Hauser, S. Kauffman, J. M. Becker, and P. J. Szaniszlo. 2001. WdChs2p, a class I chitin synthase, together with WdChs3p (class III) contributes to virulence in Wangiella (Exophiala) dermatitidis. Infect. Immun. 69:7517-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xoconostle-Casares, B., C. A. Specht, P. W. Robbins, Y. Liu, C. Leon, and J. Ruiz-Herrera. 1997. Umchs5, a gene coding for a class IV chitin synthase in Ustilago maydis. Fungal Genet. Biol. 22:199-208. [DOI] [PubMed] [Google Scholar]
- 52.Yarden, O., and C. Yanofsky. 1991. Chitin synthase 1 plays a major role in cell wall biogenesis in Neurospora crassa. Genes Dev. 5:2420-2430. [DOI] [PubMed] [Google Scholar]
- 53.Ye, X., B. Feng, and P. J. Szaniszlo. 1999. A color-selectable and site-specific integrative transformation system for gene expression studies in the dematiaceous fungus Wangiella (Exophiala) dermatitidis. Curr. Genet. 36:241-247. [DOI] [PubMed] [Google Scholar]
- 54.Ye, X., and P. J. Szaniszlo. 2000. Expression of a constitutively active Cdc42 homolog promotes development of sclerotic bodies but represses hyphal growth in the zoopathogenic fungus Wangiella (Exophiala) dermatitidis. J. Bacteriol. 182:4941-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, Z., and S. J. Gurr. 2000. Walking into the unknown: a ′step down' PCR-based technique leading to the direct sequence analysis of flanking genomic DNA. Gene 253:145-150. [DOI] [PubMed] [Google Scholar]
- 56.Zheng, L., and P. J. Szaniszlo. 1999. Cloning and use of the WdURA5 gene as a hisG cassette selection marker for potentially disrupting multiple genes in Wangiella dermatitidis. Med. Mycol. 37:85-96. [PubMed] [Google Scholar]