Abstract
Salmonella enterica serovar Typhimurium is one of the leading causes of gastroenteritis in humans. Phage typing has been used for the epidemiological surveillance of S. Typhimurium for over 4 decades. However, knowledge of the evolutionary relationships between phage types is very limited. In this study, we used single nucleotide polymorphisms (SNPs) as molecular markers to determine the relationships between common S. Typhimurium phage types. Forty-four SNPs, including 24 identified in a previous study and 20 from 6 available whole-genome sequences, were used to analyze 215 S. Typhimurium isolates belonging to 45 phage types. Altogether, 215 isolates and 6 genome strains were differentiated into 33 SNP profiles and four distinctive phylogenetic clusters. Fourteen phage types, including DT9, one of the most common phage types in Australia, were differentiated into multiple SNP profiles. These SNP profiles were distributed into different phylogenetic clusters, indicating that they have arisen independently multiple times. This finding suggests that phage typing may not be useful for long-term epidemiological studies over long periods (years) and diverse localities (different countries or continents). SNP typing provided a discriminative power similar to that of phage typing. However, 12 SNP profiles contained more than one phage type, and more SNPs would be needed for further differentiation. SNP typing should be considered as a replacement for phage typing for the identification of S. Typhimurium strains.
INTRODUCTION
Salmonella enterica serovar Typhimurium is a broad-host-range pathogen that causes gastroenteritis in humans. The phage typing scheme described by Anderson et al. (3) has been used for the routine epidemiological surveillance of S. Typhimurium infections for many years. Phage types are determined by resistance or sensitivity to a set of 34 phages, and so far, more than 300 definitive phage types (DTs) are recognized (38). Phage typing has been found to be particularly useful for tracking the spatiotemporal distribution of important pathogenic forms. For example, multidrug-resistant DT104 has caused widespread infections in humans and animals in Europe and the United States since 1994 (12, 23), but it remains rare in Australia. In contrast, DT9, DT135, and DT170/108 are common in Australia (34), while DT9 is rare in Europe and the United States. However, knowledge of the evolutionary relationships between phage types is very limited, and the genetic basis for the variation in phage sensitivity remains largely unknown. The discriminatory power of phage typing is often inadequate to track community outbreaks, especially in settings when one particular phage type is dominant. In addition, phage types can change with a few genetic modifications, such as a gain or loss of mobile elements (2, 27, 50).
Despite the extensive diversity of phage types in S. Typhimurium, population structure studies using multilocus enzyme electrophoresis (MLEE) showed that the majority of S. Typhimurium isolates are grouped together as a single clone. A previous MLEE analysis of over 300 isolates using 20 enzymes (4) found 23 electrophoretic types (ETs). The majority of isolates belong to ET Tm1, and all S. Typhimurium ETs were included with a number of other serovars in a group called the S. Typhimurium clonal complex (4). A previous four-gene multilocus sequencing typing (MLST) study of 85 S. Typhimurium isolates showed no variation (9), while a seven-gene MLST study of over 350 S. Typhimurium isolates differentiated them into 23 sequence types (STs) (42), 22 of which belonged to a single clonal complex with ST19 as the founder. ST19 is the most frequently found ST, and the ST19 clonal complex accounts for more than 90% of S. Typhimurium isolates. Therefore, seven-gene MLST is not sufficient to resolve the relationships among strains of S. Typhimurium (42).
Single nucleotide polymorphisms (SNPs) are the most valuable molecular markers for studying the evolutionary relationships of isolates in homogenous pathogenic clones (1, 10, 13, 29, 31, 33). In S. Typhimurium, SNPs were first utilized in a previous study by Hu et al. (18), which explored mutational changes by matching polymorphic amplified fragment length polymorphism (AFLP) fragments to the LT2 genome to pinpoint base changes and by sequencing the intergenic (IG) regions of selected strains. A total of 51 polymorphic sites (mainly SNPs) were used to establish the evolutionary relationships between 46 S. Typhimurium isolates of nine phage types (18).
Several S. Typhimurium whole-genome sequences are available, including one previously reported DT4 strain, LT2 (26); one DT133 strain, 14028s (19); one DT104 strain, NCTC13348 (7); one strain of an unknown phage type, D23580 (20); and unpublished genome sequences, including one DT44 strain, SL1344 (www.sanger.ac.uk), and an unnamed DT2 pigeon isolate (www.sanger.ac.uk), which offer additional SNP resources. In this study, we used 24 SNPs identified previously by Hu et al. (18) and 20 genome-wide SNPs from sequenced genomes to examine the phylogenetic relationships between S. Typhimurium isolates of 45 different phage types using real-time PCR assays based on hairpin (HP) primers.
MATERIALS AND METHODS
Bacterial strains.
A total of 215 S. Typhimurium isolates belonging to 45 phage types from various localities were used in this study (see Table S1 in the supplemental material); 37 of the selected phage types included at least 1 isolate from a nonhuman source. We selected two or more isolates from all but DT30, DT177, and DT195. The selected DTs from Australia (27 DTs) and the United Kingdom (25 DTs) have been dominant or frequently implicated in animal and human infections, and seven (DT1, DT8, DT9, DT135, DT141, DT170, and DT193) were common to both countries. DT9 and DT135 were each represented by 35 isolates. The DT135 isolates were from many countries, but the origins of the DT9 isolates were less diverse. DT9 isolates are common in Australia and New Zealand but rare elsewhere. Isolates for each DT were selected from different years and/or different localities, to ensure that they were not epidemiologically related. Chromosomal DNA was prepared by using the phenol-chloroform precipitation method (29). Reference strain LT2 was used as a control.
Genomic analyses.
Pairwise genome comparisons between S. Typhimurium LT2 (GenBank accession no AE006468 [26]) and five other S. Typhimurium genomes, SL1344 (accession no. FQ312003), NCTC13348 (DT104) (7) and a DT2 strain with no strain name released (here referred to simply as DT2) (both are available from the Sanger Centre [www.sanger.ac.uk]), 14028s (DT133) (accession no. CP001363 [19]), and D23580 (accession no. FN424405 [20]), were performed by using Artemis (5) and BLAST tools available from the Australian National Genetic Information Service (ANGIS). Fully sequenced genomes from three other S. enterica serovars, S. enterica serovar Typhi strain CT18 (accession no. AL513382 [32]), S. enterica serovar Enteritidis strain PT4 (accession no. AM933172 [47]), and S. enterica serovar Paratyphi A strain SARB42 (accession no. CP000026 [25]), were used as outgroups for phylogenetic analyses.
Real-time PCR typing of SNPs.
SNP typing was performed by using a hairpin (HP) primer real-time PCR (hpRT-PCR) assay as described previously (30, 31). Each PCR mixture contained 30 ng of chromosomal DNA; 3 μl of SYBR green (Quantace); 0.5 μl of 10 μM forward and reverse primers, respectively (see Table S2 in the supplemental material); and MilliQ water to a final volume of 10 μl. All reactions were performed with a Rotor-gene 6000 sequence detector system (Corbett Life Science, Australia). Thermal cycling conditions were as follows: stage 1 was performed at 95°C for 10 min; stage 2 was performed with 10 cycles of 72°C for 30 s, 95°C for 15 s, and 69°C for 30 s, lowering the temperature 1°C in the last step for each cycle; and stage 3 was performed at 72°C for 5 s, 95°C for 15 s, and 52°C to 60°C for 30 s, repeated 40 times. Data were collected in the last step of stage 3 for analysis.
Quality control of SNP typing was monitored by the difference in the threshold cycle (CT) values (ΔCT) between results for matched and unmatched HP primers, which were designed to have a ΔCT of >5. The assay was repeated if a lower value was obtained. Each run involved matched and unmatched primers, which effectively provided positive and a negative controls. All 44 SNPs were also tested on S. Typhimurium strain LT2, and the allelic calls obtained by hpRT-PCR were consistent with the genome data. In addition, we tested a subset of SNPs on the same set of isolates used previously by Hu et al. (18) and obtained results consistent with theirs. SNPs that showed reverse/parallel changes were also retested by hpRT-PCR and confirmed.
PCR detection of Salmonella genomic island 1 (SGI1), prophage ST104, and the allantoin operon.
The PCRs were done using a multiplex PCR targeting antibiotic resistance islands and integrase for the detection of SGI1 and ST104 (gp23, ninYZHG, and gtrA regions), respectively, as described previously by Cooke et al. (8), while for the detection of the allantoin operon (STM0517 to STM0529), PCR primers were adopted from those described previously by Garaizar et al. (11).
Bioinformatic analysis.
The relationships between the isolates were determined by using PAUP (44) to construct a maximum parsimony tree from the SNP data and to calculate the homoplasy index. Simpson's index of diversity (D) was calculated by using an in-house program, MLEECOMP (37). The confidence interval (CI) (95%) of the D values was calculated by using an online calculator (available from http://darwin.phyloviz.net). SNPT (10) was used to determine the minimum number of SNPs required for typing.
RESULTS AND DISCUSSION
Selection of SNPs.
The SNPs used for this study were selected from those reported in a previous study by Hu et al. (18) and from genome comparisons. Hu et al. (18) discovered 51 polymorphic sites, 40 of which were SNPs from analyses of 46 S. Typhimurium isolates representing nine phage types. At least one SNP was selected to cover each of the 20 branches of the tree in that paper (18), and a maximum of two were selected when there was more than one SNP on a branch. In total, 24 SNPs were selected, 16 of which were from IG regions and 8 of which were from coding regions. Of the latter, only four were from genes with a known function. Three (SNPs 2, 4, and 13) of the genes had basic cellular functions (housekeeping genes) based on the Clusters of Orthologous Groups (COG) category, including amino acid and carbohydrate metabolism as well as transport and translation. One SNP (SNP 30) was on a gene involved in cell wall biogenesis. We also incorporated SNPs from five strains, LT2 (DT4), NCTC13348 (DT104), SL1344 (DT44), D23580 (unknown phage type), and a DT2 pigeon strain, whose genomes were either published or available from the Sanger Centre. Genome comparisons of these strains revealed a total of 2,099 SNPs, with 313 to 629 SNPs per strain. We selected 20 synonymous SNPs, 5 each from the four genomes, for typing. These SNPs were uniquely present in a single genome, except for SNP 26, which was present in both the NCTC13348 and LT2 genomes. The 20 SNPs were mostly from genes in COG category C (energy production and conversion) or G (carbohydrate metabolism and transport). Categories E (amino acid metabolism and transport) and S (unknown function) were represented by one SNP each. In addition, the genes carrying the SNPs have only one SNP, making it less likely that they are under positive selection pressure.
SNP typing of S. Typhimurium isolates.
The hpRT-PCR assay was used to detect 44 selected SNPs in 215 S. Typhimurium isolates, representing 45 different phage types. The SNP data for six genome-sequenced strains (LT2, 14208s, NCTC13348, SL1344, D23580, and DT2) were also included, giving a total of 221 S. Typhimurium isolates for analysis. Of the 44 SNPs, 37 were shared by two or more isolates (parsimony informative), and 7 were unique to single-genome strains: 5 in DT2 and 1 each in strains SL1344 (DT44) and LT2 (DT4).
The 221 isolates were differentiated into 33 SNP profiles (SPs), 10 of which were unique to single isolates and 23 of which were shared by more than one isolate. The two most common SPs were SP7 and SP33, with 38 and 34 isolates, respectively. Not all isolates with the same phage type belonged to the same SP, and a single SP may contain multiple phage types. Twenty SPs contained isolates of a single phage type, 3 contained isolates with 2 phage types, and the remaining 11 contained isolates belonging to 3 to 10 different phage types. The genome strains were mostly separated into unique SPs.
The D value for SNP typing was 0.923 (95% CI = 0.907 to 0.939) in this study. SNP typing was considerably more discriminatory than MLST (D = 0.608; 95% CI = 0.562 to 0.654), based on data described previously by Sangal et al. (42), but slightly less so than phage typing (D = 0.941; 95% CI = 0.923 to 0.958), as calculated by using the data from this study. The higher D value for phage typing probably reflects the selection of isolates for a wide representation of different phage types and is not representative of findings for unselected isolates referred for epidemiological surveillance. If only the top 10 phage types (DT1, DT9, DT12a, DT44, DT108/DT170, DT126, DT135, DT135a, DT141, and DT197) in Australia are considered, the D value of SNP typing (D = 0.921) is higher than that of phage typing (D = 0.891).
However, the SNPs used in this study clearly still have limited discriminatory power, since there were many phage types grouped together within a single SP. Representative genomes from different SPs should be sequenced for a better phylogenetic coverage of their diversities.
Evolutionary relationships of the SNP profiles.
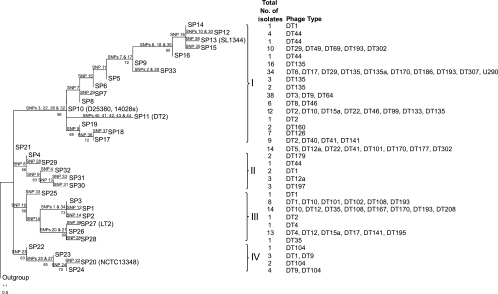
A maximum parsimony analysis gave 24 trees of equal length, one of which is shown in Fig. 1 (the tree selected is consistent with the tree generated after the removal of five conflicting SNPs, as discussed below). Four clusters (clusters I to IV) were consistently identified, each of which was supported by at least one SNP and bootstrap values greater than 50% (Table 1). The tree was rooted by using inferred ancestral SNPs based on genome sequences of three serovars, S. enterica serovars Typhi, Paratyphi A, and Enteritidis. The four clusters as well as the unclustered SP21 appear as a star phylogeny radiating out from the outgroup at the same node.
Fig 1.
Maximum parsimony tree illustrating the evolutionary relationships of the SNP profiles (SPs). For SPs containing genome strains, the strain names are in parentheses after the SP numbers. The roman numerals correspond to cluster numbers. SNPs supporting each branch are located on the node. Bootstrap values greater than 50% are displayed below each node.
Table 1.
Single nucleotide polymorphism profilesa
| Sequence or cluster | SNP profile | SNP |
|||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | ||
| Consensus | G | A | T | A | C | C | C | G | A | G | G | C | G | C | G | G | T | C | C | C | G | A | G | C | C | T | C | G | T | C | G | A | A | G | C | T | G | G | C | G | C | A | C | C | |
| Clusters | |||||||||||||||||||||||||||||||||||||||||||||
| I | 14 | · | · | C | · | · | T | · | · | · | · | A | · | · | · | A | A | C | T | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | A | · | · | · | · | · | · | · | · | · |
| 12 | · | · | C | · | · | T | · | · | · | A | A | · | · | · | A | A | C | T | · | · | · | · | · | · | · | · | · | · | · | T | · | G | · | · | A | · | · | · | · | · | · | · | · | · | |
| 13 | · | · | C | · | · | T | · | · | · | · | A | · | · | · | A | A | C | T | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | A | · | · | A | · | · | · | · | · | · | |
| 15 | · | · | C | · | · | T | · | · | · | · | A | · | · | · | A | A | C | T | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | · | · | · | · | · | · | · | · | · | |
| 16 | · | · | C | · | · | T | · | · | · | · | A | · | · | · | A | · | C | T | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | A | · | · | · | · | · | · | · | · | · | |
| 9 | · | · | C | · | · | · | · | · | · | · | A | · | · | · | A | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | · | · | · | · | · | · | · | · | · | |
| 33 | · | C | C | · | · | · | · | · | · | · | A | · | · | · | A | · | C | · | · | · | · | · | · | · | · | · | · | T | · | · | · | G | · | · | · | · | · | · | · | · | · | · | · | · | |
| 5 | · | · | C | · | · | · | A | · | · | · | A | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | · | · | · | · | · | · | · | · | · | |
| 6 | · | · | C | · | · | · | A | · | · | · | · | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | · | · | · | · | · | · | · | · | · | |
| 7 | · | · | C | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | G | · | · | · | · | · | · | · | · | · | · | · | · | |
| 8 | · | · | C | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | · | · | · | · | · | · | · | · | · | |
| 10 | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | · | · | · | · | · | · | · | · | · | |
| 11 | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | · | · | · | T | T | G | T | A | |
| 19 | · | · | C | · | G | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | · | · | · | · | · | · | · | · | · | |
| 18 | · | · | C | · | G | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | C | T | · | · | · | · | · | · | · | |
| 17 | · | · | C | · | G | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | C | · | · | · | · | · | · | · | · | |
| 21 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | |
| II | 4 | · | · | · | · | · | · | · | T | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · |
| 29 | · | · | · | · | · | · | · | T | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | C | · | T | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | |
| 32 | · | · | · | G | · | · | · | T | T | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | · | C | · | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | · | |
| 31 | · | · | · | · | · | · | · | T | T | · | · | · | A | · | · | · | · | · | · | · | · | G | · | · | · | C | · | · | · | · | · | · | T | · | · | · | · | · | · | · | · | · | · | · | |
| 30 | · | · | · | · | · | · | · | T | T | · | · | · | A | · | · | · | · | · | · | · | · | G | · | · | · | C | · | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | · | |
| III | 25 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | T | · | · | G | · | · | · | C | · | · | · | · | · | · | T | · | · | · | · | · | · | · | · | · | · | · |
| 3 | A | · | · | · | · | · | · | · | · | · | · | · | · | T | · | · | · | · | T | · | · | G | · | · | · | C | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | |
| 1 | A | · | · | · | · | · | · | · | · | · | · | A | · | T | · | · | · | · | T | · | · | G | · | · | · | C | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | |
| 2 | A | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | T | · | · | G | · | · | · | C | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | |
| 27 | · | · | · | · | · | · | · | · | · | · | · | · | · | T | · | · | · | · | T | T | A | G | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | T | · | · | · | · | · | |
| 26 | · | · | · | · | · | · | · | · | · | · | · | · | · | T | · | · | · | · | T | T | A | G | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | |
| 28 | · | · | · | · | · | · | · | · | · | · | · | · | · | T | · | · | · | · | T | T | A | G | · | · | · | C | · | T | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | |
| IV | 22 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | T | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · |
| 23 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | T | · | T | C | T | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | |
| 20 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | T | T | T | C | T | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | |
| 24 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | T | T | T | C | T | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | |
The SNP profiles were arranged according to the parsimony tree in Fig. 1. Dots indicate that the nucleotide is the same as that of the consensus sequence.
The homoplasy index for the maximum parsimony analysis was 0.17, indicating the presence of parallel or reverse changes. However, the mapping of the SNPs to the tree in Fig. 1 showed that only five SNPs appeared on different branches and therefore have undergone reverse or parallel changes, being parallel changes in two nodes (SNPs 22 and 33) or three nodes (SNP 28) or a reverse change (SNPs 7 and 14). It is interesting that SNP 28 appeared in three different nodes in clusters I, II, and III, respectively, while SNP 33 appeared in two separate nodes in clusters II and III. Reanalysis using maximum parsimony without the 5 conflicting SNPs generated a single tree with a homoplasy index of 0. A loss of the resolution of several terminal branches involving the conflicting SNPs was observed, but the major branching patterns remained unchanged.
These reverse or parallel changes are likely to have been derived by recombination within S. Typhimurium strains rather than mutations, given the level of recombination reported. Based on MLST data for S. Typhimurium (42), the ratio of recombination to mutation as the cause of SNPs is 1.69 per site, making it quite likely that the reverse or parallel changes observed are due to recombination. These reverse or parallel changes are likely to be a result of neutral events, since two of the SNPs (SNP 7 and SNP 28) were located in IG regions, and two of the three genic SNPs are synonymous.
We examined the distribution of SPs and clusters for correlations with countries of origin (Table 2). The majority of SPs or clusters were not linked with a particular locality. However, SP17 isolates, containing 4 phage types (DT2, DT40, DT41, and DT141), were all from the United Kingdom; cluster II isolates, with five single DT SPs, were all from Australia; and cluster IV isolates, except for 2 SP23 isolates of DT9, were all from the United Kingdom. SP18 contained only Australian isolates, but this may be due to sampling bias, since it contains only a single phage type, DT126, which was not represented among isolates from the United Kingdom. However, we were unable to make an inference of relationships with host species, since isolates were selected purposely with a mix of human and nonhuman isolates for a phage type, and there was a small number of isolates per phage type.
Table 2.
Summary of isolates used in this study
| Cluster | SNP profile | Total no. of isolates | Phage type(s) (DT)a | Yr of isolation | Source(s)b | Location(s)c |
|---|---|---|---|---|---|---|
| I | 14 | 1 | 1 | H | AU | |
| 12 | 4 | 44 | A, H | AU | ||
| 13 | 1 | 44 | ||||
| 15 | 10 | 29, 49, 69, 193, 302 | 2001–2009 | A, H | AU, UK | |
| 16 | 1 | 44 | H | AU | ||
| 9 | 16 | 135 | 1992 | IL | ||
| 33 | 26 | 6, 17, 29, 135, 135a, 170, 186, 193, 307, U290 | 2002–2008 | E, H | AU, UK (SG) | |
| 5 | 3 | 135 | 1991–1995 | FR, MU, UK | ||
| 6 | 2 | 135 | 1992–1998 | DE, GR | ||
| 7 | 38 | 3, 9, 64 | 1991–2004 | A, H | AU, ES, JM, RO, UK | |
| 8 | 6 | 8, 46 | 2001–2008 | H | AU, UK | |
| 10 | 12 | 10, 15a, 2, 22, 46, 99, 133, 135 | 2001–2002 | A, H | AU, UK, UK (PK), UK (Carribean) | |
| 11 | 1 | 2 | ||||
| 19 | 2 | 160 | 2000–2001 | H | UK | |
| 18 | 7 | 126 | 2008 | A, H | AU | |
| 17 | 9 | 2, 40, 41, 141 | 2001–2002 | A, H | UK, UK (ES) | |
| 21 | 14 | 5, 12a, 22, 41, 101, 170, 177, 302 | 2002–2008 | A, H | AU, UK | |
| II | 4 | 2 | 179 | 2007–2008 | AU | |
| 29 | 1 | 44 | A | AU | ||
| 32 | 2 | 1 | H | AU | ||
| 31 | 3 | 12a | A, H | AU | ||
| 30 | 3 | 197 | H | AU | ||
| III | 25 | 1 | 1 | 2002 | H | UK (IN) |
| 3 | 8 | 1, 10, 101, 102, 108, 193 | 2001–2007 | A, E, H | AU, UK | |
| 1 | 14 | 10, 108, 12, 167, 170, 193, 208, 35 | 2000–2002 | A, H | AU, UK | |
| 2 | 1 | 2 | 2002 | H | UK | |
| 27 | 1 | 4 | ||||
| 26 | 16 | 12, 15a, 17, 4, 102, 141, 195 | 2000–2009 | A, H | AU, UK | |
| 28 | 1 | 35 | 2002 | A | UK | |
| IV | 22 | 1 | 104 | 2002 | H | UK |
| 23 | 3 | 1, 9 | 1993–2002 | AE, Pacific Island | ||
| 20 | 2 | 104 | 2002 | A | UK | |
| 24 | 4 | 9, 104 | 1997–2002 | H | UK |
DTs marked in boldface type were likely to have a single origin based on SNP data in this study; see the text for details.
A, animal source; E, environmental source; H, human source.
AE, United Arab Emirates; AU, Australia; DE, Germany; ES, Spain; FR, France; GR, Greece; IL, Israel; IN, India; JM, Jamaica; MU, Mauritius; PK, Pakistan; RO, Romania; SG, Singapore; UK, United Kingdom. Countries in parentheses indicate that the isolate was obtained from a patient who had a history of travel to that country and could have been an imported case.
Multiple origins for several phage types.
Isolates from 20 of the 42 phage types with more than one isolate were typed into a single SP, suggesting that these DTs were likely to be derived from a single origin. Another eight phage types (DT12, DT29, DT35, DT46, DT102, DT104, DT135, and DT108) that were typed into 2 to 5 closely related SPs based on their relationships in the tree (Fig. 1) were also likely to have a single origin. The remaining 14 phage types, including DT9, had multiple SPs in different clusters and can be seen to have originated multiple times.
For DT9 and DT135, which are predominant phage types in Australia, we included 35 isolates representing worldwide diversity that had been used in a previous study (21). We can use DT9 and DT135 to assess the significance of phage typing for long-term epidemiology. The DT9 isolates were isolated from 1991 to 1999 in seven countries, comprising 1 isolate each from Jamaica, the Pacific Islands, Romania, Spain, and the United Arab Emirates; 1 from Australia; and 25 from the United Kingdom. These isolates were differentiated into three SPs, SP7, SP23, and SP24, with the majority belonging SP7. Only SP7 belonged to cluster I, while both SP23 and SP24 were grouped together into cluster IV with DT104 isolates, indicating two independent origins for DT9.
The DT135 isolates were isolated from 1991 to 2008; six were from Australia, while the remaining isolates were from 21 countries. The majority were either SP9 isolates (16 isolates) or SP33 isolates (12 isolates), while the remaining seven isolates belonged to SP5, SP6, and SP10, with three, two, and two isolates, respectively. The SPs differed from each other by one to two SNPs, except for SP33, which differed from the other DT135 SPs by two or more SNPs. All SPs except SP10 and SP33 were unique to DT135 isolates and belonged to cluster I. Two of the six Australian DT135 isolates fell into SP10, while the remaining four isolates were SP33 isolates. It appears that DT135 entered Australia twice as SP10 and SP33 isolates.
Similarly, DT1 is likely to have multiple origins. In the previous study by Hu et al. (18), the DT1 isolates fell into two groups closely related to DT44 and DT12a. In this study, the DT1 isolates fell into five individual SPs, SP3, SP14, SP23, SP25, and SP32, spanning all four clusters, indicating multiple origins.
A comparison of phage types of single and multiple origins showed that phage types with multiple origins tended to be those with isolates from more than one country. Of 28 phage types of a single origin, only 2 contained isolates from more than one country, while 5 of the 14 phage types with multiple origins contained isolates from more than one country. A sampling of more isolates from different countries may show that more phage types have multiple origins.
Interconvertible phage types and their genetic relationships.
Several phage types are known to be interconvertible by the gain or loss of phages or plasmids (2, 27, 50), and the relationships based on SNP typing (Fig. 1) are consistent with these observations. DT9 can be converted to DT64 by the gain of lysogenic temperate phage ST64T (27). The five DT64 isolates shared the same SNP profile (SP7) as one of the DT9 isolates, showing that the six isolates could have a common origin. It should be noted that the DT64 and DT9 isolates used in the study by Hu et al. (18) were separated by an indel of over 2,000 bp. However, since the two phage types can interconvert by the gain or loss of phage ST64T (27), the indel may not be a reliable maker for the separation of the two phage types.
Phage ST64T was also shown previously to convert DT41 isolates to DT29 (27). SNP typing separated the three DT29 isolates and the three DT41 isolates into unrelated SNP profiles, suggesting that for these isolates, the two phage types had not undergone interconversion. Phage type conversion can also be caused by the acquisition of plasmids. DT170 is convertible to DT208 by the acquisition of the FIme plasmid (2). All three DT208 isolates used in this study and two of the five DT170 isolates belonged to SP1, consistent with the interconversion of DT208 and DT170 through a gain or loss of this plasmid.
Evolution of DT135a.
DT135a is of interest because it has increased in frequency in Australia in recent years to become the second most prevalent S. Typhimurium phage type in 2007 to 2008 (35, 36). PCR typing showed that DT135 and DT135a isolates were very closely related, as they are separated by only 1 of the 22 molecular markers used (22). In this study, the three DT135a isolates were grouped together with the majority of DT135 isolates in SP33, further supporting the close relationship of the two phage types, and DT135a was most likely derived from DT135. More SNPs are required to separate the two phage types.
Evolutionary origin of multidrug-resistant epidemic phage type DT104.
Multidrug-resistant (MDR) S. Typhimurium DT104 has become widespread in humans and animals in many parts of the world, including many European countries, the United States, Canada, and Asian countries such as Japan and South Korea, since the 1990s (14). It was first reported in the 1960s and then emerged in an MDR form in the early 1980s in Britain (48). MDR DT104 isolates carry genes for multidrug resistance on 43-kb Salmonella genomic island 1 (SGI1), which encodes 44 open reading frames (28), and a prophage, ST104 (15, 46), which carries two putative toxin genes (artAB) encoding an ADP-ribosyl transferase toxin homologue (41). In addition, Matiasovicova et al. (24) found previously that all MDR DT104 isolates carry a deletion that includes the genes for the utilization of allantoin. The same deletion is found in several DT104 isolates that lack SGI1. Therefore, it was proposed that the allantoin-negative form is ancestral to the MDR DT104 isolates.
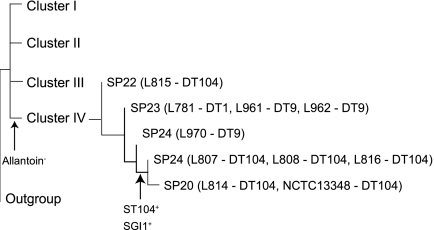
In this study, we analyzed five DT104 isolates from the United Kingdom and one epidemic MDR DT104 genome sequence. These isolates were divided into three related SPs in the same cluster, interspersed by SP23, which contains DT1 and DT9 but no DT104 isolates, and SP24, with one DT9 isolate (Fig. 1). We tested all cluster IV isolates for the presence of SGI1, ST104, and the allantoin utilization operon by PCR. All cluster IV isolates tested had lost the ability to utilize allantoin, but only DT104 isolates belonging to SP20 and SP24 carried SGI1 and prophage ST104 (Fig. 2). It is interesting that SGI1 and ST104 were gained at the same branch point. Our results suggest that MDR DT104 was indeed derived from an allantoin-negative ancestral form by gaining SGI1, as proposed previously by Matiasovicova et al. (24). However, it was less clear whether a DT104 isolate gave rise to MDR DT104. One SP24 isolate, L970, did not carry SGI1 or ST104 and could be viewed as the ancestor of MDR DT104. However, L970 is a DT9 isolate, and the only SGI1-negative DT104 isolate (SP22) was located at the base of cluster IV, while the intermediate SP23, which lies between SP22 and SP24, contains DT9 or DT1 but no DT104 isolates. It appears that the interconversion of DT104 and DT9 has occurred more than once in this cluster. It should also be noted that the current study included a small sample size of only 6 DT104 isolates, and a larger representation of DT104 isolates will provide a better view of the origin of MDR DT104.
Fig 2.
Evolution of multidrug resistance of multidrug-resistant DT104 isolates from cluster IV. Isolates within cluster IV were PCR typed for the presence of Salmonella genetic island I (SGI1), ST104, and the allantoin utilization operon to determine the branch point for the gain or loss of these 3 features. Allantoin− denotes the inability to utilize allantoin, which has been lost due to the absence of the allantoin operon (STM0517 to STM0529); SGI1+ indicates the presence of SGI1; and ST104+ signifies the presence prophage ST104. The isolates and their designated phage types are shown in parentheses, separated with a dash. Multiple isolates with the same SNP profile and genotype are separated by commas.
Value of phage typing for determination of long-term and/or worldwide epidemiology of S. Typhimurium.
Phage typing has been applied to define S. Typhimurium strains and track their spread within different host populations and their geographical distributions. However, its suitability for long-term and/or worldwide epidemiology studies over long periods (years) and diverse localities (different countries or continents) (49) is debatable in the light of our data. We have shown that some phage types are likely to have arisen more than once, and isolates may not share the same origin or be epidemiologically related. For example, DT108 and DT170 have very similar phage sensitivity patterns (40) and have now been treated as the same phage type in epidemiological data from Australia. However, our results show that the majority of DT170 isolates were in SP30 in cluster I or SP1 in cluster III, with one isolate in the unclustered SP21. Only two out of the six DT170 isolates were in SP1 with two DT108 isolates. Three DT108 isolates are also found in the closely related SP3. Thus, epidemiological comparisons based on phage types alone may not be valid. In addition, a previous study by Rabsch et al. (39) showed that among 53 archived LT2 strains in the Demerec collection, only 16 produced the expected DT4 phage type, present in the sequenced LT2 strain, while the others belonged to 11 different phage types. There are also potential issues with phage stocks that were prepared decades ago and distributed from the Enteric Reference Laboratory (43). The phage typing scheme of Anderson et al. (3) in current use has the virtue of being standardized and has served S. Typhimurium epidemiology well, but clearly, changes are needed if we are to retain a typing scheme that can be applied worldwide. Ideally, newer techniques can be incorporated into an integrated global scheme, building on the current phage typing scheme.
Concluding comments.
Phage typing has played an important role in the epidemiology and public health surveillance of food-borne infections caused by S. Typhimurium. In this study, we used SNP typing to infer the phylogenetic relationships of 221 S. Typhimurium isolates from 45 phage types. The 44 SNPs used distinguished the isolates into 33 SNP profiles and four distinct phylogenetic groups. We show that 14 phage types, including DT9 and DT135, the predominant phage types in Australia, had multiple origins. Phage typing as currently practiced does not seem suitable for the study of the long-term epidemiology of S. Typhimurium. While it is often very useful for the detection of relationships among outbreaks, there are too many cases where multiple origins of a phage type confuse the picture. The use of SNPs enables us to establish phylogenetic relationships as well as carry out typing. The SNP typing scheme devised in this study could be a valuable complement to phage typing for the study of the global epidemiology of S. Typhimurium and has the potential to replace phage typing. The SNPs selected for this study cannot differentiate all different phage types, but the power of SNP typing could be increased by incorporating more SNPs. Next-generation genome sequencing will allow a rapid expansion of SNPs for S. Typhimurium typing, as for S. Typhi (17) and Escherichia coli O157:H7 (6) strains. In this study, we used real-time PCR for typing, but the SNPs can also be used for large-scale typing by the adoption of high-throughput platforms, such as the iPlex MassArray technology (45) or the GoldenGate bead array technology (16). Establishing true relationships based on SNPs would provide a foundation for future genomic studies of differences in the prevalences and virulences of different SNP lineages or phage types.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the National Health and Medical Research Council of Australia.
We also thank Gordon Stevenson for technical assistance.
Footnotes
Published ahead of print 28 December 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Achtman M. 2008. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 62:53–70 [DOI] [PubMed] [Google Scholar]
- 2. Anderson ES, Threlfall EJ, Carr JM, McConnell MM, Smith HR. 1977. Clonal distribution of resistance plasmid-carrying Salmonella typhimurium, mainly in the Middle East. J. Hyg. 79:425–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson ES, Ward LR, Saxe MJ, de Sa JD. 1977. Bacteriophage-typing designations of Salmonella typhimurium. J. Hyg. 78:297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beltran P, et al. 1991. Reference collection of strains of the Salmonella typhimurium complex from natural populations. J. Gen. Microbiol. 137:601–606 [DOI] [PubMed] [Google Scholar]
- 5. Carver T, et al. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clawson ML, et al. 2009. Phylogenetic classification of Escherichia coli O157:H7 strains of human and bovine origin using a novel set of nucleotide polymorphisms. Genome Biol. 10:R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cooke FJ, et al. 2008. Characterization of the genomes of a diverse collection of Salmonella enterica serovar Typhimurium definitive phage type 104. J. Bacteriol. 190:8155–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cooke FJ, et al. 2007. Prophage sequences defining hot spots of genome variation in Salmonella enterica serovar Typhimurium can be used to discriminate between field isolates. J. Clin. Microbiol. 45:2590–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fakhr MK, Nolan LK, Logue CM. 2005. Multilocus sequence typing lacks the discriminatory ability of pulsed-field gel electrophoresis for typing Salmonella enterica serovar Typhimurium. J. Clin. Microbiol. 43:2215–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Filliol I, et al. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garaizar J, et al. 2002. DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J. Clin. Microbiol. 40:2074–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glynn MK, et al. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333–1338 [DOI] [PubMed] [Google Scholar]
- 13. Guard J, Shah D, Morales C, Call D. 2011. Evolutionary trends associated with niche specialization as modeled by whole genome analysis of egg-contaminating Salmonella enterica serovar Enteritidis, p 91–106 In Porowollik S. (ed), Salmonella: from genome to function. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 14. Helms M, Ethelberg S, Mølbak K, DT104 Study Group 2005. International Salmonella Typhimurium DT104 infections, 1992-2001. Emerg. Infect. Dis. 11:859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hermans AP, Abee T, Zwietering MH, Aarts HJ. 2005. Identification of novel Salmonella enterica serovar Typhimurium DT104-specific prophage and nonprophage chromosomal sequences among serovar Typhimurium isolates by genomic subtractive hybridization. Appl. Environ. Microbiol. 71:4979–4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holt KE, et al. 2010. High-throughput bacterial SNP typing identifies distinct clusters of Salmonella Typhi causing typhoid in Nepalese children. BMC Infect. Dis. 10:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holt KE, et al. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 40:987–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu H, Lan R, Reeves PR. 2006. Adaptation of multilocus sequencing for studying variation within a major clone: evolutionary relationships of Salmonella enterica serovar Typhimurium. Genetics 172:743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jarvik T, Smillie C, Groisman EA, Ochman H. 2010. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J. Bacteriol. 192:560–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kingsley RA, et al. 2009. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 19:2279–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lan R, Davison AM, Reeves PR, Ward LR. 2003. AFLP analysis of Salmonella enterica serovar Typhimurium isolates of phage types DT9 and DT135: diversity within phage types and its epidemiological significance. Microbes Infect. 5:841–850 [DOI] [PubMed] [Google Scholar]
- 22. Lan R, Stevenson G, Donohoe K, Ward L, Reeves PR. 2007. Molecular markers with potential to replace phage typing for Salmonella enterica serovar Typhimurium. J. Microbiol. Methods 68:145–156 [DOI] [PubMed] [Google Scholar]
- 23. Low JC, Angus M, Hopkins G, Munro D, Rankin SC. 1997. Antimicrobial resistance of Salmonella enterica Typhimurium DT104 isolates and investigation of strains with transferable apramycin resistance. Epidemiol. Infect. 118:97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matiasovicova J, et al. 2007. Identification of putative ancestors of the multidrug-resistant Salmonella enterica serovar Typhimurium DT104 clone harboring the Salmonella genomic island 1. Arch. Microbiol. 187:415–424 [DOI] [PubMed] [Google Scholar]
- 25. McClelland M, et al. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36:1268–1274 [DOI] [PubMed] [Google Scholar]
- 26. McClelland M, et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 27. Mmolawa PT, Willmore R, Thomas CJ, Heuzenroeder MW. 2002. Temperate phages in Salmonella enterica serovar Typhimurium: implications for epidemiology. Int. J. Med. Microbiol. 291:633–644 [DOI] [PubMed] [Google Scholar]
- 28. Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A. 2006. The genetics of Salmonella genomic island 1. Microbes Infect. 8:1915–1922 [DOI] [PubMed] [Google Scholar]
- 29. Octavia S, Lan R. 2007. Single-nucleotide-polymorphism typing and genetic relationships of Salmonella enterica serovar Typhi isolates. J. Clin. Microbiol. 45:3795–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Octavia S, Lan R. 2010. Single nucleotide polymorphism typing of global Salmonella enterica serovar Typhi isolates by use of a hairpin primer real-time PCR assay. J. Clin. Microbiol. 48:3504–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Octavia S, et al. 2011. Insight into evolution of Bordetella pertussis from comparative genomic analysis: evidence of vaccine-driven selection. Mol. Biol. Evol. 28:707–715 [DOI] [PubMed] [Google Scholar]
- 32. Parkhill J, et al. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852 [DOI] [PubMed] [Google Scholar]
- 33. Pearson T, et al. 2004. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 101:13536–13541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Powling J. 2005. National enteric pathogens surveillance scheme, vol non-human annual report 2004. Microbiological Diagnostic Unit, University of Melbourne, Victoria, Australia [Google Scholar]
- 35. Powling J. 2008. National enteric pathogens surveillance scheme, vol human annual report 2008. Microbiological Diagnostic Unit, University of Melbourne, Victoria, Australia [Google Scholar]
- 36. Powling J. 2007. National enteric pathogens surveillance scheme, vol human annual report 2007. Microbiological Diagnostic Unit, University of Melbourne, Victoria, Australia [Google Scholar]
- 37. Pupo GM, Lan R, Reeves PR, Baverstock P. 2000. Population genetics of Escherichia coli in a natural population of native Australian rats. Environ. Microbiol. 2:594–610 [DOI] [PubMed] [Google Scholar]
- 38. Rabsch W. 2007. Salmonella typhimurium phage typing for pathogens. Methods Mol. Biol. 394:177–211 [DOI] [PubMed] [Google Scholar]
- 39. Rabsch W, Helm RA, Eisenstark A. 2004. Diversity of phage types among archived cultures of the Demerec collection of Salmonella enterica serovar Typhimurium strains. Appl. Environ. Microbiol. 70:664–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ross IL, Heuzenroeder MW. 2005. Discrimination within phenotypically closely related definitive types of Salmonella enterica serovar Typhimurium by the multiple amplification of phage locus typing technique. J. Clin. Microbiol. 43:1604–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saitoh M, et al. 2005. The artAB genes encode a putative ADP-ribosyltransferase toxin homologue associated with Salmonella enterica serovar Typhimurium DT104. Microbiology 151:3089–3096 [DOI] [PubMed] [Google Scholar]
- 42. Sangal V, et al. 2010. Evolution and population structure of Salmonella enterica serovar Newport. J. Bacteriol. 192:6465–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmieger H. 1999. Molecular survey of the Salmonella phage typing system of Anderson. J. Bacteriol. 181:1630–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Swofford DL. 1998. PAUP: phylogenetic analysis using parsimony, 4.0 beta ed. Sinauer Associates, Sunderland, MA [Google Scholar]
- 45. Syrmis MW, et al. 2011. Comparison of a multiplexed MassARRAY system with real-time allele-specific PCR technology for genotyping of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 17:1804–1810 [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 46. Tanaka K, et al. 2004. Molecular characterization of a prophage of Salmonella enterica serotype Typhimurium DT104. J. Clin. Microbiol. 42:1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thomson NR, et al. 2008. Comparative genome analysis of Salmonella enteritidis PT4 and Salmonella gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 18:1624–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Threlfall EJ. 2000. Epidemic Salmonella typhimurium DT104—a truly international multiresistant clone. J. Antimicrob. Chemother. 46:7–10 [DOI] [PubMed] [Google Scholar]
- 49. Tibayrenc M. 1996. Towards a unified evolutionary genetics of microorganisms. Annu. Rev. Microbiol. 50:401–429 [DOI] [PubMed] [Google Scholar]
- 50. Willshaw GA, Threlfall EJ, Ward LR, Ashley AS, Rowe B. 1980. Plasmid studies of drug-resistant epidemic strains of Salmonella typhimurium belonging to phage types 204 and 193. J. Antimicrob. Chemother. 6:763–773 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.