Abstract
The frequency of Escherichia coli O157 genotypes among bovine, food, and human clinical isolates from The Netherlands was studied. Genotyping included the lineage-specific polymorphism assay (LSPA6), the Shiga-toxin-encoding bacteriophage insertion site assay (SBI), and PCR detection and/or subtyping of virulence factors and markers [stx1, stx2a/stx2c, q21/Q933, tir(A255T), and rhsA(C3468G)]. LSPA6 lineage II dominated among bovine isolates (63%), followed by lineage I/II (35.6%) and lineage I (1.4%). In contrast, the majority of the human isolates were typed as lineage I/II (77.6%), followed by lineage I (14.1%) and lineage II (8.2%). Multivariate analysis revealed that the tir(A255T) SNP and the stx2a/stx2c gene variants were the genetic features most differentiating human from bovine isolates. Bovine and food isolates were dominated by stx2c (86.4% and 65.5%, respectively). Among human isolates, the frequency of stx2c was 36.5%, while the frequencies of stx2a and stx2a plus stx2c were 41.2% and 22.4%, respectively. Bovine isolates showed equal distribution of tir(255A) (54.8%) and tir(255T) (45.2%), while human isolates were dominated by the tir(255T) genotype (92.9%). LSPA6 lineage I isolates were all genotype stx2c and tir(255T), while LSPA6 lineage II was dominated by tir(255A) (86.4%) and stx2c (90.9%). LSPA6 lineage I/II isolates were all genotype tir(255T) but showed more variation in stx2 types. The results support the hypothesis that in The Netherlands, the genotypes primarily associated with human disease form a minor subpopulation in the bovine reservoir. Comparison with published data revealed that the distribution of LSPA6 lineages among bovine and human clinical isolates differs considerably between The Netherlands and North America.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) O157 is considered a serious pathogen due to its low infectious dose, the severe clinical symptoms (especially among children), and the potential for food- and waterborne outbreaks (7, 22, 35). STEC O157 can be transmitted to humans by direct contact with the ruminant reservoir or indirectly via ingestion of contaminated food or water (33). STEC O157 has several virulence genes that play a crucial role in the development of clinical symptoms. These include the Shiga toxin genes 1 and 2 (stx1 and stx2), several genes located on the chromosomal locus of enterocyte effacement (LEE), which codes for a specialized epithelium attachment system responsible for the characteristic attaching and effacing (A/E) phenotype, and (putative) virulence factors encoded on various genomic pathogenicity islands (20, 25).
In The Netherlands, the prevalences of STEC O157 at the herd level are on average 8% for dairy herds and 13% for veal herds (2), with seasonal peaks from 30% to 70% during the summer months (12, 19). The notification rates of STEC O157 disease cases in The Netherlands (including outbreaks) for 2007, 2008, and 2009 were 0.50, 0.27, and 0.34 cases per 100,000 inhabitants per year, respectively (14). It can be considered paradoxical that the number of human STEC O157 infections is rather low for a pathogen with a low infectious dose when herd-level prevalence is relatively high. This could, at least partly, be explained if only a subset of STEC O157 isolates present in the bovine reservoir are characterized by unique transmissibility and/or virulence characteristics (6).
To assess the public health risk associated with STEC O157, it is important to understand the genotypic diversity among STEC O157 isolates. Distinguishing STEC O157 isolates that constitute a high risk to human health from isolates that have a lower association with clinical symptoms is an important aspect of risk-based monitoring and surveillance. Recently, several PCR typing assays have been reported discriminating between bovine and human clinical STEC O157 isolates. The lineage-specific polymorphism assay LSPA6 uses six loci derived from octamer-based genome scanning to classify STEC O157 isolates into three major lineages (23, 43), which have previously been shown to display a nonrandom distribution among human and bovine isolates in North America (38, 43, 45) and Japan (26). A typing assay based on Shiga toxin-encoding bacteriophage insertion sites clustered STEC O157 isolates with nonrandom distribution among bovine and human clinical isolates (3, 37). Additional genetic markers found to occur at different relative frequencies among clinical and bovine isolates include variants of the stx2-Q antiterminator junction alleles (Q933 and q21) (27), the single nucleotide polymorphism (SNP) in the tir gene [tir(255T) and tir(255A)] (6), and stx2 gene variants (4, 9, 13). Manning et al. (34) developed a subtyping scheme based on 32 SNPs which separated STEC O157 isolates into 9 distinct evolutionary clades, with clade 8 isolates being more virulent than other isolates.
The main objective of the present study was to genetically characterize and compare bovine, food, and human clinical STEC O157 isolates from The Netherlands using various genotyping methods and multivariate statistics. The goal was to determine whether different genotypes occur at different frequencies among isolates from these different sources and to identify the genetic features most differentiating the isolates from these sources. A set of 73 bovine, 29 food, and 85 human clinical isolates were used. The most distinctive genetic features that separated bovine and human clinical isolates were identified using multivariate statistical analyses. The results showed clear nonrandom distributions of genotypes among bovine, food, and human clinical isolates. In addition, comparison with published data revealed significant differences in the distribution of LSPA6 lineages among bovine and human isolates between The Netherlands versus North America and Japan.
MATERIALS AND METHODS
Isolates and growth conditions.
A set of 187 STEC O157 isolates (73 bovine, 29 food, and 85 human clinical isolates) was obtained from the collection of the Food and Consumer Product Safety Authority (Zutphen, The Netherlands) and the National Institute for Public Health and The Environment (Bilthoven, The Netherlands). All isolates possessed the genes encoding Shiga toxin 2 (stx2), intimin (eae), and hemolysin (hlyA). The bovine isolates were isolated from 2002 to 2009 during national surveys and were maintained in cryovials (Microbak, Pro-Lab). The clinical human isolates were strains isolated from patients with STEC symptoms and sent in by hospitals for confirmation in the period 1999 to 2010. The food strains (23 meat isolates and 6 leafy green isolates) were isolated during national surveys by the Food and Consumer Product Safety Authority in the Netherlands. All clinical human isolates were maintained at room temperature in Mueller-Hinton agar. Isolates were propagated on blood agar or nutrient agar (Oxoid), and DNA was extracted using the Chelex-100 (Bio-Rad, Hercules, CA) resin-based technique. One colony of each isolate was transferred into 300 μl 10% Chelex-100 solution, which was subsequently heated for 5 min at 56°C to resuspend the cells. The tubes were briefly cooled at room temperature and mixed for 15 s before heating for 15 min at 98 to 99°C for lysis of the bacteria. After cooling at room temperature, the lysates were centrifuged for 5 min at 13,000 rpm, and up to 200 μl of the supernatant was transferred to a clean tube and stored at −20°C.
Genetic profiling.
All isolates were characterized by two alternative molecular genotyping assays (the lineage-specific polymorphism assay and the Shiga toxin-encoding bacteriophage insertion site assay) and four additional genetic markers: stx2a/stx2c, q21/Q933, tir(A255T), and clade 8 rhsAb(C3468G). Table 1 provides an overview of the primers and probes used. STEC O157 EDL 933 (ATCC 43895) was used as a control strain for all assays.
Table 1.
Oligonucleotides used in this study
| Assay | Target | Type | Sequence (5′→3′)a | Reference |
|---|---|---|---|---|
| Clade 8_rhsA | rhsA | Primer | CCACAATACAGAACGACAGA | This study |
| Primer | ACCTTGCGGATCTACATTTA | |||
| LSPA6 | folD-sfmA | Primer | NED-TACGTAGGTCGAAGGG | 43 |
| Primer | CCAGATTTACAACGCC | |||
| Z5935 | Primer | FAMc-GTGTTCCCGGTATTTG | 43 | |
| Primer | CTCACTGGCGTAACCT | |||
| yhcG | Primer | VIC-CTCTGCAAAAAACTTACGCC | 43 | |
| Primer | CAGGTGGTTGATCAGCG | |||
| rbsB | Primer | VIC-AGTTTAATGTTCTTGCCAGCC | 43 | |
| Primer | ATTCACCGCTTTTTCGCC | |||
| rtcB | Primer | FAM-GCGCCAGATCGATAAAGTAAG | 43 | |
| Primer | GCCGTTGTAAACGTGATAAAG | |||
| arp-iclR | Primer | NED-GCTCAATCTCATAATGCAGCC | 43 | |
| Primer | CACGTATTACCGATGACCG | |||
| q21/Q933 | q21-stx2a | Primer | GAAATCCTCAATGCCTCGTTG | 27 |
| Primer | CCGAAGAAAAACCCAGTAACAG | |||
| Q933-stx2a | Primer | CGGAGGGGATTGTTGAAGGC | 27 | |
| Primer | CCGAAGAAAAACCCAGTAACAG | |||
| SBI | stx1 | Primer | CGCTTTGCTGATTTTTCACA | 37 |
| Primer | GTAACATCGCTCTTGCCACA | |||
| stx2 | Primer | GTTCCGGAATGCAAATCAGT | 37 | |
| Primer | CGGCGTCATCGTATACACAG | |||
| yehV-LJb | Primer | CACCGGAAGGACAATTCATC | 37 | |
| Primer | CCGACCTTTGTACGGATGTAA | |||
| yehV-RJb | Primer | AACAGATGTGTGGTGAGTGTCTG | 37 | |
| Primer | CGAATCGCTACGGAATAGAGA | |||
| wrbA-LJb | Primer | AAGTGGCGTTGCTTTGTGAT | 37 | |
| Primer | AGGAAGGTACGCATTTGACC | |||
| wrbA-RJb | Primer | GATGCACAATAGGCACTACGC | 37 | |
| Primer | ATCGTTCGCAAGAATCACAA | |||
| stx2a/stx2c | stx2a, stx2c | Primer | GTGGTGAGAGCGAGCGAC | This study |
| Primer | GTCATTATTAAACTGCACTTCAG | |||
| tir(A255T) | tir | Primer | TGGCGGCGTCTGAGATAAC | 5 |
| Primer | GAGTATCGAGCGGACCATGATC | |||
| tir(A255) | Probe | Cy5-ACTGAATGATGGATTTG-BHQ2 | ||
| tir(T255) | Probe | FAM-CTGAATGAAGGATTTG-BHQ1 |
The reverse primer has homology with stx2a as well as stx2c.
LJ is left junction, and RJ is right junction.
FAM, 6-carboxyfluorescein.
LSPA6 was performed in essence as described previously (43), using the LSPA6 primer sequences listed in Table 1. Amplicons were generated in two multiplex PCRs: one PCR mixture contained the folD-sfmA, rtcB, and yhcG primers, and the other PCR mixture contained the arp-iclR, rbsB, and Z5935 primers. After amplification (GeneAmp PCR system 9700 in 9600 mode; Applied Biosystems), the products were separated on a capillary sequencer (3130 genetic analyzer; Applied Biosystems) in the presence of an internal marker (GeneScan 600 LIZ size standard; Applied Biosystems). Raw data were analyzed using BioNumerics 6.1 (Applied Maths) to determine fragment sizes. Arranged in the order folD-sfmA, rbsB, Z5935, rtcB, arp-iclR, and yhcG, each allele was assigned a number as described previously (43). Isolates showing the LSPA6 genotype 111111 were classified as LSPA6 lineage I (LSPA6 LI), while those with LSPA6 genotype 211111 were classified as LSPA6 lineage I/II (LSPA6 LI/II). Unique alleles (aberrant amplicon size) were assigned new numbers. All deviations from the genotypes 111111 and 211111 were classified as LSPA6 lineage II (LSPA6 LII) (42, 43, 45).
The Shiga toxin-encoding bacteriophage insertion site (SBI) assay consists of six PCRs that amplify the Shiga toxin genes and their genomic insertion site junctions of the Shiga toxin-encoding bacteriophages of STEC O157 (3, 37). The presence of intact or occupied chromosomal insertion site regions (yehV and wrbA) was determined for all isolates. The PCR results of the amplification of the bacteriophage-yehV right and left junctions (yehV-R and yehV-L, respectively) and the bacteriophage-wrbA left and right junctions (wrbA-R and wrbA-L, respectively) were binary interpreted as the sites being occupied (amplification) or intact (no amplification). Based on the yehV-R, yehV-L, wrbA-R, and wrbA-L PCR results in combination with the presence of stx1 and/or stx2, different SBI genotypes were assigned.
The q21/Q933 assay targets two different allelic variations of the antiterminator gene Q upstream of the prophage stx2 region (27). Allele Q933 is responsible for a strong antiterminator activity resulting in relatively high expression levels of stx2, while allele q21 represents a weak antiterminator activity resulting in lower stx2 expression levels (41). The assay was conducted as described by Lejeune et al. (27).
The tir(A255T) assay targets an A255→T nucleotide polymorphism in the gene coding for the translocated intimin receptor and was found to provide a clear discrimination between human and bovine isolates (6). The assay was conducted as described by Bono et al. (5) on a LightCycler 480 real-time PCR system (Roche Diagnostics Nederland B.V., Almere, The Netherlands) with different dyes attached to the probes (Table 1).
The clade 8_rhsA assay targets one of the single nucleotide polymorphisms (SNPs) possible in the rhsA gene (SNP position 3468) (29). A PCR fragment with a length of 635 bp was generated with the rhsA primers indicated in Table 1. The 50-μl PCR mixture contained 1× PCR buffer (Invitrogen BV, Breda, The Netherlands), 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.2 μM each primer, 1 U Taq DNA polymerase (Invitrogen BV, Breda, The Netherlands), and 2 μl DNA template (approximately 20 ng). PCR restriction fragment analysis was performed on 5 μl of the rhsA PCR product using 10 U of either HaeII or Sau96I (New England BioLabs, Ipswich, MA) and the recommended buffer at 37°C for 2 h. The restriction fragments were separated in a 2% agarose gel. The size of DNA fragments was estimated in comparison to a marker: a 100-bp DNA ladder (New England BioLabs, Ipswich, MA). With clade 8 isolates [rhsA(3468C)], 2 fragments will be generated after digestion with HaeII (lengths, 525 and 110 bp), whereas Sau96I will result in 3 bands on the gel (lengths, 296, 273, and 66 bp). Non-clade 8 isolates [rhsA(3468G)] will produce the following results: HaeII, 269, 256, and 110 bp; and Sau96I, 339 and 296 bp.
The stx2a/stx2c assay is based on the amplification of a part of the upstream region of the stx2 operon and the A and B subunit genes, followed by restriction fragment analysis. The PCR fragment with a length of 1,566 bp with stx2a and 1,560 bp with stx2c was generated in a 50-μl PCR mixture containing 1× PCR buffer (Invitrogen BV, Breda, The Netherlands), 2 mM MgCl2, 0.2 mM dNTPs, 0.2 μM each stx2/stx2c primers (Table 1), 1 U Taq DNA polymerase (Invitrogen BV, Breda, The Netherlands), and 2 μl DNA template (approximately 20 ng). PCR-restriction fragment analysis was performed on 5 μl of the generated PCR product using 10 U of either HincII or ScaI (New England BioLabs, Ipswich, MA) and the recommended buffer at 37°C for 2 h. The restriction fragments were separated in a 1.5% agarose gel. The size of the DNA fragments was estimated in comparison to a marker: a 100-bp DNA ladder (New England BioLabs, Ipswich, MA). The following fragments were generated with HincII: stx2a, 1,163, 262, 79, and 62 bp; and stx2c, 1,157, 324, and 79 bp. The following fragments were generated with ScaI: stx2a, 1,566 bp; and stx2c, 1,432 and 128 bp.
Data analysis.
Differences in frequencies of genetic markers (denoted in binary values 0 and 1) between isolates from different sources (bovine, food, and human) were statistically evaluated using the chi-square test (IBM SPSS Statistics, version 19). In order to identify the genetic features most differentiating between isolates of different origins, multinomial logistic regression analysis was performed (IBM SPSS Statistics, version 19) with the data on the human clinical isolates as the reference category. Principal component analysis (PCA) was performed, and biplots (showing observation and variables) were created using SAS Stat studio 3.1 (SAS Institute, Inc., Cary, NC).
RESULTS
Lineage specific polymorphism assay.
In total, 9.1% of STEC O157 isolates from The Netherlands were typed as LSPA6 lineage I, 55.1% as lineage I/II, and 35.8% as lineage II (Table 2). The distribution of the LSPA6 lineages was significantly associated with the isolation source (χ2 = 73.3, df = 3, P < 0.001). LSPA6 lineage II dominated among bovine isolates (63.0%), followed by lineage I/II (35.6%) and lineage I (1.4%). The majority of the human isolates were typed as lineage I/II (77.6%), followed by lineage I (14.1%) and lineage II (8.2%). The distribution of LSPA6 lineages among food isolates was intermediate between the compositions of bovine and human isolates (lineage II, 48.3%; lineage I/II, 37.9%; lineage I, 13.8%). The diversity of LSPA6 lineages was higher among bovine isolates than food and human isolates, with totals of 17 different LSPA6 genotypes among bovine isolates, 8 different genotypes among the food isolates, and 5 genotypes among human isolates (Table 2).
Table 2.
Distribution of LSPA6 lineages among STEC O157 strains from bovine, food, and human clinical cases from The Netherlands
| Genotypea | Lineage | No. (%) of strains with lineage shown: |
|||
|---|---|---|---|---|---|
| Total | Bovine (n = 73) | Food (n = 29) | Clinical (n = 85) | ||
| 111111 | I | 17 (9.1) | 1 (1.4) | 4 (13.8) | 12 (14.1) |
| 211111 | I/II | 103 (55.1) | 26 (35.6) | 11 (37.9) | 66 (77.6) |
| 211112 | II | 7 (3.7) | 5 (6.8) | 1 (3.4) | 1 (1.2) |
| 212221 | II | 20 (10.7) | 11 (15.1) | 8 (27.6) | 1 (1.2) |
| 212222 | II | 20 (10.7) | 13 (17.8) | 2 (6.9) | 5 (5.9) |
| 212231b | II | 4 (2.1) | 4 (5.5) | 0 | 0 |
| 212232b | II | 3 (1.6) | 2 (2.7) | 1 (3.4) | 0 |
| 212321b | II | 1 (0.5) | 1 (1.4) | 0 | 0 |
| 212322b | II | 2 (1.1) | 2 (2.7) | 0 | 0 |
| 213111b | II | 1 (0.5) | 1 (1.4) | 0 | 0 |
| 214222b | II | 2 (1.1) | 2 (2.7) | 0 | 0 |
| 215232b | II | 1 (0.5) | 1 (1.4) | 0 | 0 |
| 216222b | II | 1 (0.5) | 1 (1.4) | 0 | 0 |
| 222242b | II | 2 (1.1) | 2 (2.7) | 0 | 0 |
| 224242b | II | 1 (0.5) | 1 (1.4) | 0 | 0 |
| 212111b | II | 1 (0.5) | 0 | 1 (3.4) | 0 |
| 215222b | II | 1 (0.5) | 0 | 1 (3.4) | 0 |
The character string in the genotype indicates the allele number at the folD-sfmA, rbsB, Z5935, rtcB, arp-iclR, and yhcG loci.
Genotypes based on newly observed fragments.
Shiga toxin-encoding SBI assay.
Characterization of the bacteriophage insertion site genotypes revealed that genotypes 1, 3, 5, and 6 accounted for 93% of the bovine isolates and food isolates and for 72% of the clinical isolates in The Netherlands (Table 3). The distribution of the SBI genotypes was associated with the isolation source (χ2 = 54.6, df = 8, P < 0.001). The Dutch bovine STEC O157 isolates were dominated by SBI genotypes 5 (40.8%) and 1 (32.9%), while the clinical isolates were dominated by genotype 1 (49.7%). The food isolates showed a distribution of SBI genotypes intermediate between those observed for bovine and human clinical isolates. SBI genotypes 1 and 21 were found in a significantly higher frequency among clinical isolates compared to bovine isolates (χ2 = 81, df = 1, P < 0.001, and χ2 = 14, df = 1, P < 0.001, respectively), while genotype 5 was found in a significantly higher frequency among bovine isolates (χ2 = 45, df = 1, P < 0.001). In relation to the bovine and human isolates, genotypes 1 and 5 were found at intermediate frequencies among the food isolates (37.9% and 20.7%, respectively).
Table 3.
Distribution of Shiga toxin-encoding SBI genotypes among bovine, food, and human clinical STEC O157 isolates from The Netherlands
| Genotypea | Genotyping data |
No. (%) of isolates |
||||
|---|---|---|---|---|---|---|
| stxb | yehVc | wrbAd | Bovine (n = 73) | Food (n = 29) | Human (n = 85) | |
| 1 | stx2 | Occupied | Intact | 24 (32.9) | 11 (37.9) | 42 (49.7) |
| 3 | stx1 + stx2 | Occupied | Occupied | 1 (1.3) | 3 (10.3) | 5 (5.9) |
| 5 | stx2 | Variant-R | Intact | 30 (40.8) | 6 (20.7) | 3 (3.5) |
| 6 | stx1 + stx2 | Variant-R | Intact | 13 (18.4) | 6 (20.7) | 12 (14.2) |
| 11 | stx2 | Intact | Intact | 1 (1.3) | 1 (3.5) | 1 (1.2) |
| 16 | stx1 + stx2 | Occupied | Intact | 2 (2.6) | 1 (3.5) | 6 (7.1) |
| 21 | stx2 | Occupied | Variant-L | 2 (2.6) | 1 (3.5) | 9 (10.1) |
| NTe | 0 (0) | 0 | 7 (8.2) | |||
Stx-encoding bacteriophage insertion site genotypes based on patterns of stx genes and Stx-encoding bacteriophage insertion sites.
PCR detection of stx1 and stx2 genes.
“Occupied” indicates detection of the yehV-Stx1-encoding bacteriophage left and right junctions. “Intact” indicates the detection of neither junction. “Variant-R” indicates that the left junction was detected (occupied) but the right junction was not detected (intact).
“Occupied” indicates bilateral detection of the wrbA-Stx2-encoding bacteriophage junctions. “Intact” indicates the detection of neither junction. Variant-L indicates that the left junction was not detected (intact) but the right junction was detected (occupied).
NT, nontypeable.
q21/Q933 and stx2a/stx2c assays.
The distributions of the Q allele variant (q21 and Q933) and the stx2 variant (stx2a and stx2c) were significantly associated with the isolation source (χ2 = 55.9, df = 4, P < 0.001, and χ2 = 55.7, df = 3, P < 0.001, respectively). Bovine and food isolates primarily harbored the stx2c gene variant (86.4% and 65.5%, respectively) (Table 4). The distribution of stx2a/stx2c genotypes among human isolates was less skewed, with frequencies of 36.5% for stx2c, 22.4% for stx2a, and 22.4% for stx2a plus stx2c. The occurrence of the Q gene variants was correlated with the stx2 subtype (stx2a and stx2c), with Q933 mostly associated with stx2a and q21 with stx2c. Therefore, the distribution of the Q gene variants highly resembled the distribution of the stx2 variants (Table 4).
Table 4.
Distribution of virulence factors among STEC O157 strains of bovine, food, and human clinical origin from The Netherlands
| Genotype | No. (%) of strains with virulence factor |
|||
|---|---|---|---|---|
| Total | Bovine (n = 73) | Food (n = 29) | Human (n = 85) | |
| stx2a | 50 (26.7) | 5 (6.8) | 8 (27.6) | 35 (41.2) |
| stx2c | 113 (60.5) | 63 (86.4) | 19 (65.5) | 31 (36.5) |
| stx2astx2c | 24 (12.8) | 5 (6.8) | 2 (6.9) | 19 (22.4) |
| q21 | 113 (60.5) | 62 (84.9) | 18 (62.1) | 33 (38.8) |
| Q933 | 44 (23.5) | 7 (9.6) | 8 (27.6) | 29 (34.1) |
| q21 Q933 | 23 (12.3) | 1 (1.4) | 2 (6.9) | 20 (23.5) |
| Neither q21 nor Q933 | 7 (3.7) | 3 (4.1) | 1 (3.4) | 3 (3.5) |
| tir(255A) | 57 (30.5) | 40 (54.8) | 11 (37.9) | 6 (7.1) |
| tir(255T) | 130 (69.5) | 33 (45.2) | 18 (62.1) | 79 (92.9) |
| rhsA(3468C) | 76 (40.6) | 30 (41.1) | 13 (44.8) | 33 (38.8) |
| rhsA(3468G) | 111 (59.4) | 43 (58.9) | 16 (55.2) | 52 (61.2) |
tir(A255T) polymorphism assay.
The distribution of the tir(255A) and tir(255T) alleles was associated with the isolation source (χ2 = 49.0, df = 2, P < 0.001). While tir(255A) and tir(255T) were approximately equally distributed among bovine isolates (respectively, 54.8% and 45.2%), the tir(255T) allele was dominant among food isolates (62.1%) and was overrepresented among human clinical isolates (92.9%) (Table 4).
Clade 8_rhsA assay.
The distribution of STEC O157 clade 8 isolates [rhsA(3468C) variant] was not associated with isolation source (χ2 = 0.76, df = 2, P = 0.686). The frequencies of clade 8 isolates were approximately equal among the bovine (41.0%), food (44.8%), and human clinical isolates (38.8%). Clade 8 isolates were not specifically associated with one of the LSPA6 lineages.
Multivariate analyses of population structure.
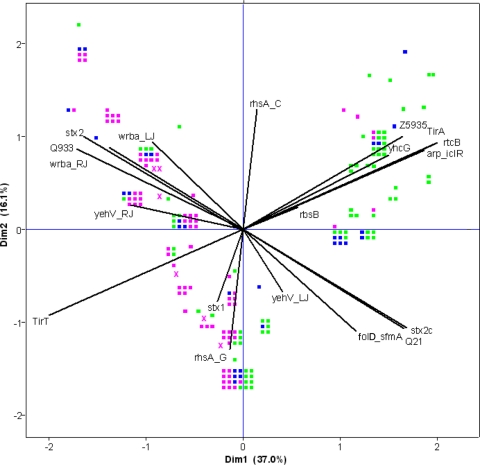
Multivariate statistical techniques were applied in order to gain insight into relationships between the genetic variables investigated and to elucidate the most distinctive genetic features differentiating bovine, food, and human clinical STEC O157 isolates. Principal component analysis was used as an ordination technique and revealed the discrimination of bovine and human isolates based on the studied genetic markers (Fig. 1). Although most of the markers contributed considerably to the observed clustering, the allelic variants of the tir gene [Tir(255A) and Tir(255T)] and the stx2 subtypes (stx2a/stx2c) explained most of the observed clustering. The ordination plot of the PCA showed that the presence of the tir variants was correlated to the values of the LSPA6 markers Z5935, rtcB, arp-iclR, and yhcG. Indeed, all LSPA6 lineage I and I/II isolates harbored the tir(255T) variant, and 88% of the lineage II isolates contained tir(255A). The presence of stx2a and/or stx2c was highly correlated to the Q933 and q21 gene variants: i.e., 96% of the isolates carrying stx2c were positive for the q21 variant, and 95% of the isolates carrying the stx2a variant were positive for the Q933 allelic variant. The presence/absence of the stx1 gene and the clade 8 marker rhsA(3468C) were negatively correlated with each other, and both markers were uncorrelated to all other markers tested.
Fig 1.
Ordination plot showing the results of the PCA for all genetic markers tested on STEC O157 isolates from bovine (green squares), food (blue squares), and human clinical cases (pink squares) from The Netherlands. Isolates clustering together have similar genetic compositions. The strength of the contribution of a specific genetic feature to the explained variation is indicated by the projected length of the line on the horizontal and vertical axes. Dim1 and Dim2 represent the first two dimensions of the PCA.
Multinomial logistic regression analysis was conducted to statistically identify the genetic features differentiating bovine, clinical and food isolates. Univariate analysis revealed significant associations (P < 0.05) of stx2a/stx2c, q21/Q933, tir(A255T), folD-sfmA, Z5935, yhcG, rtcB, yehV-RJ, and wrbA-RJ with the isolation source (Table 5). However, with the multivariate analysis, only the stx2a/stx2c and tir(A255T) variants were significantly (P = 0.001 and P < 0.0001, respectively) associated with the isolation source and were subsequently identified as the genetic features most differentiating between STEC O157 isolates from the bovine reservoir, food, and humans (Table 5).
Table 5.
Result of univariate and multivariate multinomial regression analysis of gene presence among bovine, food, and clinical human STEC O157 isolates from The Netherlandsd
| Variable | Univariable analysisa |
Multivariable analysisa,b |
||||||
|---|---|---|---|---|---|---|---|---|
| Bovine |
Food |
Bovine |
Food |
|||||
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| stx1 | 0.68 (0.33–1.40) | 0.294 | 1.34 (0.54–3.29) | 0.526 | ||||
| stx2a | 0.13 (0.06–0.27) | <0.0001 | 0.33 (0.14–0.81) | 0.015 | 0.25 (0.11–0.54) | 0.001 | 0.54 (0.21–1.41) | 0.208 |
| stx2c | 5.08 (2.30–11.24) | <0.0001 | 2.02 (0.81–5.08) | 0.133 | ||||
| q21 | 3.74 (1.73–8.13) | 0.001 | 1.41 (0.57–3.47) | 0.454 | ||||
| Q933 | 0.13 (0.06–0.28) | <0.0001 | 0.41 (0.17–0.98) | 0.044 | ||||
| tir(255A) | 14.71 (5.68–37.03) | <0.0001 | 8.06 (2.63–24.39) | <0.0001 | ||||
| tir(255T) | 0.07 (0.03–0.18) | <0.0001 | 0.12 (0.04–0.38) | <0.0001 | 0.12 (0.04–0.31) | <0.0001 | 0.16 (0.05–0.53) | 0.003 |
| rhsA(3468C) | 1.21 (0.65–2.27) | 0.554 | 1.28 (0.55–3.00) | 0.570 | ||||
| rhsA(3468G) | 0.83 (0.44–1.55) | 0.554 | 0.78 (0.33–1.83) | 0.570 | ||||
| folD-sfmA | 12.35 (1.56–97.00) | 0.017 | 1.03 (0.30–3.47) | 0.003 | ||||
| Z5935 | 12.73 (5.02–32.26) | <0.0001 | 9.51 (3.50–25.83) | <0.0001 | ||||
| yhcG | 0.123 (0.09–0.32) | <0.0001 | 0.37 (0.10–1.3) | <0.0001 | ||||
| rtcB | 14.12 (3.53–6.06) | <0.0001 | 7.43 (2.49–22.18) | <0.0001 | ||||
| rbsB | NAc | NA | ||||||
| yehV-LJ | 0.30 (0.02–1.05). | 0.056 | 0.37 (0.10–1.3) | 0.120 | ||||
| yehV-RJ | 3.68 (1.91–7.09) | 0.002 | 1.69 (0.71–4.06) | 0.237 | ||||
| wrbA-LJ | 4.69 (0.53–41.06) | 0.163 | 0.39 (0.01–1.57) | 0.185 | ||||
| wrbA-RJ | 8.50 (2.43–29.73) | 0.001 | 1.34 (0.48–3.72) | 0.567 | ||||
The odds ratios (ORs) indicate whether a gene or allelic variant is more (>1) or less (<1) likely to be associated with the bovine or food isolates compared to the human isolates. (The human isolates serve as the reference in the regression analysis.) 95% CI, 95% confidence interval. The P values shown represent the P values of the ORs.
Model fit, P < 0.0001; likelihood ratio tests, stx2a, chi-square = 13.74, P = 0.002; tir(T255), chi-square = 23.39, P < 0.0001.
NA, not available. The OR could not be calculated because all human and food isolates showed the same allelic type (type 1) in the rbsB locus.
Significant values are indicated in boldface.
The distribution of genotypes defined by the different combinations of the tir(A255T) SNP and the stx2a/stx2c gene variations markedly differed between the isolation sources and between the LSPA6 genotypes (Table 6). The frequency of tir(255A) plus stx2c was clearly lower among human isolates relative to bovine isolates (respectively, 4.7% and 53.4%, an 11-fold difference). In contrast, genotype tir(255T) stx2astx2c was clearly more present among human isolates than among bovine ones (respectively, 22.4% and 1.4%, 16-fold difference). Also genotype tir(255T) stx2a was higher in numbers among human isolates (37.6%) than among bovine isolates (11.0%) (3-fold difference). The frequencies of genotypes tir(255A) stx2a stx2c and tir(255T) stx2c were more or less equal between the different isolation sources (Table 6).
Table 6.
Frequencies of STEC O157 tir(255A)/tir(255T) and stx2a/stx2c genotypes based on isolation source and LSPA6 lineage
| Genotype | Frequency (%) of genotype bya: |
|||||
|---|---|---|---|---|---|---|
| Isolation source |
Lineage |
|||||
| Bovine | Food | Human | LSPA6 I | LSPA6 I/II | LSPA6 II | |
| tir(255A) stx2a | 0 | 0 | 0 | 0 | 0 | 0 |
| tir(255A) stx2c | 53.4 | 37.9 | 4.7 | 0 | 0 | 80.3 |
| tir(255A) stx2astx2c | 2.7 | 0 | 2.4 | 0 | 0 | 6.1 |
| tir(255T) stx2a | 11.0 | 31.0 | 37.6 | 100 | 31.7 | 3.0 |
| tir(255T) stx2c | 31.5 | 24.1 | 32.9 | 0 | 51.0 | 10.6 |
| tir(255T) stx2astx2c | 1.4 | 6.9 | 22.4 | 0 | 17.3 | 0 |
Shown are the percentages of the total number of isolates in each group.
LSPA6 lineage I isolates were all genotype tir(255T) stx2c, while LSPA6 lineage II was dominated by the tir(255A) stx2c genotype (80.3%) (Table 6). Although LSPA6 lineage I/II isolates were all tir(255T), they showed more variation in combination with the stx2a/stx2c gene variants (Table 6).
No differences were found in the frequencies of genotypes defined by the different combinations of the tir(A255T) SNP and the stx2a/stx2c gene variations between isolates from persons that were hospitalized and those from persons who were not. The same applied for hemolytic-uremic syndrome (HUS) versus non-HUS patients. The seven HUS isolates were characterized as follows: LSPA6 I/II, stx2a stx2c tir(255T) (n = 2); LSPA6 I/II, stx2a tir(255T) (n = 3); LSPA6 I/II, stx2c tir(255T) (n = 1); and LSPA6 II, stx2c tir(255A) (n = 1). Among the food isolates, six were isolated from fresh produce. Five of these isolates were characterized by LSPA6 LII, tir(255A), and stx2c, while one strain was characterized by LSPA6 LI/II, tir(255T), and stx2c.
DISCUSSION
In this study, major differences were revealed in the frequencies of STEC O157 genotypes among isolates from the bovine reservoir, food, and humans using LSPA6 and SBI genotyping methods and PCR typing of virulence factors. The results demonstrated that STEC O157 genotypes and genetic polymorphisms occurring at high frequency among human clinical isolates [LSPA6 lineage I/II, stx2a, Q933, and tir(255T)] occurred at relatively low numbers among bovine isolates. Similarly, genotypes occurring at high frequency among bovine isolates [LSPA6 lineage II, tir(255A), stx2c, q21, and SBI genotype 5] were less prevalent among human clinical isolates. These observations support the hypothesis that only a fraction of STEC O157 strains in the bovine reservoir are associated with causing disease in humans. Food isolates mostly showed genotype and marker frequencies intermediate between those observed among bovine and human clinical isolates, indicating a possible role for food as a selection agent for potentially human-associated genotypes. Multivariate statistical analysis showed that the most distinctive genetic features differentiating human from bovine isolates were tir(255T) and stx2a. Genotypes defined by the combination of these two markers were strongly correlated to the LSPA6 lineage and nonrandomly distributed among bovine and human isolates.
The results clearly showed different STEC O157 population structures among bovine, food, and human clinical isolates based on LSPA6 genotype distribution. While the LSPA6 LII genotypes dominated among bovine isolates of STEC O157, they formed a minority among human isolates. In turn, human isolates were dominated by LSPA6 LI/II (211111). LSPA6 LI (111111) was found to be a relatively rare genotype in The Netherlands (1.4%, 13.8%, and 14.1% among bovine, food, and human clinical isolates, respectively). Data from North America, where the STEC O157 disease incidence is higher than that in the Netherlands, showed different LSPA6 genotypic distributions among bovine and human clinical isolates. Originally, Kim et al. (23) described the existence of two distinct lineages of STEC O157 from the Untied States: lineage I was composed mostly of clinical isolates (36/44), and lineage II consisted mostly of cattle isolates (25/32). However, Yang et al. (43) reported the dominance of LSPA6 LI among both U.S. cattle and human clinical isolates. Similarly, in Canada the majority of cattle and human clinical isolates were typed as LSPA6 LI (38, 45). In Japan, LSPA6 LI was dominant among human isolates (69%), but all three LSPA6 lineages were considerably represented among cattle isolates (26). The present study showed that the occurrence of LSPA6 genotypes in The Netherlands is clearly different from those in North America and Japan. The most striking differences are the low frequency of LSPA6 LI isolates and the high frequency of LSPA6 LI/II isolates among human clinical isolates. Earlier postulated statements on the increased capability of LSPA6 LI isolates to cause disease in humans and the rapid spread of this lineage into a previously unoccupied ecological niche (30) do not seem to be supported by the Dutch LSPA6 genotypic distribution.
In contrast to earlier published results from the United States on the distribution of SBI genotypes among bovine and human isolates (3), the results of the present study do not show a higher diversity among STEC O157 isolates from the bovine reservoir compared to human isolates. Seven different (untypeable isolates excluded) SBI genotypes were observed among bovine isolates as well as among food and human clinical isolates. Besser et al. (3) observed four different SBI genotypes among human isolates from the United States, but 93% of all isolates were represented by genotypes 1 (30%) and 3 (62%), which together also comprised nearly 60% of the bovine isolates. In sharp contrast, in The Netherlands, SBI genotype 3 seemed of minor importance. Similar to the United States, in The Netherlands, SBI genotype 1 could be identified as a risk genotype since it comprised 50% of the human clinical isolates. Although dominant among Dutch bovine isolates, SBI genotype 5 was found to be of little clinical importance. In contrast, the clinical association of SBI genotype 1 is associated with increased expression of virulence (LEE and pO157) genes compared to the bovine-associated genotype 5 (40). In turn, bovine genotype 5 is associated with increased acid survival (40). The capacity to withstand extreme acidity is critical for STEC O157 isolates in order to successfully colonize cattle. In addition, increased expression of the central activator of the glutamate decarboxylase acid resistance system negatively regulates expression of LEE (39).
Clade 8 isolates of STEC O157 were suggested to be more virulent than isolates from other clades (34). Recent studies demonstrated that U.S. clade 8 isolates belonged primarily to LSPA6 genotype 211111 (LI/II) (24, 29). However, in the present study, no specific association of clade 8 isolates with a particular LSPA6 lineage in The Netherlands was observed. In Sweden, clade 8 STEC O157 isolates were overrepresented among isolates from cattle farms associated with human cases compared with STEC O157 isolates isolated in prevalence studies (10). In The Netherlands, clade 8 isolates were not more prevalent among human clinical isolates compared to bovine isolates and are therefore not suitable as marker for identifying isolates with increased risk of causing clinical infections.
The Q933 variant of the antiterminator gene Q, located upstream of the stx2 gene, resulting in relatively high expression of the stx2 gene, was initially reported as a useful marker for (potential) human risk among bovine isolates since only 44% of the bovine isolates and 83% of the human clinical isolates were positive for Q933 (27). In The Netherlands, the frequency of Q933 increased from 10% among bovine isolates to 34% among human clinical isolates. In addition, the frequency of isolates carrying both gene variants (q21 and Q933) increased from 1% among bovine isolates to 24% among human clinical isolates. Very strong correlations were observed between q21 and stx2c and between Q933 and stx2a. Consequently, like with the Q gene variants, bovine isolates were dominated by isolates carrying only stx2c (86%), while this frequency was only 37% among clinical isolates. Similar patterns were reported by recent Norwegian (18) and Japanese (26) studies. Epidemiological studies have revealed that stx2a is more strongly associated with clinical isolates and causes more severe symptoms than stx2c (1, 4, 21, 36). In addition, the effective dose to inhibit protein synthesis in Vero cells has been reported to be 100 times lower for stx2a than stx2c, which correlated with a 50% lethality dose for mice 154 times lower than that for stx2a (16).
Intimin (encoded by eae) and its translocated intimin receptor (encoded by tir) are bacterial proteins that mediate adhesion between mammalian cells and attaching and effacing (A/E) pathogens like STEC O157 (20). The results of the tir(A255T) polymorphism assay confirm earlier findings of an overrepresentation of the tir(255T) variant among human clinical isolates of STEC O157 isolates and an approximately equal distribution of both variants among bovine isolates (6, 18). The reason for the specific association of human clinical isolates with tir(255T) is unknown. It might be possible that isolates carrying the tir(255A) allele are shed for shorter periods of time by cattle and/or survive less well in manure, soil, water, or food. Another possibility is that STEC O157 tir(255A) isolates are less virulent for humans than STEC O157 tir(255T) isolates. It was originally suggested that the central role of tir in human infection points toward a possible functional discrimination between both alleles (6). Adherence to epithelial cells is a major function of the Tir protein; allele polymorphisms could affect adherence and consequently the association with human infection (6). The SNP in the tir gene is located near the amino terminus of the Tir protein, which anchors host cytoskeletal components (such as actin) that are needed to form the characteristic A/E lesions on the host cell surface upon bacterial adhesion (32). However, more investigation will be necessary to delineate the structure-function relationships of these tir(A255T) polymorphisms and its relationship to the clinical outcome of infection.
Multivariate statistics revealed that the tir(A255T) single nucleotide polymorphism and the stx2a/stx2c gene variation were the most differentiating genetic features between bovine, food, and human isolates from The Netherlands. Bovine isolates compromised mostly stx2c genotypes [tir(255A) stx2c and tir(255T) stx2c)], while human isolates were dominated by tir(255T) genotypes [tir(255T) stx2a, tir(255T) stx2c, and tir(255T) stx2a stx2c]. The stx2a gene variation was the most distinctive feature in human isolates compared to bovine isolates in Japan (26). Other studies showed a strong association between LSPA6 genotypes and the stx2a/stx2c genotypes, with a strong bias of stx2a toward LSPA6 LI and of stx2c to LSPA6 LII (26, 45). The results of the present study are consistent with those studies and showed that LSPA6 LI isolates consisted solely of genotype tir(255T) stx2c, while LSPA6 LII was dominated by tir(255A) stx2a. As shown previously (26), LSPA6 LI/II isolates showed more diversity in tir(A255T) stx2a/stx2c genotypes, which is in agreement with the suggestion that LSPA6 LI/II is an intermediate lineage between LI and LII (43).
The results of the present study support the hypothesis that the STEC O157 genotypes which are primarily associated with human disease form a minor subpopulation in the bovine reservoir. This might argue for more focused regulatory attention on the subset of STEC O157 genotypes dominant among clinical isolates. It is of interest that the distribution of LSPA6 lineages among bovine and human isolates is very different in The Netherlands (this study) than those in North America (38, 45) and Japan (26). To the best of our knowledge, no other studies have been reported on the distribution of LSPA6 lineages in European STEC O157 populations. There are clear geographical differences in STEC O157 disease incidence, which is generally higher in North America, the United Kingdom, and Sweden than those in most continental European countries (8). These differences might be related to STEC O157 farm prevalence (28), the environmental load of STEC O157 due to super shedders (8), and/or the proportions of human populations living in areas with high cattle density (15, 17). In addition, disease incidence might also be related to the differences in STEC O157 genetic population structures. The higher disease incidence in North America compared to The Netherlands might be (partly) explained by the difference in LSPA6 lineage I frequency, which is much higher in North America (38, 43, 45). LSPA6 lineage I is highly correlated with the presence of the more virulent gene variants like stx2a and tir(255T) and may therefore result in a higher disease rate. Indeed, LSPA6 LI isolates showed higher levels of epithelial adherence and Shiga toxin production than LI and LI/II strains (30, 44).
Another explanation for the geographic differences in disease incidence might be that humans are primarily exposed to STEC O157 by other pathways in different geographic regions (direct contact with animals and/or feces, environmental transmission, and food transmission) and that isolates of different LSPA6 lineages differ in their capacity to be transmitted by these routes. Although a significant longer survival in manure-amended soil of human STEC O157 isolates compared to animal isolates was demonstrated previously, no relationship could be identified between the virulence profile and survival and between the LSPA6 and survival (11). Likewise, no differences with respect to survival in manure were observed between LSPA6 lineage I and II isolates (31). The underlying reason for the variety in population structure in different geographic regions with respect to LSPA6 lineage is still unclear but might be related to specific selection pressures encountered in the various types of dairy and veal production systems and their surrounding environments. Future research is required to reveal the mechanisms responsible for the shaping of STEC O157 populations and the relationship to differences in disease incidence in different geographic regions. This can ultimately be used for the development of intervention strategies.
ACKNOWLEDGMENTS
This work was financed by the Dutch Ministry of Economic Affairs, Agriculture and Innovation, and the Dutch Food and Consumer Product Safety Authority.
We thank El Bouw for technical assistance.
Footnotes
Published ahead of print 21 December 2011
REFERENCES
- 1. Andersson T, et al. 2011. Modeling gene associations for virulence classification of verocytotoxin-producing E. coli (VTEC) from patients and beef. Virulence 2:41–53 [DOI] [PubMed] [Google Scholar]
- 2. Berends IMGA, et al. 2008. Prevalence of VTEC O157 in dairy and veal herds and risk factors for veal herds. Prev. Vet. Med. 87:301–310 [DOI] [PubMed] [Google Scholar]
- 3. Besser TE, et al. 2007. Greater diversity of Shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl. Environ. Microbiol. 73:671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beutin L, et al. 2007. Identification of human-pathogenic strains of Shiga toxin-producing Escherichia coli from food by a combination of serotyping and molecular typing of Shiga toxin genes. Appl. Environ. Microbiol. 73:4769–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bono JL. 2009. Genotyping Escherichia coli O157:H7 for its ability to cause disease in humans. Curr. Protoc. Microbiol. 14:5A.3.1-5A.3.10 [DOI] [PubMed] [Google Scholar]
- 6. Bono JL, et al. 2007. Association of Escherichia coli O157:H7 tir polymorphisms with human infection. BMC Infect. Dis. 7:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caprioli A, Morabito S, Brugere H, Oswald E. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36:289–311 [DOI] [PubMed] [Google Scholar]
- 8. Chase-Topping M, Gally D, Low C, Matthews L, Woolhouse M. 2008. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat. Rev. Microbiol. 6:904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eklund M, Scheutz F, Siitonen A. 2001. Clinical isolates of non-O157 Shiga toxin-producing Escherichia coli: serotypes, virulence characteristics, and molecular profiles of strains of the same serotype. J. Clin. Microbiol. 39:2829–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eriksson E, Söderlund R, Boqvist S, Aspan A. 2011. Genotypic characterization to identify markers associated with putative hypervirulence in Swedish Escherichia coli O157:H7 cattle strains. J. Appl. Microbiol. 110:323–332 [DOI] [PubMed] [Google Scholar]
- 11. Franz E, Hoek HAM, Bouw E, Aarts HJM. 2011. Variability of Escherichia coli O157 strain survival in manure-amended soil in relation to strain origin, virulence profile, and carbon nutrition profile. Appl. Environ. Microbiol. 77:8088–8096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franz E, Klerks MM, De Vos OJ, Termorshuizen AJ, Van Bruggen AHC. 2007. Prevalence of Shiga toxin-producing Escherichia coli stx1, stx2, eaeA, and rfbE genes and survival of E. coli O157:H7 in manure from organic and low-input conventional dairy farms. Appl. Environ. Microbiol. 73:2180–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friedrich AW, et al. 2002. Escherichia coli harboring shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74–84 [DOI] [PubMed] [Google Scholar]
- 14. Friesema IHM, et al. 2011. Intensieve surveillance van Shiga-toxineproducerende Escherichia coli (STEC) in Nederland, 2009. Infectieziektebulletin 22:22–29 [Google Scholar]
- 15. Friesema IHM, Van De Kassteele J, De Jager CM, Heuvelink AE, Van Pelt W. 2011. Geographical association between livestock density and human Shiga toxin-producing Escherichia coli O157 infections. Epidemiol. Infect. 139:1081–1087 [DOI] [PubMed] [Google Scholar]
- 16. Fuller CA, Pellino CA, Flagler MJ, Strasser JE, Weiss AA. 2011. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 79:1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haack JP, et al. 2003. Escherichia coli O157 exposure in Wyoming and Seattle: serologic evidence of rural risk. Emerg. Infect. Dis. 9:1226–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haugum K, Brandal LT, Løbersli I, Kapperud G, Lindstedt BA. 2011. Detection of virulent Escherichia coli O157 strains using multiplex PCR and single base sequencing for SNP characterization. J. Appl. Microbiol. 110:1592–1600 [DOI] [PubMed] [Google Scholar]
- 19. Heuvelink AE, et al. 1998. Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. J. Clin. Microbiol. 36:3480–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaper JB, Nataro JP, Mobley HLT. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 21. Käppeli U, Hächler H, Giezendanner N, Cheasty T, Stephan R. 2011. Shiga toxin-producing Escherichia coli O157 associated with human infections in Switzerland, 2000–2009. Epidemiol. Infect. 139:1097–1104 [DOI] [PubMed] [Google Scholar]
- 22. Karmali MA, Gannon V, Sargeant JM. 2010. Verocytotoxin-producing Escherichia coli (VTEC). Vet. Microbiol. 140:360–370 [DOI] [PubMed] [Google Scholar]
- 23. Kim J, Nietfeldt J, Benson AK. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. U. S. A. 96:13288–13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laing CR, et al. 2009. In silico genomic analyses reveal three distinct lineages of Escherichia coli O157:H7, one of which is associated with hyper-virulence. BMC Genomics 10:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. LeBlanc JJ. 2003. Implication of virulence factors in Escherichia coli O157:H7 pathogenesis. Crit. Rev. Microbiol. 29:277–296 [DOI] [PubMed] [Google Scholar]
- 26. Lee KI, et al. 2011. Multivariate analyses revealed distinctive features differentiating human and cattle isolates of Shiga toxin-producing Escherichia coli O157 in Japan. J. Clin. Microbiol. 49:1495–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lejeune JT, Abedon ST, Takemura K, Christie NP, Sreevatsan S. 2004. Human Escherichia coli O157:H7 genetic marker in isolates of bovine origin. Emerg. Infect. Dis 10:1482–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lejeune JT, Hancock D, Wasteson Y, Skjerve E, Urdahl AM. 2006. Comparison of E. coli O157 and Shiga toxin-encoding genes (stx) prevalence between Ohio, U.S.A. and Norwegian dairy cattle. Int. J. Food Microbiol. 109:19–24 [DOI] [PubMed] [Google Scholar]
- 29. Liu K, Knabel SJ, Dudley EG. 2009. rhs genes are potential markers for multilocus sequence typing of Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 75:5853–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lowe RMS, et al. 2009. Escherichia coli O157:H7 strain origin, lineage, and Shiga toxin 2 expression affect colonization of cattle. Appl. Environ. Microbiol. 75:5074–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lowe RMS, et al. 2010. Factors influencing the persistence of Escherichia coli O157:H7 lineages in feces from cattle fed grain versus grass hay diets. Can. J. Microbiol. 56:667–675 [DOI] [PubMed] [Google Scholar]
- 32. Luo Y, et al. 2000. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405:1073–1077 [DOI] [PubMed] [Google Scholar]
- 33. Mainil JG, Daube G. 2005. Verotoxigenic Escherichia coli from animals, humans and foods: who's who? J. Appl. Microbiol. 98:1332–1344 [DOI] [PubMed] [Google Scholar]
- 34. Manning SD, et al. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105:4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paton JC, Paton AW. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Persson S, Olsen KEP, Ethelberg S, Scheutz F. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45:2020–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shaikh N, Tarr PI. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sharma R, et al. 2009. Escherichia coli O157:H7 lineages in healthy beef and dairy cattle and clinical human cases in Alberta, Canada. J. Food Prot. 72:601–607 [DOI] [PubMed] [Google Scholar]
- 39. Shin S, et al. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, per. Mol. Microbiol. 41:1133–1150 [DOI] [PubMed] [Google Scholar]
- 40. Vanaja SK, Springman AC, Besser TE, Whittam TS, Manning SD. 2010. Differential expression of virulence and stress fitness genes between Escherichia coli O157:H7 strains with clinical or bovine-biased genotypes. Appl. Environ. Microbiol. 76:60–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wagner PL, et al. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183:2081–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Whitworth J, et al. 2010. Diverse genetic markers concordantly identify bovine origin Escherichia coli O157 genotypes underrepresented in human disease. Appl. Environ. Microbiol. 76:361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang Z, et al. 2004. Identification of common subpopulations of non-sorbitol-fermenting, β-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl. Environ. Microbiol. 70:6846–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang Y, et al. 2010. Lineage and host source are both correlated with levels of Shiga toxin 2 production by Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 76:474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ziebell K, et al. 2008. Genotypic characterization and prevalence of virulence factors among Canadian Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 74:4314–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]