Abstract
Herpes simplex virus 1 (HSV-1) esophagitis diagnosis is routinely based on the endoscopic findings confirmed by histopathological examination of the esophagitis lesions. Virological diagnosis is not systematically performed and restricted to viral culture or to qualitative PCR assay from esophagitis biopsy specimens. The aim of this study was to assess the interest of quantitative real-time PCR assay in HSV-1 esophagitis diagnosis by comparing the results obtained to those of histological examination associated with immunohistochemical staining, which is considered the “gold standard.” From 53 esophagitis biopsy specimens, the PCR assay detected HSV-1 in 18 of 19 histologically proven to have herpetic esophagitis and in 9 of 34 that had esophagitis related to other causes, demonstrating sensitivity, specificity, positive predictive value, and negative predictive value of 94.7%, 73%, 66.7%, and 96%, respectively. Interestingly, HSV-1 was not detected in 16 specimens without the histological aspect of esophagitis. The viral loads normalized per μg of total extracted DNA in each biopsy specimen detected positive by HSV PCR were then compared and appeared to be significantly higher in histopathologically positive herpetic esophagitis (median = 2.9 × 106 ± 1.1 × 108) than in histopathologically negative herpetic esophagitis (median = 3.1 × 103 ± 6.2 × 103) (P = 0.0009). Moreover, a receiver operating characteristics analysis revealed that a viral load threshold greater than 2.5 × 104 copies would allow an HSV-1 esophagitis diagnosis with a sensitivity and specificity of 83.3% and 100%, respectively. In conclusion, this work demonstrated that HSV quantitative PCR results for paraffin-embedded esophageal tissue was well correlated to histopathological findings for an HSV-1 esophagitis diagnosis and could be diagnostic through viral load assessment when histopathological results are missing or uncertain.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is the second most common infectious etiological cause of esophagitis after Candida albicans (15, 18, 22). HSV-1 esophagitis is well documented in immunocompromised patients, whereas this clinical entity is rare in immunocompetent patients (2, 4, 5, 9). HSV-1 esophagitis may represent a primary infection in particular in the immunocompetent host but is commonly due to a reactivation of a latent infection in the immunocompromised host (7).
Friable mucosa, numerous ulcers, and whitish exudates commonly involving the distal or the midesophagus are classical endoscopic aspects in HSV-1 esophagitis (10, 11, 14, 19). These findings require confirmation by histological examination completed with immunohistochemical staining of the biopsy specimens from the ulcer edges, which still remains the “gold standard” for HSV-1 esophagitis diagnosis (13, 14, 19). Virological diagnosis has been reported to optimize the diagnostic sensitivity of HSV-1 esophagitis (2, 16, 19). However, it is not systematically performed and is restricted to viral culture or to qualitative PCR assay from esophageal biopsy specimens (2, 19).
During the last 20 years, HSV-1 DNA detection by PCR assay has been demonstrated to be more sensitive than tissue viral culture in diagnosing HSV-1 esophagitis (3, 17, 20). However, the high sensitivity of the molecular technique could lead to the detection of low viral loads potentially associated with asymptomatic viral shedding that cannot be related to the clinical findings (1). Thus, the utility of HSV-1 DNA detection by PCR assay remains to be assessed in establishing a correct diagnosis of HSV-1 esophagitis. In this context, the aim of this study was to compare the results obtained from 69 esophageal biopsy specimens analyzed by HSV quantitative real-time PCR assay to the classical histopathological examination including HSV immunohistochemical staining, considered the gold standard. Moreover, the interest of HSV-1 quantitation in esophageal specimens for the herpetic esophagitis diagnosis was assessed.
MATERIALS AND METHODS
Clinical specimens.
A total of 69 paraffin-embedded esophageal biopsy specimens from 69 patients collected between 1997 and 2007 by the Department of Pathology of the Reims University Hospital were retrospectively investigated. Histological examinations after hematein phloxine safron (HPS) and periodic acid-Schiff (PAS) staining were systematically performed by two pathologists. Histological lesions corresponding to viral cytopathic effect were noted. Immunohistochemical staining was performed in all cases on 4-μm-thick paraffin sections with a polyclonal rabbit anti-herpes simplex virus 1 and 2 antibody (DakoCytomation, Glostrup, Denmark) without antigenic restoration using BenchMark XT (Ventana, Tucson, AZ). Sections were incubated for 1 h at 37°C with the antibody at a 1:100 dilution followed by staining with a streptavidin-biotin peroxidase kit and were then revealed with an ultraView Universal DAB detection kit (Ventana, Tucson, AZ) and counterstained with hematoxylin for 1 min.
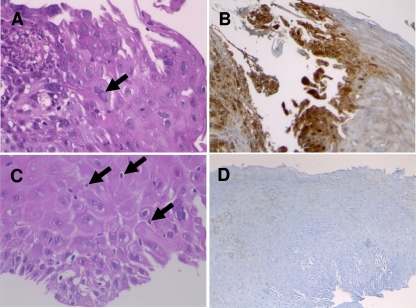
Among the 69 biopsy specimens selected, 19 (patient male-to-female sex ratio, 2.8; median age in years, 60 ± 17.4) were diagnosed as esophagitis related to herpetic infection on the basis of typical herpetic histological changes (presence of plurinuclear squamous cells with vitreous aspect of the cores and/or intranuclear inclusion bodies) and positive immunohistochemical staining (Fig. 1). These samples were designated histopathologically positive herpetic (HPH) esophagitis. Another 34 (male-to-female sex ratio, 2.4; median age in years, 66 ± 17.5) were considered nonherpetic esophagitis, since biopsy specimens of esophagitis lesions presented neither histological evidence of HSV infection nor positive HSV immunohistochemical staining (Fig. 1). These samples were designed as histopathologically nonherpetic (HNH) esophagitis. HNH esophagitis cases were related to reflux esophagitis (n = 27), candidal esophagitis (n = 2), and postirradiation esophagitis (n = 5). Finally, 16 samples were collected from patients (male-to-female sex ratio, 1.7; median age in years, 58 ± 14.3) who did not present with esophagitis lesions at the endoscopic examination; this status was confirmed by the absence of histological evidence of esophagitis and negative HSV immunohistochemical staining of the esophageal biopsy specimens.
Fig 1.
Histopathological aspect of herpetic (A and B) and nonherpetic (C and D) esophagitis biopsy specimens. (A) HSV esophagitis: typical multinucleated epithelial cells with ground-glass nuclei and margination of the chromatin (black arrow). HPS stain; magnification ×400. (B) Immunohistochemical detection of HSV glycoproteins (in brown) in epithelial cells. Magnification, ×100. (C) Non-HSV esophagitis: neither multinucleated nor ground-glass nuclei; intraepithelial exocytosis of lymphocytes (black arrows). HPS; magnification, ×400. (D) Negative immunohistochemical detection of HSV antigens. Magnification, ×100.
HSV quantitative real-time PCR assay.
After the dewaxing phase, the whole block of esophageal tissue selected according to the histological examination results was subjected to proteinase K (600 milli-Anson units/ml) (Merck Novagen, Darmstadt, Germany) digestion in extraction buffer containing 20 mM Tris-HCl, pH 8.3 (Sigma-Aldrich, St. Louis, MO), and 0.5% SDS (Sigma-Aldrich, St. Louis, MO) for 30 min in a water bath at 56°C. DNA for real-time PCR assay was then extracted from the supernatant obtained after centrifugation (1 min at 6, 000 × g) using EasyMAG (bioMérieux, Marcy l'Etoile, France) according to the manufacturer's instructions. A glyceraldehyde 3-phosphate dehydrogenase (GAPDH) real-time PCR assay was performed on the 69 samples investigated and demonstrated the ability to amplify at least a 600-base-pair amplimer of the DNA extracted from formalin-fixed, paraffin-embedded archival biopsy specimens (13).
The TaqMan HSV quantitative real-time PCR assay integrating PCR inhibitor detection was performed using an iCycler IQ (Bio-Rad, Marnes-la-Coquette, France) as described previously (6). The oligonucleotide primers used for amplification were constructed to detect the HSV-1 and HSV-2 glycoprotein B gene. Thus, HSV detected in the esophageal specimens were typed with the multiplex endpoint PCR kit Herpes Consensus Generic (Argène, France), screening for HSV-1, HSV-2, varicella-zoster virus (VZV), cytomegalovirus (CMV), Epstein-Barr virus (EBV), and human herpesvirus 6 (HHV-6). Moreover, the HSV-1 DNA-positive biopsy specimens were independently tested five times each in five successive experiments in order to assess the repeatability as well as three times each within the same experiment to assess the reproducibility of the viral quantitation from paraffin-embedded esophageal tissue. The mean value of the viral load for each sample assessed through the repeated analyses was then used for statistical comparison between HPH and HNH esophagitis samples.
For each sample, total extracted DNA was quantified by optical density at 260/280 nm using a Picodrop Microliter UV/Vis spectrophotometer (Saffron Walden, United Kingdom). HSV-1 viral load levels were then normalized and expressed as the number of HSV-1 DNA copies per microgram of total extracted DNA.
Statistical analysis.
Quantitative variables were described as median ± standard deviation and qualitative data as number and percentage. The accuracy of the quantitative real-time PCR assay to diagnose HSV-1 esophagitis was determined by sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) using the histopathological examination as the gold standard. The reproducibility and the repeatability of HSV-1 quantitative measurements in paraffin-embedded esophageal tissue were assessed through intraclass correlation coefficient (ICC) determination. Viral load levels of HPH and HNH esophagitis samples were compared using the Wilcoxon test. A receiver operating characteristics (ROC) analysis was performed to find the best viral load threshold separating the HPH esophagitis from the HNH esophagitis. Results were considered statistically significant for two-sided P values of <0.05. All statistical analyses were performed using the SAS system release 9.0 (SAS Institute Inc.).
RESULTS
Comparison of HSV DNA detection to histopathological examination for HSV esophagitis diagnosis.
Among the 69 esophageal specimens tested, the HSV quantitative real-time PCR assay detected HSV DNA in 27. HSV was detected only in esophagitis biopsy specimens, whereas the 16 esophageal specimens obtained from patients presenting without any lesion of esophagitis remained negative. The multiplex endpoint PCR kit Herpes Consensus Generic (Argène, France) identified HSV-1 in all 27 HSV DNA-positive specimens.
Table 1 shows the overall results obtained by both quantitative real-time PCR assay and histopathological examination from the 53 esophagitis biopsy specimens investigated. HSV was not detected in only one HPH esophagitis sample, whereas 9 of the 34 HNH esophagitis samples tested positive for HSV DNA by PCR assay. In summary, the HSV quantitative real-time PCR assay demonstrated, by comparison to the routinely used histopathological examination considered the gold standard, a sensitivity of 94.7% (range, 89.4 to 99.9), a specificity of 73.5% (58.7 to 88.3), a PPV of 66.7% (55.6 to 77.8), and an NPV of 96.1% (88.7 to 100) for HSV-1 esophagitis diagnosis.
Table 1.
Comparison of esophagitis biopsy specimen testing results by histopathological examination and quantitative PCR assay
| Quantitative PCR assay result | No. with result of histopathological examination |
|
|---|---|---|
| Positive | Negative | |
| Positive | 18 | 9 |
| Negative | 1 | 25 |
| No. of specimens tested | 19 | 34 |
Viral load quantitation in HPH and HNH esophagitis biopsy specimens.
Viral load was then determined in each HSV-1 DNA-positive biopsy specimen. Repeated analyses demonstrated ICCs of 0.98266 (0.93555 to 0.99710) and 0.94109 (0.75580 to 0.99317), demonstrating the reproducibility and repeatability, respectively, of HSV-1 quantitative measurements in paraffin-embedded esophageal tissue.
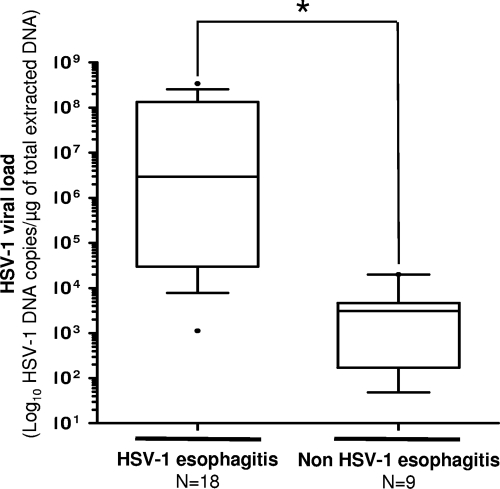
Viral loads measured in the 18 HPH esophagitis biopsy specimens positive by HSV PCR assay were then compared to those quantified in the 9 HNH esophagitis biopsy specimens also positive for HSV-1 DNA detection. The viral load levels were from 1.3 × 103 to 3.4 × 108 (median, 2.9 × 106 ± 1.1 × 108) and from 48 to 2.0 × 104 (median, 3.1 × 103 ± 6.2 × 103) HSV-1 DNA copies/μg of total extracted DNA in HPH and HNH HSV-1 DNA-positive esophagitis biopsy specimens, respectively (Fig. 2). HSV-1 viral loads were significantly higher in HPH esophagitis biopsy specimens by comparison to HNH esophagitis samples (P = 0.0009).
Fig 2.
Box plots showing HSV viral load levels (y axis) in cases of HSV or non-HSV esophagitis (x axis). The lower and upper ends of the box are the 25th and 75th quartiles, respectively. The line across the middle of each box identifies the median value of the HSV viral load in the esophageal specimens tested. The asterisk indicates a statistically significant result (P < 0.05).
ROC analysis.
A receiver operating characteristics (ROC) analysis was also performed to find out the best viral load threshold enabling separation of the HPH and HNH esophageal biopsy specimens positive by the HSV PCR assay. The area under the ROC curve was 0.951 (range, 0.870 to 1.032), confirming that the histopathological results were well correlated to the viral load quantified by HSV quantitative real-time PCR assay in the clinical samples. The best cutoff value was 2.5 × 104 HSV-1 DNA copies/μg of total extracted DNA, demonstrating sensitivity and specificity of 83.3% and 100%, respectively, for HSV-1 esophagitis diagnosis by comparison to results of histopathological examination considered the reference method.
DISCUSSION
HSV-1 esophagitis diagnosis is based on the endoscopic findings confirmed by histopathological examination of the esophagitis lesions (10, 11, 12, 14, 19). The place of the virological diagnosis in an HSV-1 esophagitis diagnosis remains unclear. However, previously published series revealed that histopathological examination alone may miss the diagnosis whereas adding tissue viral culture enhances the diagnostic sensitivity (2, 19). To optimize HSV-1 esophagitis diagnosis, HSV-1 DNA detection by PCR assay has been described as a more sensitive, rapid, and easier diagnostic tool than viral culture, allowing an efficient etiological diagnosis in order to initiate early antiviral treatment (9). The aim of the present study was to assess the advantage of the esophageal biopsy specimen testing by quantitative real-time HSV PCR assay by comparing the results obtained to those of histopathological examination, which is considered the gold standard.
The data obtained from 53 esophagitis biopsy specimens demonstrated a good sensitivity of the PCR assay of paraffin-embedded tissue, since HSV-1 DNA was detected in 18 of the 19 HPH esophagitis samples. Thus, histopathological and molecular virological results appeared to be well correlated for the diagnosis of HSV-1 esophagitis. Furthermore, the high negative predictive value of the HSV-1 DNA detection suggested that HSV-1 esophagitis could be excluded in the case of a negative PCR result. Only one case of HPH esophagitis was not detected by the quantitative real-time HSV PCR assay (Table 1). The discrepant result could be likely explained by a false-negative result of the PCR assay due to incomplete removal of protein-nucleic acid cross-links from extracted DNA or by alteration of viral nucleic acid from paraffin-embedded tissue despite a positive GAPDH PCR assay result for all of the esophageal biopsy specimens investigated (8, 19). This discrepant sample highlighted the main drawback of performing PCR with paraffin-embedded tissues, where many inhibitors are encountered, which could lead to false-negative PCR results.
In contrast, the real-time PCR assay showed a low positive predictive value for correct diagnosis of HSV-1 esophagitis. Indeed, HSV-1 DNA was detected in 9 of the 34 HNH esophagitis samples, highlighting that qualitative virus detection alone is inadequate to confirm HSV-1-related esophagitis (Table 1). The reference method used for this comparison was the histopathological examination, which is the only method allowing assessment of viral cytopathic effect and virus replication in the esophageal biopsy specimens. No other reference method is available, since the endoscopic aspect is not specific to HSV infection; clinical improvement in spite of the lack of antiviral medication was previously reported in many studies, and virus detection either by viral culture or PCR assay can be linked to asymptomatic viral shedding with latent HSV reactivation, which is frequently found, in particular, in the immunocompromised host and could not be considered HSV-1 esophagitis (1, 2, 5). Besides, HSV-1 esophagitis may appear to be more frequent when using the PCR assay, which is more sensitive in detecting HSV in esophagitis biopsy specimens than previously reported when only histology and immunohistochemical staining were used (21). Moreover, histopathological examination may miss the herpetic esophagitis diagnosis, since HSV-1 replication within the esophagus mucosa could be focal. In this case, HSV DNA-positive detection with no histological evidence of herpetic esophagitis could be observed. This hypothesis is supported by the detection of HSV-1 DNA in HNH esophagitis biopsy specimens whereas esophageal biopsy specimens without histological lesions of esophagitis remained negative. The lack of a reference method other than histopathological examination for HSV-1 esophagitis diagnosis avoids a conclusion of the clinical significance of HSV-1 DNA detection in esophagitis biopsy specimens when immunohistochemical staining remains negative.
To address this question and to discriminate HSV-1 esophagitis from asymptomatic viral shedding, quantitation of the viral load in esophageal biopsy specimens could be of great interest. Indeed, viral loads measured in HPH esophagitis biopsy specimens in this study (18 specimens out of 19) were significantly higher than those quantified in HNH esophageal biopsy specimens in which HSV-1 DNA was detected (9 specimens out of 34) (Fig. 2). As shown by the ROC analysis, viral load levels assessed by the HSV quantitative real-time PCR assay appeared to be well correlated to the histopathological examination results of the esophageal biopsy specimens. The cutoff viral load level determined through the ROC analysis (2.5 × 104 copies/μg of total extracted DNA) allowed the HSV-1 esophagitis diagnosis with a sensitivity and specificity of 83.3% and 100%, respectively. These data suggest that a viral load greater than 2.5 × 104 copies/μg of total extracted DNA should be a good threshold for HSV-1 esophagitis diagnosis when histological and immunohistochemical staining results are missing or uncertain. Nevertheless, the clinical relevance of low viral load detection in esophagitis biopsy specimens without histological evidence of HSV-1 infection remains to be determined in further studies.
In conclusion, this work demonstrated that HSV-1 DNA detection from paraffin-embedded esophageal tissue was well correlated to histopathological findings for HSV-1 esophagitis diagnosis. This method could be diagnostic in cases of high viral load assessed in esophageal biopsy specimens. However, due to the high sensitivity of the PCR, viral quantitation is mandatory to differentiate HSV-1 esophagitis from asymptomatic viral shedding associated with negative histopathological examination of esophageal biopsy specimens. Further studies using larger numbers of HPH and HNH esophagitis biopsy specimens will be required to confirm the viral load threshold associated with HSV-1 esophagitis.
ACKNOWLEDGMENTS
This study was supported by a grant for clinical and virological research (EA-4303/IFR53) from the Medical University and School of Medicine of Reims, France. Fanny Renois is supported by an official grant from the French Army department (Bourse DGA: Délégation Générale de l'Armement, Ministère de la Défense, Topic: Microbiology, infectious diseases, 2009–2012).
None of the authors of the present article have a commercial or other association that might pose a conflict of interest (e.g., pharmaceutical stock ownership, consultancy). No extra funding was used.
Footnotes
Published ahead of print 14 December 2011
REFERENCES
- 1. Borges MC, Colares JK, Lima DM, Fonseca BA. 2009. Advantages and pitfalls of the polymerase chain reaction in the diagnosis of esophageal ulcers in AIDS patients. Dig. Dis. Sci. 54:1933–1939 [DOI] [PubMed] [Google Scholar]
- 2. Canalejo Castrillero E, García Durán F, Cabello N, García Martínez J. 2010. Herpes esophagitis in healthy adults and adolescents: report of 3 cases and review of the literature. Medicine 89:204–210 [DOI] [PubMed] [Google Scholar]
- 3. Coyle PV, Desai A, Wyatt D, McCaughey C, O'Neill HJ. 1999. A comparison of virus isolation, indirect immunofluorescence and nested multiplex polymerase chain reaction for the diagnosis of primary and recurrent herpes simplex type 1 and type 2 infections. J. Virol. Methods 83:75–82 [DOI] [PubMed] [Google Scholar]
- 4. Dekeyser S, El Nady M, Diaz E, Cassagnou M, Descamps D. 2009. Diagnosis of herpetic esophagitis in the immunocompetent subject by PCR (Herpès Consensus Générique-Argène). Report of six cases. Pathol. Biol. (Paris) 57:101–106 (In French.) [DOI] [PubMed] [Google Scholar]
- 5. Elliott SY, Kerns FT, Kitchen LW. 1993. Herpes esophagitis in immunocompetent adults: report of two cases and review of the literature. W. V. Med. J. 89:188–190 [PubMed] [Google Scholar]
- 6. Frobert E, et al. 2008. Genotypic detection of acyclovir-resistant HSV: characterization of 67 ACV-sensitive and 14 ACV-resistant viruses. Antiviral Res. 79:28–36 [DOI] [PubMed] [Google Scholar]
- 7. Galbraith JC, Shafran SD. 1992. Herpes simplex esophagitis in the immunocompetent patient: report of four cases and review. Clin. Infect. Dis. 14:894–901 [DOI] [PubMed] [Google Scholar]
- 8. Gazziero A, Guzzardo V, Aldighieri E, Fassina A. 2009. Morphological quality and nucleic acid preservation in cytopathology. J. Clin. Pathol. 62:429–434 [DOI] [PubMed] [Google Scholar]
- 9. Genereau T, Rozenberg F, Bouchaud O, Marche C, Lortholary O. 1997. Herpes esophagitis: a comprehensive review. Clin. Microbiol. Infect. 3:397–407 [DOI] [PubMed] [Google Scholar]
- 10. Geraci G, et al. 2009. Herpes simplex esophagitis in immunocompetent host: a case report. Diagn. Ther. Endosc. 2009:717183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Greenson J, Beschorner W, Boitnott J, Yardley J. 1991. Prominent mononuclear cell infiltrate is characteristic of herpes esophagitis. Hum. Pathol. 22:541–549 [DOI] [PubMed] [Google Scholar]
- 12. Itoh T, et al. 2003. Herpes simplex esophagitis from 1307 autopsy cases. J. Gastroenterol. Hepatol. 18:1407–1411 [DOI] [PubMed] [Google Scholar]
- 13. Jacques J, et al. 2008. Human Bocavirus quantitative DNA detection in French children hospitalized for acute bronchiolitis. J. Clin. Virol. 43:142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee B, Caddy G. 2007. A rare cause of dysphagia: herpes simplex esophagitis. World J. Gastroenterol. 13:2756–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maguire A, Sheahan K. 22 August 2011. Pathology of oesophagitis. Histopathology. doi:10.1111/j.1365-2559.2011.03855.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 16. McBane RD, Jr, Gross JB. 1991. Herpes esophagitis: clinical syndrome, endoscopic appearance, and diagnosis in 23 patients. Gastrointest. Endosc. 37:600–603 [DOI] [PubMed] [Google Scholar]
- 17. Nahass GT, et al. 1992. Comparison of Tzanck smear, viral culture, and DNA diagnostic methods in detection of herpes simplex and varicella-zoster infection. JAMA 268:2541–2544 [PubMed] [Google Scholar]
- 18. Ponticelli C, Passerini P. 2005. Gastrointestinal complications in renal transplant recipients. Transpl. Int. 18:643–650 [DOI] [PubMed] [Google Scholar]
- 19. Ramanathan J, Rammouni M, Baran J, Khatib R. 2000. Herpes simplex virus esophagitis in the immunocompetent host: an overview. Am. J. Gastroenterol. 95:2171–2176 [DOI] [PubMed] [Google Scholar]
- 20. Slomka MJ, Emery L, Munday PE, Moulsdale M, Brown DW. 1998. A comparison of PCR with virus isolation and direct antigen detection for diagnosis and typing of genital herpes. J. Med. Virol. 55:177–183 [PubMed] [Google Scholar]
- 21. Wilcox CM, Straub RF, Clark WS. 1995. Prospective evaluation of oropharyngeal findings in human immunodeficiency virus-infected patients with esophageal ulceration. Am. J. Gastroenterol. 90:1938–1941 [PubMed] [Google Scholar]
- 22. Yee J, Wall SD. 1994. Infectious esophagitis. Radiol. Clin. North Am. 32:1135–1145 [PubMed] [Google Scholar]