Abstract
Salmonella enterica serovar Stanley (S. Stanley) is a common serovar in Southeast Asia and was the second most common serovar implicated in human salmonellosis in Thailand in the years 2002 to 2007. In contrast, this serovar is relatively uncommon in Europe. The objective of this study was to characterize a collection of S. Stanley strains isolated from Thai (n = 62), Danish (n = 39), and French (n = 24) patients to gain a broader understanding of the genetic diversity, population dynamics, and susceptibility to antimicrobials. All isolates were characterized by pulsed-field gel electrophoresis and antimicrobial susceptibility testing. The molecular mechanisms of resistance to extended-spectrum cephalosporins and plasmid-mediated resistance to quinolones were characterized by PCR and sequencing. Plasmid profiling, replicon typing, and microarray analysis were used to characterize the genetic mechanisms of antimicrobial resistance in 10 extended-spectrum cephalosporinase-producing isolates. Considerable genetic diversity was observed among the isolates characterized with 91 unique XbaI pulsed-field gel electrophoresis (PFGE) patterns, including 17 distinct clusters consisting of two to seven indistinguishable isolates. We found some of the S. Stanley isolates isolated from patients in Europe were acquired during travel to Southeast Asia, including Thailand. The presence of multiple plasmid lineages carrying the extended-spectrum cephalosporinase-encoding blaCMY-2 gene in S. Stanley isolates from the central part of Thailand was confirmed. Our results emphasize that Thai authorities, as well as authorities in other countries lacking prudent use of antimicrobials, should improve the ongoing efforts to regulate antimicrobial use in agriculture and in clinical settings to limit the spread of multidrug-resistant Salmonella isolates and plasmids among humans and pigs in Thailand and abroad.
INTRODUCTION
Salmonella enterica is a common cause of human gastroenteritis and bacteremia worldwide (14, 28, 31, 36), and a wide variety of animals, particularly food animals, have been identified as reservoirs for nontyphoidal Salmonella (27). The majority of human Salmonella infections are caused by a limited number of the more than 2,600 Salmonella serovars described to date, and the prevalence of specific Salmonella serovars differs by geographical region (21, 23, 29, 34). In developed countries, Salmonella enterica serovar Typhimurium and S. Enteritidis are the most commonly reported causes of human salmonellosis, while in Thailand a recent study showed that Salmonella serovar Stanley was the second most common serovar, accounting for 11% of all human salmonellosis cases from 2002 to 2007 (23). A steady increase of Salmonella serovar Stanley infections has also been observed. This serovar accounted for 1.9% (n = 64) of salmonellosis cases in 1993 and 10.1% of cases (n = 141) in 2003 (23). Salmonella serovar Stanley is a serovar generally associated with pig and pork production but is neither host restricted nor host adapted (16, 33).
Shifts in the prevalences of specific Salmonella strain types and serovars in human and animal populations may follow the movement of bacterial isolates through international travel, human migration, and food and livestock trade. The growth of international travel is a public health concern, as an increasing numbers of travelers have the potential to acquire infections abroad and introduce imported bacterial isolates into their home communities (1, 2, 22, 30, 35).
In Denmark, Salmonella serovar Stanley was the fifth most common (n = 44) serovar among the 3,657 confirmed human salmonellosis cases reported to the Statens Serum Institut (SSI) in 2008, and 24 (55%) of these cases were travel related and primarily acquired in Thailand (17).
In France, Salmonella serovar Stanley was ranked 36th (n = 25) among the 10,378 serotyped Salmonella isolates from humans registered at the Institut Pasteur (IP) in 2008. Travel information was available for 13 of these patients and showed that five (38%) patients had traveled to Southeast Asia in the 2 weeks before the onset of illness.
Extended-spectrum cephalosporins (ESC) and fluoroquinolones (FQ) are the drugs of choice for treatment of severe Salmonella infections. Accordingly, these drugs have been classified by the World Health Organization as critically important antimicrobials (15), and limiting the occurrence of resistance (R) to extended-spectrum cephalosporins and fluoroquinolones is a public health priority. An emerging issue in resistance to extended-spectrum cephalosporins and fluoroquinolones is the presence of resistance genes on mobile genetic elements such as plasmids that are easily transferred among bacterial isolates (6). In Salmonella spp., a large number of plasmid-borne genes coding for enzymes inactivating extended-spectrum cephalosporins have been described to date, including the extended-spectrum β-lactamase gene group consisting of blaTEM, blaSHV, blaCTX-M, and blaCMY, also known as extended-spectrum cephalosporinases (5). Among the plasmid-mediated quinolone resistance (PMQR) genes described to date, qnr genes represent the largest group and have been found worldwide (32).
An increased occurrence of resistance to extended-spectrum cephalosporins and decreased susceptibility to fluoroquinolones in Salmonella isolates causing human infections have been reported in Southeast Asian countries (29). In Denmark, a recent study showed that Salmonella isolates acquired abroad exhibited higher levels of antimicrobial resistance than domestically acquired isolates, thus posing possible treatment problems (17).
In this study, we characterized a collection of clinical Salmonella serovar Stanley isolates from Thailand, Denmark, and France to (i) determine the occurrence of antimicrobial resistance, (ii) investigate the genetic mechanisms of resistance to critically important antimicrobials, and (iii) examine the genotypes circulating in Thailand and their possible link to travel-related infections in patients from Europe.
MATERIALS AND METHODS
Bacterial isolates.
Salmonella serovar Stanley isolates from clinical cases reported in Thailand, Denmark, and France in 2008 were included in this study.
In Bangkok (BKK), Thailand, the WHO National Salmonella and Shigella Center (NSSC) receives all presumptive Salmonella isolates from diagnostic laboratories throughout Thailand and records available epidemiological information. Of 276 Salmonella serovar Stanley isolates (all specimen types) received at the NSSC in 2008, 50 isolates were selected based on the geographical zones (1 to 13) of isolation (4). Within each zone, isolates were randomly selected to proportionally represent the distribution of Salmonella serovar Stanley isolates per zone as determined from 2002 to 2007 (23). Additional isolates were selected based on a preliminary antibiogram obtained at the NSSC using disk diffusion testing (10, 11). Of the total of 276 isolates, 193 were available for antimicrobial susceptibility testing. Groups of 11 and 63 Salmonella serovar Stanley isolates displaying a phenotype compatible with the presence of extended-spectrum cephalosporinase-encoding genes (10, 11) and plasmid-mediated quinolone-resistance (PMQR) genes (8), respectively, were also included in the study.
A total of 124 Salmonella serovar Stanley isolates were dispatched from NSSC to the National Food Institute (DTU-Food), Kgs. Lyngby, Denmark, for further analyses.
In Denmark, the Statens Serum Institut (SSI) receives data on all laboratory-confirmed Salmonella cases, including epidemiological information. All regional clinical laboratories submit Salmonella isolates to the SSI (>95% coverage). In Denmark, a total of 44 Salmonella serovar Stanley cases were registered in 2008. Of these, 39 Salmonella serovar Stanley isolates were available for this study.
In France, approximately 65% of all human Salmonella isolates recovered in clinical practice are reported to the French National Reference Centre at the Institut Pasteur (IP) by approximately 1,400 hospital and private clinical laboratories. A total of 25 Salmonella serovar Stanley isolates were collected during 2008, but only 24 isolates were viable and were shipped to DTU-Food, Denmark, for further analyses.
Serotyping.
All isolates were serotyped by using slide agglutination in the country of origin. O and H antigens were characterized by agglutination with hyperimmune sera (S & A Reagents Laboratory, Ltd., Bangkok, Thailand; Statens Serum Institut, Copenhagen, Denmark; Bio-Rad, Marnes la Coquette, France; and WHO Collaborative Centre on Reference and Research on Salmonella, Institut Pasteur, Paris, France), and serotypes were assigned according to the Kauffmann-White scheme (19).
Antimicrobial susceptibility testing.
MIC determinations were performed on all isolates included in the study at DTU-Food, Denmark (for isolates from Thailand and France), and SSI, Denmark (for isolates from Denmark), by using a commercially prepared, dehydrated panel (Sensititre; TREK Diagnostic Systems Ltd., East Grinstead, England). The following antimicrobials and interpretative criteria for resistance (R) were used: ampicillin (AMP; R > 8 mg/liter); amoxicillin-clavulanic acid (AMC; R ≥ 32 mg/liter); apramycin (APR; R > 32 mg/liter); cefotaxime (CTX; R > 0.5 mg/liter); ceftiofur (XNL; R > 2 mg/liter); chloramphenicol (CHL; R > 16 mg/liter); ciprofloxacin (CIP) (R low level, 0.064 to 1 mg/liter; R high level, >1 mg/liter); colistin (COL; R > 2 mg/liter); florfenicol (FFN; R > 16 mg/liter); gentamicin (GEN; R > 2 mg/liter); nalidixic acid (NAL; R > 16 mg/liter); neomycin (NEO; R > 4 mg/liter); spectinomycin (SPT; R > 64 mg/liter); streptomycin (STR; R > 16 mg/liter); sulfamethoxazole (SMX; R > 256 mg/liter); tetracycline (TET; R > 8 mg/liter); and trimethoprim (TMP; R > 2 mg/liter). Presumptive extended-spectrum cephalosporinase producers were also tested for susceptibility to cefazolin (CFZ; R ≥ 4 mg/liter), cefepime (FEP; R ≥ 32 mg/liter), cefoxitin (FOX; R > 8 mg/liter), cefpodoxime (CPD; R ≥ 8 mg/liter), ceftazidime (CAZ; R > 2 mg/liter), ceftriaaxone (CRO; R ≥ 4 mg/liter), cephalothin (CEF; R > 16 mg/liter), imipenem (IPM; R > 1 mg/liter), and meropenem (MEM; R > 0.125 mg/liter) by using the method described above. Interpretation of antimicrobial susceptibility test results was performed by using epidemiological cutoff values according to EUCAST recommendations (http://www.eucast.org) for all antimicrobials except for AMC, CRO, CFZ, CPD, FEP, SMX, and SPT, for which Clinical and Laboratory Standards Institute (CLSI) clinical breakpoints were used (11–13), since EUCAST values have not been available to date. Due to the lack of epidemiologic cutoff values in the EUCAST system and CLSI clinical breakpoints, APR results were interpreted according to research results from DTU. Quality control was performed by using reference strain Escherichia coli ATCC 25922 according to CLSI guidelines (11, 13).
Detection of extended-spectrum cephalosporinase-encoding genes and plasmid-mediated quinolone resistance genes.
Based on MIC results, Salmonella serovar Stanley isolates exhibiting a phenotype compatible with the presence of extended-spectrum cephalosporinase-encoding genes or PMQR genes were selected for PCR amplification of blaCTX-M, blaSHV, blaTEM, blaCMY-1,and blaCMY-2 genes or of qnrA, qnrB, qnrC, qnrD, qnrS, qepA, and aac(6′)-lb genes, respectively. The primers and conditions used were described previously (20, 24). Obtained amplicons were purified by using a GFX PCR DNA kit (GE Healthcare, Chalfont St. Giles, United Kingdom) according to the manufacturer's recommendations and then sequenced (Macrogen Inc., Seoul, South Korea) by using the same primers as in the PCR analysis. Sequence analysis was performed using Vector NTI suite 9 software (InforMax Inc., Bethesda, MD). The obtained nucleotide sequences and the derived amino acid sequences were compared with previously described sequences obtained from the GenBank database (http://www.ncbi.nlm.nih.gov/) and www.lahey.org/studies/, respectively.
Pulsed-field gel electrophoresis.
All isolates from Denmark and France and a subset of 62 Thai isolates selected on the basis of the geographical origin and MIC determination results were genotyped by pulsed-field gel electrophoresis (PFGE) using XbaI according to the CDC PulseNet protocol (32a). The electrophoresis of the French and Thai isolates was performed at DTU-Food, Denmark, in a Chef DR III system (Bio-Rad Laboratories, Hercules, CA) using 1% SeaKem Gold agarose in 0.5× Tris-borate-EDTA (TBE) buffer. Running conditions consisted of one pulse of 2.2 to 63.8 s for 20 h at 6 V/cm on a 120-degree angle in 14°C TBE buffer. The PFGE of the Danish isolates was performed at SSI, Denmark, using the same protocol and equipment. Subsequently, a bundle file containing the PFGE analysis of the Danish isolates was sent to DTU-Food. Comparative analysis of the PFGE profiles of all isolates was performed by using Bionumerics software version 4.6 (Applied Maths, Sint-Martens-Latem, Belgium) and the Dice correlation coefficient for band matching with a 0.9% position tolerance and an optimization at 0.9%. The PFGE patterns were not uploaded to PulseNet due to the lack of a database in Europe.
Plasmid characterization.
Ten Thai blaCMY-2-positive Salmonella serovar Stanley isolates representing different PFGE lineages and geographical regions of isolation were selected for plasmid analysis. Plasmid DNA was extracted with a Plasmid Mini Kit (Qiagen, Hilden, Germany) and transformed into electrocompetent E. coli DH10B (Invitrogen, Taastrup, Denmark). Transformants were selected on BHI agar (Becton Dickinson, Franklin Lakes, NJ) plates supplemented with cefotaxime (2 μg/ml), and the presence of blaCMY-2 was confirmed on all transformants by colony PCR using the primers and conditions described above.
PFGE with S1 nuclease (Promega, Madison WI) digestion was performed as described previously (35) on (i) transformants (to ensure that only one plasmid had been transferred into the recipient and to estimate the approximate size of blaCMY-2-harboring plasmids) and (ii) donors (to verify that the transferred plasmid was actually present in the donor). Plasmid incompatibility groups were determined by PCR-based replicon typing (7). To examine the presence of additional antimicrobial resistance genes on the blaCMY-2-harboring plasmids, transformants were used for MIC determinations as described above and for microarray analysis as described previously (35). Since the recipient strain DH10B is resistant to streptomycin, this experiment was unable to determine whether the blaCMY-2-harboring plasmids also carried streptomycin resistance genes.
RESULTS
Epidemiological information on Salmonella serovar Stanley cases from Thailand.
Based on data from all 13 regional zones in 2008, Salmonella serovar Stanley was identified in 272 (11%) out of 2,446 Salmonella isolates from the specimen types, namely, stool/rectal swab/rectum and blood samples. The distribution of Salmonella serovar Stanley-positive samples varied according to specimen type and region of isolation. The majority of isolates, 260 (96%), were obtained from stool/rectal swab/rectum samples, and 12 (4%) isolates were obtained from blood samples. Salmonella serovar Stanley-positive stool/rectal swabs were detected throughout Thailand except in zone 6 and were variously distributed among the different regional zones. Of note, the highest numbers of cases were described in zones 1 and 4 (n = 41 in each zone) and in BKK (Bangkok) (n = 63) (data not shown). Salmonella serovar Stanley-positive blood samples were detected in zone 2 (n = 3), 4 (n = 4), 5 (n = 1), 7 (n = 2), and BKK (n = 2), thus covering the central part of Thailand from east to west (data not shown).
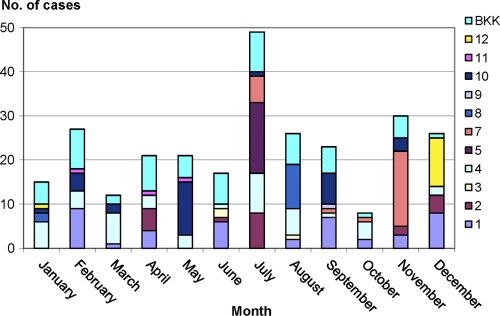
Information on age was available for 211 (76%) cases, and those patients ranged from 3 days to 94 years of age. The majority of cases (n = 134 [64%]) occurred in children less than 5 years of age, with a median of 1 year. The cases (n = 275 [99%]) were evenly distributed by gender (males = 144 [52%], females = 131 [48%]) (data not shown). Information on infection onset was available for 275 (99%) cases and revealed that the number of patients was relatively stable throughout the year, with the exception of July and October, the months that showed the highest (n = 49, i.e., 18%) and lowest (n = 8, i.e., 3%) occurrences of infections, respectively (Fig. 1).
Fig 1.
Seasonal variation in numbers per regional zone of Salmonella serovar Stanley infections among Thai patients during 2008.
Epidemiological information on Salmonella serovar Stanley cases from Denmark.
The 39 Danish Salmonella serovar Stanley-infected patients suffered from gastroenteritis. Their ages ranged from one to 71 years, and the median was 28 years. Information on gender showed that 20 (51%) and 19 (49%) patients were females and males, respectively. Twenty-four (62%) cases were travel associated, and those patients reported travel to Thailand (n = 19, i.e., 79%), Vietnam (n = 2, i.e., 8%), Philippines (n = 2, i.e., 8%), and Singapore (n = 1, i.e., 4%)(Fig. 2, 3, and 4) (Table 1). Four (10%) patients did not report traveling prior to illness onset, and travel data could not be retrieved for the 11 (28%) remaining cases. Twelve (92%) of the 13 cases reported during the months of December, January, and February were travel related. These 3 months are both the peak travel period for Danish travelers to visit Asia and the months when the highest number of Salmonella serovar Stanley infections are reported in Thailand.
Fig 2.
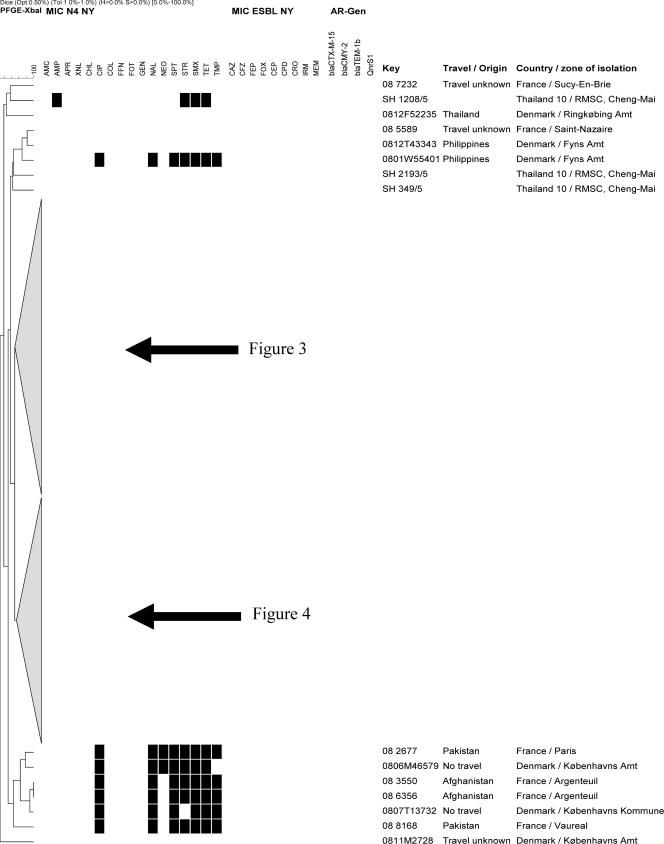
Dendrogram showing the genotypic relatedness of Salmonella enterica serovar Stanley isolates based on XbaI-PFGE fingerprints.
Fig 3.
Dendrogram showing the genotypic relatedness of Salmonella enterica serovar Stanley isolates based on XbaI-PFGE fingerprints. The dotted arrow indicates an isolate encoding a qnrS1 gene; the black arrows indicate the 20 isolates with 78% similarity and encoding extended-spectrum cephalosporinase genes.
Fig 4.
Dendrogram showing the genotypic relatedness of Salmonella enterica serovar Stanley isolates based on XbaI-PFGE fingerprints. Black squares represent the isolates classified as exhibiting resistance. The codes of the antimicrobials are as follows: ampicillin, AMP; amoxicillin-clavulanic acid, AMC; apramycin, APR; cefalothin, CEP; cefazolin, CFZ; cefepime, FEP; cefoxitin, FOX; cefpodoxime, CPD; cefotaxime, CTX; ceftazidime, CAZ; ceftiofur, XNL; ceftriaxone, CRO; chloramphenicol, CHL; ciprofloxacin, CIP; colistin, COL; florfenicol, FFN; gentamicin, GEN; imipenem, IRM; meropenem, MEM; nalidixic acid, NAL; neomycin, NEO; spectinomycin, SPT; streptomycin, STR; sulfamethoxazole, SMX; tetracycline, TET; trimethoprim, TMP.
Table 1.
Occurrence of antimicrobial resistance among Salmonella serovar Stanley isolates from Thai, Danish, and French patients during 2008
| Country | Origin | Total no. of isolates | No. (%) of isolates resistant to various antimicrobial agents at the indicated breakpoint(s) (μg/ml)a |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC ≥32 | AMP >8 | APR >32 | CTX >0.5 | CHL >16 | CIP |
COL >2 | FFN >16 | GEN >2 | NAL >16 | NEO >4 | SPT >64 | STR >16 | SMX >256 | TET >8 | TMP >2 | XNL >2 | ||||
| 0.064–1 | >1 | |||||||||||||||||||
| Denmark | Travelb | 24 | 1 (4) | 7 (29) | 0 | 1 (4) | 3 (13) | 1 (4) | 0 | 0 | 1 (4) | 1 (4) | 1 (4) | 0 | 4 (4) | 7 (29) | 7 (29) | 11 (46) | 1 (4) | 1 (4) |
| Denmark | No travel | 4 | 1 (25) | 1 (25) | 0 | 1 (25) | 1 (25) | 2 (50) | 0 | 0 | 1 (25) | 0 | 2 (50) | 1 (25) | 2 (50) | 2 (50) | 3 (75) | 3 (75) | 1 (25) | 1 (25) |
| Denmark | Unknown | 11 | 2 (18) | 6 (55) | 0 | 2 (18) | 3 (27) | 0 | 0 | 0 | 2 (18) | 2 (18) | 0 | 0 | 3 (27) | 3 (27) | 3 (27) | 6 (55) | 0 | 2 (18) |
| France | Travelc | 10 | 0 | 1 (10) | 0 | 0 | 1 (10) | 4 (40) | 0 | 0 | 0 | 0 | 4 (40) | 1 (10) | 5 (50) | 5 (50) | 5 (50) | 6 (60) | 4 (40) | 0 |
| France | No travel | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| France | Unknown | 12 | 0 | 1 (8) | 0 | 0 | 1 (8) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (8) | 2 (17) | 2 (17) | 3 (25) | 1 (8) | 0 |
| Thailand | Zone 1 | 7 | 1 (14) | 3 (43) | 0 | 1 (14) | 1 (14) | 0 | 0 | 0 | 1 (14) | 1 (14) | 0 | 0 | 1 (14) | 2 (29) | 2 (29) | 4 (57) | 0 | 1 (14) |
| Thailand | Zone 2 | 4 | 0 | 2 (50) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (25) | 3 (75) | 3 (75) | 2 (50) | 0 | 0 |
| Thailand | Zone 4 | 15 | 12 (80) | 14 (93) | 0 | 12 (80) | 11 (73) | 0 | 0 | 0 | 10 (67) | 3 (20) | 0 | 0 | 4 (27) | 14 (93) | 14 (93) | 14 (93) | 0 | 12 (80) |
| Thailand | Zone 5 | 4 | 0 | 3 (75) | 0 | 0 | 1 (25) | 1 (25) | 0 | 0 | 0 | 0 | 0 | 0 | 1 (25) | 1 (25) | 1 (25) | 2 (50) | 0 | 0 |
| Thailand | Zone 7 | 6 | 0 | 3 (50) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (50) | 0 | 0 |
| Thailand | Zone 8 | 2 | 0 | 2 (100) | 0 | 0 | 1 (50) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (50) | 1 (50) | 1 (50) | 2 (100) | 0 | 0 |
| Thailand | Zone 9 | 1 | 0 | 1 (100) | 0 | 0 | 1 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 | 0 |
| Thailand | Zone 10 | 5 | 0 | 2 (40) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (20) | 1 (20) | 2 (40) | 0 | 0 |
| Thailand | Zone 12 | 3 | 0 | 2 (67) | 0 | 0 | 1 (33) | 0 | 0 | 0 | 1 (33) | 0 | 0 | 0 | 0 | 2 (67) | 1 (33) | 3 (100) | 0 | 0 |
| Thailand | Zone BKK | 15 | 6 (40) | 10 (67) | 0 | 7 (47) | 6 (40) | 0 | 0 | 0 | 6 (40) | 7 (47) | 0 | 0 | 6 (40) | 7 (47) | 7 (47) | 9 (60) | 1 (7) | 7 (47) |
| Total | 125 | 23 (18) | 58 (46) | 0 | 24 (19) | 31 (25) | 8 (6) | 0 | 0 | 22 (18) | 14 (11) | 7 (6) | 2 (2) | 30 (24) | 51 (41) | 51 (41) | 71 (57) | 8 (6) | 24 (19) | |
Abbreviations: AMP, ampicillin; AMC, amoxicillin-clavulanic acid; APR, apramycin; XNL, ceftiofur; CHL, chloramphenicol; CIP, ciprofloxacin; COL, colistin; FFN, florfenicol; GEN, gentamicin, CTX, cefotaxime; NAL, nalidixic acid; NEO, neomycin; SPT, spectinomycin; STR, streptomycin; SMX, sulfamethoxazole; TET, tetracycline; TMP, trimethoprim.
Thailand (n = 18), Vietnam (n = 2), Philippines (n = 2), and Singapore (n = 1).
Pakistan (n = 2), Thailand (n = 2), Cambodia (n = 1), Asia (n = 1), Mauritius (n = 2), and Afghanistan (n = 2).
Epidemiological information on Salmonella serovar Stanley cases from France.
The 25 French Salmonella serovar Stanley-positive patients reported gastroenteritis. Their ages ranged from 1 to 82 years, and the median was 7 years. Information on gender showed that 14 (56%) and 11 (44%) patients were females and males, respectively. Eleven (44%) cases were associated with travel to Thailand (n = 3, i.e., 27%), Pakistan (n = 2, i.e., 18%), Mauritius (n = 2, i.e., 18%), Afghanistan (n = 2, i.e., 18%), Cambodia (n = 1, i.e., 9%), and an unspecified Southeastern Asian country (n = 1, i.e., 9%) (Fig. 2 to 4) (Table 1). Two (8%) patients did not report traveling prior to illness onset, and travel data were not available for the 12 (48%) remaining cases. Major seasonal variations in the occurrence of infection were not observed; however, peaks in the number of cases occurred in April (n = 5, i.e., 20%) and in September (n = 4, i.e., 16%).
Antimicrobial susceptibility testing.
The occurrence of antimicrobial resistance among the Salmonella serovar Stanley isolates from Thai patients differed according to the regional zones of isolation (Table 1). The highest levels of antimicrobial resistance were observed in isolates from zone 1, 4, and BKK; those isolates were resistant to up to 12 of the 17 tested antimicrobials, including extended-spectrum cephalosporins. In zone 4, 12 (80%) isolates were resistant to CTX and XNL, which represents the highest percentage of extended-spectrum cephalosporin-resistant isolates observed among the different regional zones (Table 1). In addition, one (25%) isolate (SH1796) from zone 5 showed reduced susceptibility to CIP (MIC = 0.5 μg/ml) and intermediate resistance to NAL (MIC = 16 mg/liter) (Fig. 3, Table 1), thus displaying a phenotype compatible with the presence of PMQR genes. All Thai isolates were susceptible to APR, COL, NAL, and NEO (Table 1).
The 24 isolates from Danish patients with travel history were resistant to up to 13 out of 17 tested antimicrobials, whereas the 11 isolates from patients with unknown travel history were resistant to up to 11 antimicrobials (Table 1). Of note, resistance to CTX and XNL was observed in one isolate from a patient with travel history, one isolate from a patient with no travel history, and two isolates from patients with unknown travel association (Table 1). Additionally, three isolates showed reduced susceptibility to CIP (MIC = 0.064 to 1 μg/ml) and resistance to NAL (MIC > 16 μg/ml) (Table 1). All isolates from Danish patients were susceptible to APR (approved for veterinary use only) and COL.
The isolates from French patients with documented (n = 10) and unknown (n = 12) travel history were resistant to up to 10 and 7 antimicrobials, respectively, while the isolates from French patients with no travel history (n = 2) were pansusceptible and of another clonal lineage (Table 1, Fig. 2 to 4). Extended-spectrum cephalosporin resistance was not detected among the French isolates. However, low-level ciprofloxacin resistance was observed in four isolates associated with travel to Pakistan and Afghanistan (Table 1).
All isolates (n = 24; Table 1) resistant to CTX and XNL were tested for susceptibility to additional β-lactams. All Danish (n = 4) and all but one of the Thai (n = 19) isolates exhibited resistance to FOX (Table 2), which could indicate the presence of AmpC-type β-lactamase-encoding genes. The FOX-susceptible Thai isolate was resistant to FEP (Table 2), thus indicating the presence of extended-spectrum β-lactamase-encoding genes. All tested isolates were susceptible to carbapenems (Table 2).
Table 2.
Occurrence of β-lactam resistance among extended-spectrum cephalosporinase-producing Salmonella serovar Stanley isolates from Danish and Thai patients during 2008
| Country | Origin | Total no. of isolates | No. (%) of isolates resistant to the indicated β-lactam at the indicated breakpoint (μg/ml)a |
No. (%) of isolates with indicated β-lactam resistance gene |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ >2 | CFZ ≥4 | FEP ≥32 | FOX >8 | CEP >16 | CPD ≥8 | CRO ≥4 | IRM >1 | MEM >0.125 | blaCTX-M-15 | blaCMY-2 | blaTEM-1b | |||
| Denmark | Travelb | 1 | 1 (100) | 1 (100) | 0 | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 | 0 | 0 | 1 (100) | 0 |
| Denmark | No travel | 1 | 1 (100) | 1 (100) | 0 | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 | 0 | 0 | 1 (100) | 0 |
| Denmark | Unknown | 2 | 2 (100) | 2 (100) | 0 | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 0 | 0 | 0 | 2 (100) | 0 |
| Thailand | Zone 1 | 1 | 1 (100) | 1 (100) | 0 | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 | 0 | 0 | 1 (100) | 0 |
| Thailand | Zone 4 | 12 | 11 (92) | 12 (100) | 0 | 12 (100) | 12 (100) | 11 (92) | 11 (92) | 0 | 0 | 0 | 12 (100) | 8 (67) |
| Thailand | Zone BKK | 7 | 7 (100) | 7 (100) | 1 (14) | 6 (86) | 7 (100) | 7 (100) | 7 (100) | 0 | 0 | 1 (14) | 6 (86) | 2 (29) |
| Total | 24 | 23 (96) | 24 (100) | 1 (4) | 23 (96) | 24 (100) | 23 (96) | 23 (96) | 0 | 0 | 1 (4) | 23 (96) | 10 (42) | |
Abbreviations: CAZ, ceftazidime; CFZ, cefazolin; FEP, cefepime; FOX, cefoxitin; CEP, cephalothin; CPD, cefpodoxime; CRO, ceftriaxone; IRM, imipenem; MEM, meropenem.
Thailand.
Identification of extended-spectrum cephalosporinase-encoding genes and plasmid-mediated quinolone resistance genes.
PCR and sequencing of the 24 extended-spectrum cephalosporin-resistant isolates showed that 23 (96%) isolates harbored a blaCMY-2 gene and one (4%) isolate harbored a blaCTX-M-15 gene, thus showing an exact correlation between the observed phenotype and genotype (Table 2). In Thailand, the overall prevalence of extended-spectrum cephalosporin-resistant isolates was 10.4% (20 resistant out of 193 tested).
PCR and sequencing performed on the one Thai isolate displaying a phenotype compatible with the presence of PMQR genes (reduced susceptibility to CIP and intermediate resistance to NAL [MIC = 16 mg/liter]) demonstrated the presence of a qnrS1 gene (highlighted in Fig. 3).
Pulsed-field gel electrophoresis typing.
A total of 125 isolates, including 62 Thai isolates (50 proportionally representing the different regional zones, 11 resistant to extended-spectrum cephalosporins based on disk diffusion screening performed in Thailand, 1 harboring qnrS1), 39 isolates from Denmark, and 24 isolates from France, were genotyped by PFGE. A total of 91 unique XbaI PFGE patterns and 17 clusters were observed (Fig. 2 to 4).
Analysis of the genetic background of isolates harboring extended-spectrum cephalosporinase-encoding genes showed that four Danish isolates and one, nine, and six Thai isolates from regional zones 1, 4, and BKK, respectively, clustered in a group defined by 78% similarity (highlighted in Fig. 3). These isolates showed highly similar antimicrobial susceptibility profiles and harbored the blaCMY-2 gene (Fig. 3). Also, one isolate from BKK harboring a blaCTX-M-15 gene belonged to this PFGE lineage (Fig. 3). The remaining blaCMY-2-harboring isolates (n = 3) originating from zone 4 and showing a slightly different antimicrobial resistance profile clustered in a different branch of the dendrogram (Fig. 4).
Interestingly, indistinguishable PFGE patterns and antimicrobial resistance profiles were observed in isolates from two Thai patients (SH 3045 and SH 3099), a Danish patient (0804T69015) infected in Thailand, and a French patient (08 9717) with unknown travel history (Fig. 4). Likewise, indistinguishable PFGE patterns and antimicrobial resistance profiles, which in these cases consisted of susceptibility to all tested antimicrobials, were observed in isolates from a Thai patient (SH 584) and two Danish patients reporting travel to Singapore (0810F36699) and Vietnam (0811T40304), respectively. Of note, these PFGE patterns were highly similar (88% similarity) to those observed in isolates from a Danish patient reporting travel to Thailand (0804F2922) and a French patient reporting travel to Cambodia (08 2649) (Fig. 4), which also showed susceptibility to all tested antimicrobials (Fig. 4).
Isolates from French patients reporting travel to Pakistan and Afghanistan showed PFGE patterns characterized by 72% similarity and belonged to a PFGE lineage different from those of the other isolates included in the study (Fig. 2).
Plasmid characterization.
The blaCMY-2 gene was transferred from 9 out of 10 donor isolates as determined by colony PCR performed on the transformants. The blaCMY-2-positive isolate (SH 1727) that failed to transfer the extended-spectrum cephalosporinase-encoding gene was not examined further. The blaCMY-2-harboring plasmids ranged in size from approximately 80 to 195 kb and were identified as IncI1 (n = 1), IncA/C (n = 6), and IncA/C + IncY (n = 2) (Table 3). Resistance to up to six additional antimicrobials was cotransferred by eight (89%) plasmids (Table 3). Of importance, five plasmids showing the same replicon type (IncA/C), size (approximately 140 kb), and antimicrobial resistance profile (AMP, AMC, CHL, FFN, CTX, GEN, SMX, SPT, TET, and XNL) were isolated from genetically related isolates (belonging to the above-mentioned PFGE cluster defined by 78% similarity) collected from different geographical zones (1, 4, and BKK), different hospitals within a zone (BKK/Ramathibordi Hospital, BKK/Taksin Hospital, and BKK/Chalemkongpracharak Hospital), and different times (March, April, May, June, and October of 2008) (Table 3). Analysis of the genes cotransferred with the blaCMY-2 showed that all these five plasmids harbored aadA1, floR, sul1, sul2, and tetA (Table 3). In addition, the blaCMY-2-positive plasmids from SH1216 and SH777 also contained aac3-(IV) and aac3-(IV) + cmlA, respectively (Table 3).
Table 3.
Genetic and phenotypic traits of blaCMY-2-harboring Salmonella serovar Stanley isolates from Thai patients
| Isolate | Plasmid characterizationa |
Epidemiological informationd |
||||
|---|---|---|---|---|---|---|
| Replicon typing | Plasmid size (kb) | Transferred resistancesb | Transferred resistance gene(s) | Regional zone/hospital of isolation | Isolation date | |
| SH 646 | A/C | 140 | AMP, AMC, CHL, CTX, FFN, GEN,c SMX, SPT, TET, XNL | aadA1, blaCMY-2, floR, sul1, sul2, tetA | 4/Ratchaburi | 18 March 2008 |
| SH 777 | A/C | 140 | AMP, AMC, CHL, CTX, FFN, GEN,c SMX, SPT, TET, XNL | aac3-(IV), aadA1, blaCMY-2, cmlA, floR, sul1, sul2, tetA | BKK/Taksin | 08 April 2008 |
| SH 1005 | A/C | 140 | AMP, AMC, CHL, CTX, FFN, GEN,c SMX, SPT, TET, XNL | aadA1, blaCMY-2, floR, sul1, sul2, tetA | BKK/Ramathibordi | 01 May 2008 |
| SH 1216 | A/C | 140 | AMP, AMC, CHL, CTX, FFN, GEN,c SMX, SPT, TET, XNL | aac3-(IV), aadA1, blaCMY-2, floR, sul1, sul2, tetA | BKK/Chalemkongpracharak | 03 June 2008 |
| SH 2452 | A/C | 140 | AMP, AMC, CHL, CTX, FFN, GEN,c SMX, SPT, TET, XNL | aadA1, blaCMY-2, floR, sul1, sul2, tetA | 1/Mongkutwattana | 13 October 2008 |
| SH 1602 | A/C | 195 | AMP, AUG, CHL, CTX, FFN, SMX, TET, XNL | blaCMY-2, blaTEM-1, floR, sul2, tetA | 4/Ratchaburi | 22 July 2008 |
| SH 1982 | A/C, Y | 152 | AMP, AUG, CHL, CTX, FFN, SMX, TET, XNL | blaCMY-2, floR, sul2, tetA | 4/Ratchaburi | 19 August 2008 |
| SH 2272 | A/C, Y | 172 | AMP, AUG, CHL, CTX, FFN, SMX, TET, XNL | blaCMY-2, blaTEM-1, floR, sul1, tetA | 4/Ratchaburi | 16 September 2008 |
| SH 1712 | I1 | 80 | AMP, AUG, CTX, XNL | blaCMY-2 | 4/Ratchaburi | 29 July 2008 |
Determination was done on plasmids obtained from the transformants.
AMP, ampicillin; AMC, amoxicillin-clavulanic acid; CHL, chloramphenicol; CTX, cefotaxime; FFN, florfenicol; GEN, gentamicin; SMX, sulfamethoxazole; SPT, spectinomycin; STR, streptomycin; TET, tetracycline; TMP, trimethoprim; XNL, ceftiofur.
Not all the possible genes conferring aminoglycoside resistance were present on the microarray used in this experiment.
All isolates originated from stool specimens.
DISCUSSION
In this study, we characterized Salmonella serovar Stanley isolates from clinical cases reported in Thailand, Denmark, and France in 2008. Overall, we observed a wide range of genetic diversity and high levels of resistance to antimicrobials, including critically important antimicrobials such as extended-spectrum cephalosporins. The epidemiological data from Thailand are consistent with what have been described by Hendriksen et al., emphasizing that Salmonella serovar Stanley mainly causes diarrhea in both genders, with no specific season of high incidence (23). Country-based differences could be identified mainly in relation to the numbers of occurrences of cases, ages of infected patients, and antimicrobial resistance profiles. In 2008, Salmonella serovar Stanley was the 2nd, 5th, and 36th most prevalent cause of human salmonellosis in Thailand, Denmark, and France, respectively. The majority (62%) of the observed cases in Denmark were related to documented travel to Southeast Asia, whereas only 20% of Salmonella serovar Stanley-infected patients in France were linked to documented travel to Southeast Asia. A higher occurrence of antimicrobial resistance was detected in isolates from Denmark and Thailand compared to that seen with isolates from France (Table 1). The occurrence of a high proportion of multidrug-resistant Salmonella serovar Stanley isolates from Southeast Asia was also reported previously (26, 29, 34). Of the Danish and Thai isolates tested for MIC, resistance to extended-spectrum cephalosporins was observed in 10% and 32%, respectively, and resistance to fluoroquinolones was observed in 8% and 2%, respectively. The finding that the proportion of resistant isolates in Denmark was higher than the proportion of resistant isolates in France is most likely linked with larger-scale importation of food from Southeast Asia to Denmark and more-frequent travel from Denmark to destinations within this region. Of note, the prevalence of resistance to extended-spectrum cephalosporins and fluoroquinolones among Thai isolates might have been underestimated, since some of the isolates included in the study were selected based on the results of a disk diffusion screening, which was not reliable for the detection of extended-spectrum cephalosporin and fluoroquinolone resistance. A subset of isolates that were defined as intermediately resistant to extended-spectrum cephalosporins and were included in the study as representative of the regional zones and not as potential extended-spectrum cephalosporinase producers were shown to harbor extended-spectrum cephalosporinase-encoding genes, and the detection of PMQR genes by disk diffusion is notoriously difficult (8).
At the genotypic level, we observed both broad genetic diversity among the Salmonella serovar Stanley isolates from Thailand and a partial cluster with isolates associated with travel in Vietnam, Cambodia, Singapore, and the Philippines, supporting the results of previous studies which showed that Salmonella serovar Stanley is endemic in Southeast Asia and the Western Pacific region (3, 18, 25, 29). In addition, the presence of indistinguishable PFGE patterns and antimicrobial susceptibility profiles among isolates from Thai patients and Danish and French patients reporting travel to Asia strongly indicates that at least some European cases were travel related.
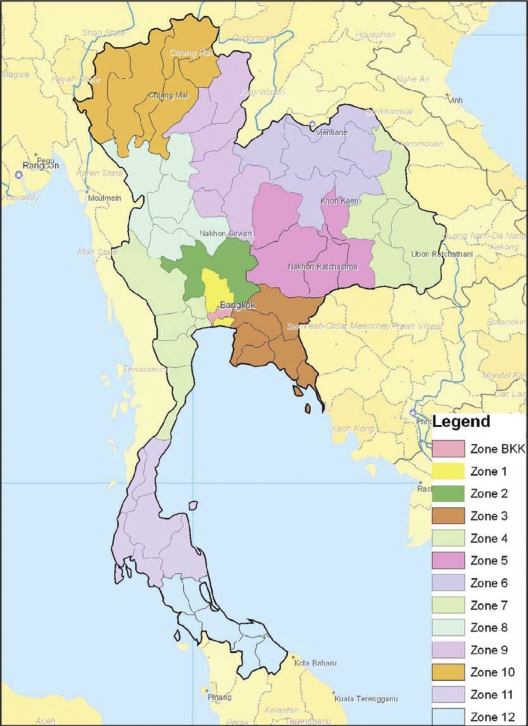
Several studies linked Salmonella serovar Stanley with pigs and pig farming in Thailand (9, 16). Hendriksen et al. observed a higher frequency of human infections caused by pig-related serovars, including Salmonella serovar Stanley, in the central regions of Thailand, where pig farming is intensive compared to the remaining regions of the country (23). The present study also showed that Salmonella serovar Stanley is predominantly found in the central region of Thailand (zones 1, 4, and BKK) (Fig. 5). Of note, the extended-spectrum cephalosporinase-producing isolates detected in this study were isolated in zones 1, 4, and BKK. Analysis of the molecular mechanisms of resistance performed on 10 (50%) extended-spectrum cephalosporin-resistant isolates revealed that the blaCMY-2 gene was carried on five different plasmids (Table 3). Despite this variability, the same plasmid type was found in five isolates from different zones and different hospitals within a zone (Table 3); interestingly, that plasmid was highly similar to a plasmid described previously in a study of Salmonella choleraesuis isolates causing severe clinical cases of bacteremia in zone 4 and BKK in Thailand (35). This plasmid was identified as an IncA/C of approximately 140 kb and carried genes conferring resistance to chloramphenicol, florfenicol, gentamicin, sulfametoxazole, spectinomycin, and tetracycline. IncA/C plasmids harboring blaCMY-2 are widespread in Salmonella spp. (6). However, the presence of highly similar plasmids detected over different times in different hospitals and in different geographical areas indicates that horizontal genetic exchange contributes greatly to the epidemiology of this plasmid. Horizontal plasmid mobility is more likely than clonal spread, since these IncA/C plasmid-harboring isolates, though characterized by 78% similarity of the PFGE band patterns, still exhibited a degree of variability. The presence of a particularly successful plasmid able to appear in multiple isolates and serovars is worrisome. The remaining four plasmid types carrying blaCMY-2 were isolated in zone 4 in Ratchaburi Hospital. The presence of multiple plasmid lineages in the same hospital in a relatively short time period (Table 3) could indicate the presence of selective pressure for blaCMY-2. However, pressure that is selective for other antimicrobial resistance genes should also be considered, since all plasmids except one carried genes conferring resistance to additional antimicrobials (Table 3).
Fig 5.
Zone distribution in Thailand. (Map courtesy of the National Institute of Health, Thailand; modified with permission.)
Our results show that pig-associated Salmonella serovars with extended-spectrum cephalosporin resistance are emerging in the human population in Thailand. We cannot establish whether the blaCMY-2-harboring plasmids were acquired by Salmonella serovar Stanley isolates in pigs or in humans. In Thailand, there is strong selective pressure for antimicrobial resistance both in farming and in the community, since antimicrobials are easily obtainable over the counter. The government adopted the Codex Alimentarius code of practice for “Control of Use of Veterinary Drugs” (CAC/RCP 38-1993) (www.codexalimentarius.net) and is trying to strengthen the awareness of farmers concerning antimicrobial resistance, recognizing that “Thailand is the kitchen of the world.” However, only highly productive farms need to follow this regulation to obtain export certification.
Our study highlighted the finding that there is a high occurrence of clinical cases in Thailand caused by Salmonella serovar Stanley, which is a pig-associated serovar. We observed broad genetic diversity among the isolates characterized, and we identified the presence of multiple plasmid lineages carrying extended-spectrum cephalosporinase-encoding genes in isolates from the central part of Thailand, which is characterized by a relatively confined coexistence of humans and pigs. Of note, we detected a low level of plasmid-mediated quinolone resistance determinants in Salmonella serovar Stanley isolates from Thailand, but we stress that improved methods for detection of PMQR genes could have given different results and should therefore be implemented in Thailand. Interestingly, we showed that at least some of the Salmonella serovar Stanley isolates collected from patients in Europe were acquired during travel to Southeast Asia, including Thailand. Our results emphasize that Thai authorities should improve the ongoing efforts to regulate antimicrobial use in agriculture and in clinical settings to limit the spread of multidrug-resistant Salmonella isolates and plasmids among humans and pigs in Thailand and abroad.
ACKNOWLEDGMENTS
We are grateful to Matthew Mikoleit, Centers for Disease Control and Prevention, Enteric Disease Epidemiology Branch, Atlanta, GA, for a critical review and Tune Øst-Jacobsen for outstanding technical assistance. We thank all the corresponding laboratories of the French National Reference Centre for Salmonella.
This work was supported by the World Health Organization Global Foodborne Infections Network (www.who.int/gfn) and by the Center for Genomic Epidemiology at the Technical University of Denmark (funded by grant 09-067103/DSF from the Danish Council for Strategic Research). The French National Reference Centre for Salmonella is funded by the Institut Pasteur and the Institut de Veille Sanitaire.
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Aarestrup FM, et al. 2007. International spread of multidrug-resistant Salmonella Schwarzengrund in food products. Emerg. Infect. Dis. 13:726–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Archambault M, et al. 2006. Molecular characterization and occurrence of extended-spectrum beta-lactamase resistance genes among Salmonella enterica serovar Corvallis from Thailand, Bulgaria, and Denmark. Microb. Drug Resist. 12:192–198 [DOI] [PubMed] [Google Scholar]
- 3. Bangtrakulnonth A, et al. 2004. Salmonella serovars from humans and other sources in Thailand, 1993–2002. Emerg. Infect. Dis. 10:131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bangtrakulnonth A, et al. 2008. Shigella from humans in Thailand during 1993 to 2006: spatial-time trends in species and serotype distribution. Foodborne Pathog. Dis. 5:773–784 [DOI] [PubMed] [Google Scholar]
- 5. Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 54:969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 8. Cavaco LM, Aarestrup FM. 2009. Evaluation of quinolones for use in detection of determinants of acquired quinolone resistance, including the new transmissible resistance mechanisms qnrA, qnrB, qnrS, and aac(6′)Ib-cr, in Escherichia coli and Salmonella enterica and determinations of wild-type distributions. J. Clin. Microbiol. 47:2751–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chuanchuen R, Pathanasophon P, Khemtong S, Wannaprasat W, Padungtod P. 2008. Susceptibilities to antimicrobials and disinfectants in Salmonella isolates obtained from poultry and swine in Thailand. J. Vet. Med. Sci. 70:595–601 [DOI] [PubMed] [Google Scholar]
- 10. Clinical and Laboratory Standards Institute 2000. Performance standards for antimicrobial disk susceptibility tests, M2-A7, 7th ed (Approved standard.) Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11. Clinical and Laboratory Standards Institute 2008. Performance standards for antimicrobial susceptibility testing, M100-S16, 18th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 12. Clinical and Laboratory Standards Institute 2006. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 31-A3, 3rd ed (Approved standard.) Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 13. Clinical and Laboratory Standards Institute 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, M07-A7, 7th ed (Approved standard.) Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 14. Coburn B, Grassl GA, Finlay BB. 2007. Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 85:112–118 [DOI] [PubMed] [Google Scholar]
- 15. Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM. 2009. World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin. Infect. Dis. 49:132–141 [DOI] [PubMed] [Google Scholar]
- 16. Dorn-In S, et al. 2009. A cross-sectional study of Salmonella in pre-slaughter pigs in a production compartment of northern Thailand. Prev. Vet. Med. 88:15–23 [DOI] [PubMed] [Google Scholar]
- 17. Ethelberg S, Muller L, Molbak K, Nielsen EM. 2010. Salmonella and campylobacter infections in 2008. Ugeskr. Laeger 172:1451–1455 (In Danish.) [PubMed] [Google Scholar]
- 18. Galanis E, et al. 2006. Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg. Infect. Dis. 12:381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grimont PAD, Weil Fx. (ed). 2007. Antigenic formulae of the Salmonella serovars, 9th ed WHO Collaborating Center for Reference and Research on Salmonella, Institut Pasteur, Paris, France [Google Scholar]
- 20. Hasman H, Mevius D, Veldman K, Olesen I, Aarestrup FM. 2005. Beta-lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56:115–121 [DOI] [PubMed] [Google Scholar]
- 21. Hendriksen RS, et al. 2011. Global monitoring of serotype distribution based on quality assured data from the WHO Global Salmonella Survey Country Data Bank, 2001-2007. 2010. Foodborne Pathog. Dis. 8:887–900 [DOI] [PubMed] [Google Scholar]
- 22. Hendriksen RS, et al. 2008. Antimicrobial resistance and molecular epidemiology of Salmonella Rissen from animals, food products, and patients in Thailand and Denmark. Foodborne Pathog. Dis. 5:605–619 [DOI] [PubMed] [Google Scholar]
- 23. Hendriksen RS, et al. 2009. Risk factors and epidemiology of the ten most common Salmonella serovars from patients in Thailand: 2002-2007. Foodborne Pathog. Dis. 6:1009–1019 [DOI] [PubMed] [Google Scholar]
- 24. Hendriksen RS, et al. 2009. Emergence of multidrug-resistant Salmonella Concord infections in Europe and the United States in children adopted from Ethiopia, 2003-2007. Pediatr. Infect. Dis. J. 28:814–818 [DOI] [PubMed] [Google Scholar]
- 25. Herikstad H, Motarjemi Y, Tauxe RV. 2002. Salmonella surveillance: a global survey of public health serotyping. Epidemiol. Infect. 129:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang KY, et al. 2007. Molecular epidemiology and antimicrobial susceptibility of Salmonella enterica serotype Stanley isolates in Taiwan. J. Microbiol. Immunol. Infect. 40:411–418 [PubMed] [Google Scholar]
- 27. Humphrey TJ. 2000. Public-health aspects of Salmonella infections, p 245–263 In Wray C, Wray A. (ed), Salmonella in domestic animals. CABI Publishing, Wallingford, United Kingdom [Google Scholar]
- 28. Jones TF, et al. 2008. Salmonellosis outcomes differ substantially by serotype. J. Infect. Dis. 198:109–114 [DOI] [PubMed] [Google Scholar]
- 29. Lee HY, et al. 2009. High rate of reduced susceptibility to ciprofloxacin and ceftriaxone among nontyphoid Salmonella clinical isolates in Asia. Antimicrob. Agents Chemother. 53:2696–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Le Hello S, et al. 2011. International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J. Infect. Dis. 204:675–684 [DOI] [PubMed] [Google Scholar]
- 31. Newell DG, et al. 2010. Food-borne diseases—the challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 139(Suppl. 1):S3–S15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poirel L, Cattoir V, Nordmann P. 2008. Is plasmid-mediated quinolone resistance a clinically significant problem? Clin. Microbiol. Infect. 14:295–297 [DOI] [PubMed] [Google Scholar]
- 32a. Ribot EM, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 33. Sanguankiat A, et al. 2010. A cross-sectional study of Salmonella in pork products in Chiang Mai, Thailand. Foodborne Pathog. Dis. 7:873–878 [DOI] [PubMed] [Google Scholar]
- 34. Sirichote P, et al. 2010. Serotypes and antimicrobial resistance of Salmonella enterica in the lower central region of Thailand, 2001-2006. Southeast Asian J. Trop. Med. Public Health 41:1405–1415 [PubMed] [Google Scholar]
- 35. Sirichote P, et al. 2010. Molecular characterization of extended-spectrum cephalosporinase-producing Salmonella enterica serovar Choleraesuis isolates from patients in Thailand and Denmark. J. Clin. Microbiol. 48:883–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Voetsch AC, et al. 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin. Infect. Dis. 38(Suppl. 3):S127–S134 [DOI] [PubMed] [Google Scholar]