Abstract
In this study, we investigated the correlation between the microbiological characteristics of Clostridium difficile clinical isolates and the recurrence of C. difficile-associated disease (CDAD). Twenty C. difficile isolates recovered from 20 single infection cases and 53 isolates from 20 recurrent cases were analyzed by pulsed-field gel electrophoresis (PFGE) and PCR ribotyping, and the cytotoxicity, antimicrobial susceptibility, and sporulation/germination rates of the isolates were examined. Recurrent cases were divided into relapse or reinfection cases by the results of C. difficile DNA typing. Among the 20 recurrent cases, 16 cases (80%) were identified to be relapse cases caused by the initial strain and the remaining 4 cases (20%) were identified to be reinfection cases caused by different strains. All 73 isolates were susceptible to both vancomycin and metronidazole, but resistance against clindamycin, ceftriaxone, erythromycin, and ciprofloxacin was found in 87.7%, 93.2%, 87.7%, and 100% of the isolates, respectively. No correlations between DNA typing group, cytotoxicity, and sporulation rate of isolates and infection status, i.e., single, relapse, or reinfection, were observed. However, the isolates recovered from relapse cases showed a significantly higher germination rate when incubated in medium lacking the germination stimulant sodium taurocholate. These results indicate that the germination ability of C. difficile may be a potential risk factor for the recurrence of CDAD.
INTRODUCTION
Clostridium difficile is a Gram-positive, obligately anaerobic, spore-forming bacillus which is the causative pathogen of pseudomembranous colitis (PMC) and is also associated with a large proportion of inpatient cases of antibiotic-associated diarrhea (AAD) (5, 22, 35). The main virulence factors of C. difficile are the two large clostridial glucosylating toxins, toxin A (TcdA) and toxin B (TcdB), which have enterotoxic and cytotoxic activity, respectively. These genes are located within a pathogenicity locus (PaLoc) along with other toxin production-related genes. Most pathogenic strains isolated in cases of C. difficile-associated disease (CDAD) produce both toxins (A+, B+ strains). Regarding other toxin variant strains, isolates that produce toxin B but not toxin A (A−, B+) have been reported worldwide, but naturally occurring isolates that produce toxin A but not toxin B (A+, B−) have not previously been reported. Some isolates produce an additional binary toxin (CDT), but its role in CDAD is not well understood (10, 26, 27, 35). Recently, outbreaks due to an emerging hypervirulent strain of C. difficile, strain BI/NAP1/027, which produces a binary toxin, have been reported in North America and Europe (28, 35).
Oral antibiotics, such as vancomycin or metronidazole, are commonly used and effective in the treatment of CDAD. However, recurrent cases of CDAD (rCDAD) are common and result in a prolonged duration of hospitalization and increasing treatment costs (36, 40). Recurrence rates are generally 10 to 35%, but patients with known recurrent CDAD may exhibit rates higher than this (4, 9, 29).
Risk factors for recurrent CDAD are continuous use of antibiotics, older age, hypoalbuminemia, diabetes mellitus, use of antacids, and stool colonization with vancomycin-resistant enterococci (VRE) (8, 12, 24, 37). Abnormal flora and/or host immune response is also a possible reason for recurrence (40).
Recurrences are caused by the persistence of the same strain of C. difficile that caused the first episode and/or the reinfection with a new strain from the environment. It has been reported that the relapse rates due to the same strain were 25 to 87.5% (2, 4, 9, 32, 41, 45). However, it is important to note that these may include an external reinfection by the original strain.
Various studies have reported the molecular and microbiological characterization of isolates from CDAD and/or rCDAD patients. In most of these studies, the DNA and toxin type were investigated, and in some, the antimicrobial susceptibility and/or cytotoxicity of C. difficile clinical isolates was also tested as a phenotypic characteristic (9, 23, 30, 32, 33, 42). However, reports for other phenotypic characteristics such as sporulation rate are very limited.
Sporulation and germination are two of the most important factors in the pathogenicity of C. difficile and the recurrence of CDAD. It is presumed that the toxin production of C. difficile is related to sporulation and germination (3, 16) and that sporulation allows persistence of C. difficile in the intestinal tract and the environment (4, 32, 35). Spores are resistant to antibiotics, and it is reasonable to assume that sporulation contributes to the spread and survival of C. difficile. A rapid germination rate may allow rapid growth of C. difficile when antibiotic levels are greatly reduced or absent.
In this study, we performed DNA typing of clinical isolates of C. difficile and examined cytotoxicity, antibiotic susceptibility, and sporulation/germination rates to investigate the correlation between microbiological characteristics and the recurrence of CDAD.
MATERIALS AND METHODS
C. difficile strains.
Seventy-three clinical isolates of C. difficile were used in this study to compare genotypic and/or phenotypic characteristics between strains isolated from single infection cases and those isolated from recurrent cases. A single detection of C. difficile from one patient during the observation period (more than 1.5 years) was regarded to be a single infection case, and multiple detections from one patient with an interval of 2 or more weeks was regarded to be a recurrent case. Twenty strains from 20 single infection cases and 53 strains from 20 recurrent cases were isolated from inpatients in the following two facilities: Tokyo Metropolitan Geriatric Hospital, Tokyo, Japan (TMG strains, 2002 to 2005, 3 strains from 3 single infection cases and 7 strains from 3 recurrent cases), and Kyorin University Hospital, Tokyo, Japan (KY strains, 2004 and 2005, 17 strains from 17 single infection cases and 46 strains from 17 recurrent cases).
Identification.
C. difficile isolates were identified by PCR assay using primer set B (CCGTCAATTCMTTTRAGTTT, where M is A or C and R is A or G) and PG-48 (CTCTTGAAACTGGGAGACTTGA), derived from the C. difficile 16S rRNA gene according to the procedure previously described, with slight modifications (13, 20). Briefly, a single colony of C. difficile grown on Gifu anaerobic medium (GAM) agar (Nissui Medical Co., Tokyo, Japan) was suspended in 100 μl of TE (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). The composition of GAM agar is as follows: peptone, 1%; soybean peptone, 0.3%; proteose peptone, 1%; digested blood powder, 1.35%; yeast extract, 0.5%; meat extract, 0.22%; liver extract powder, 0.12%; glucose, 0.3%; potassium dihydrogen phosphate, 0.25%; sodium chloride, 0.3%; soluble starch, 0.5%; l-cysteine monohydrochloride, 0.03%; sodium thioglycolate, 0.03%; and agar, 1.5%. The pH was 7.1, and the agar was sterilized at 115°C for 15 min. The suspension was boiled for 10 min and centrifuged at 10,000 × g for 5 min. The resultant supernatant was used as template DNA.
PCR ribotyping.
PCR ribotyping was performed by the method previously described, with slight modification, and primers CTGGGGTGAAGTCGTAACAAGG (positions 1445 to 1466 of the 16S rRNA gene) and GCGCCCTTTGTAGCTTGACC (positions 20 to 1 of the 23S rRNA gene) (18, 39). Briefly, the volume of the PCR mixture was downscaled to 50 μl, and the amplified PCR products were concentrated to a final volume of approximately 10 μl, followed by heating at 75°C for 90 to 120 min, before electrophoresis in 3% Metaphor agarose (Lonza Rockland Inc., Basel, Switzerland) at a constant voltage of 120 V for 4 h. Isolates with patterns differing by one or more bands were assigned to different PCR ribotypes, and the difference in faint bands was ignored.
PFGE.
Pulsed-field gel electrophoresis (PFGE) analysis was performed by the method previously described with slight modifications (6, 18, 19). One milliliter of C. difficile overnight culture was inoculated into 4 ml of tryptone-yeast extract-glucose medium and incubated at 37°C for 5 h in an anaerobic chamber (10% CO2, 10% H2, and N2 to balance), and an arbitrary volume of the culture was centrifuged at 5,000 × g for 5 min to obtain an appropriate size of pellet. The pellet was washed with 1 ml of TES buffer (50 mM Tris [pH 8.0], 5 mM EDTA, 50 mM NaCl) twice, embedded in an agarose plug, and incubated at 37°C for 1 h with lysozyme-lysostaphin, followed by incubation with proteinase K at 50°C for 18 h using a GenePath group 1 reagent kit (Bio-Rad Laboratories, Inc., CA) according to the manufacturer's instructions. DNA in the plug was digested with SmaI for 18 h at 25°C.
Electrophoresis was performed with a contour-clamped homogeneous electric field (CHEF) Mapper system (Bio-Rad Laboratories, Inc., CA) and a GenePath gel kit (Bio-Rad Laboratories, Inc., CA) with program 16 (6 V/cm, 120° angle, 5.3- to 49.9-s switch time, nonlinear ramp, 14°C, 19.7-h run time). If a smear band was obtained, the sample was again subjected to electrophoresis by adding it with thiourea to a 1.0% agarose gel and running buffer at a final concentration of 200 μM to prevent DNA degradation during electrophoresis (25).
DNA fragments were stained with ethidium bromide and photographed using a GelDoc system (Bio-Rad Laboratories, Inc., CA). Major PFGE types were defined by more than three fragment differences, and these major types were subtyped by three or less than three fragment differences (43). Dendrogram analysis was carried out using Fingerprinting II software (Bio-Rad Laboratories, Inc., CA).
Using the results of PFGE analysis, recurrent cases were divided into relapse cases and reinfection cases. If the strain isolated at the time of recurrence had a PFGE type identical to that of the initial strain, it was deigned to be a relapse case, but if the type was different, it was regarded to be a reinfection.
Toxin detection.
The toxin type of C. difficile strains was analyzed by PCR as previously described (18, 20, 21). The template DNA was prepared by the boiling method described above. Two primer sets, NK3 (GGAAGAAAAGAACTTCTGGCTCACTCAGGT) and NK2 (CCCAATAGAAGATTCAATATTAAGCTT) and NK11 (TGATGCTAATAATGAATCTAAAATGGTAAC) and NK9 (CCACCAGCTGCAGCCATA), were used for the detection of the toxin A gene, and primer set NK104 (GTGTAGCAATGAAAGTCCAAGTTTACGC) and NK105 (CACTTAGCTCTTTGATTGCTGCACCT) was used for the detection of the toxin B gene.
Cytotoxicity assay.
Cytotoxicity was determined by the method described by Kamiya and Borriello (15). Briefly, African green monkey kidney (Vero) cells were grown in Eagle's minimum essential medium (MEM; Sigma-Aldrich, Inc., MO) supplemented with 8% fetal calf serum (Sigma-Aldrich, Inc., MO) in 96-well microplates, and the cell culture medium was removed and replaced with 100 μl of fresh maintenance medium (Eagle's MEM containing 0.5% calf serum) before assay. The culture supernatant of C. difficile in brain heart infusion (BHI) broth (incubated at 37°C for 24 h) was subjected to 2-fold serial dilution with maintenance medium. The cell culture medium was then removed and replaced with 100 μl of each diluted sample. The cytotoxicity of the sample was determined to be the highest dilution resulting in 100% cell rounding after incubation for 24 h.
Antimicrobial susceptibility testing.
MICs of vancomycin, metronidazole, clindamycin, ceftriaxone, erythromycin, and ciprofloxacin were measured by Etest (AB Biodisk, Solna, Sweden). A cotton swab dipped into a 1.0 McFarland standard-matched suspension of the test isolate was used to inoculate a prereduced GAM agar plate. Etest strip and MIC value measurements were performed according to the manufacturer's instructions. Antibiotic resistance was defined as follows, in accordance with the interpretative criteria described in the manufacturer's package insert and previous reports (1, 48): vancomycin, ≥32 mg/liter; metronidazole, ≥32 mg/liter; clindamycin, ≥ 8 mg/liter; ceftriaxone, ≥ 64 mg/liter; erythromycin, ≥8 mg/liter; and ciprofloxacin, ≥4 mg/liter.
Spore formation rate.
The spore formation rate of C. difficile was determined by microscopic observation. A few colonies of C. difficile grown on a GAM agar plate at 37°C for 5 days were suspended in an appropriate volume of sterile saline, and the numbers of spores and vegetative cells were counted under a phase-contrast microscope using a bacterium-counting chamber. The spore formation rate (in percent) was calculated by the following formula: (number of spores per ml/number of total cells [spores and vegetative cells] per ml) × 100.
Germination rate.
The germination rate of C. difficile was evaluated by the methods described previously (17, 31). To count the number of spores, a suspension of C. difficile cells was heated at 70°C for 10 min to kill all the vegetative cells. The heat-treated suspension was then subjected to 10-fold serial dilution with sterile saline and inoculated onto GAM agar or BHI agar (BD, NJ) with or without supplementation of sodium taurocholate (Wako Pure Chemical Industries, Ltd., Osaka, Japan). After incubation of the plates at 37°C for 48 h in an anaerobic chamber, the colonies were counted and the germination rate (in percent) was calculated by the following formula: (number of CFU per ml/microscopic counts of spores per ml) × 100.
Statistics.
For the statistical comparison of the spore formation rate and germination rate, Tukey-Kramer's test and Steel-Dwass's test were used for parametric and nonparametric data, respectively. The normality of a distribution and homogeneity of variances were evaluated by chi-square goodness-of-fit test and Bartlett's test.
RESULTS
Genotyping.
We carried out PCR ribotyping and PFGE analysis to determine the infection status of recurrent cases and to investigate the correlation between genotype, phenotype, and infection status.
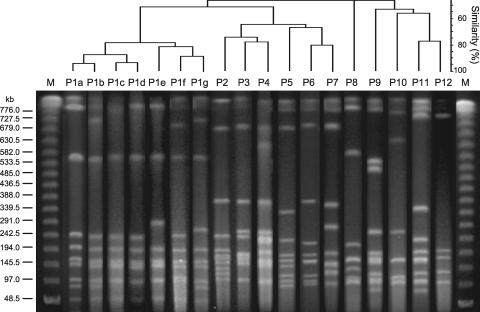
All 73 strains were typeable and resolved into 11 ribotypes (Fig. 1 and Table 1) and 12 major PFGE types (Fig. 2 and Table 1). Major ribotypes of C. difficile isolates were R2 (31 strains, 42.5%), R1 (14 strains, 19.2%), and R4 (13 strains, 17.8%). The DNA degradation during PFGE assay observed in some isolates completely resolved by addition of thiourea at a final concentration of 200 μM. Forty-nine strains were assigned to PFGE major type P1 (67.1%), and 11 strains were assigned to type P3 (15.1%), and these 2 types accounted for more than 80% of the total (Table 1). Type P1 strains were recovered from 28 out of 40 patients (70.0%), and type P3 strains were recovered from 8 out of 40 (20.0%).
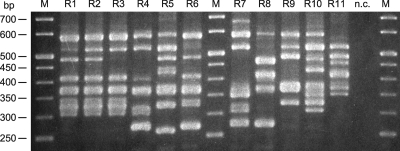
Fig 1.
PCR ribotype patterns of C. difficile isolates. Lanes R1 to R11, representative pattern of each ribotype; lanes M and n.c., 50-bp size markers and negative control, respectively.
Table 1.
Typing of C. difficile isolates
| Major PFGE type and subtype | PCR ribotype | Toxin-producing type | No. of C. difficile isolates (no. of patients) |
|
|---|---|---|---|---|
| TMGa | KYb | |||
| P1 | ||||
| a | R1 | A+, B+ | 5 (3) | |
| b | R2 | A+, B+ | 1 (1) | 20 (8) |
| R3 | A+, B+ | 3 (1) | ||
| c | R1 | A+, B+ | 9e,f (7) | |
| R2 | A+, B+ | 1c (1) | 2 (2) | |
| d | R2 | A+, B+ | 2 (1) | |
| e | R2 | A+, B+ | 2 (1) | |
| f | R2 | A+, B+ | 3d (2) | |
| g | R3 | A+, B+ | 1 (1) | |
| Subtotal | 7 (5) | 42 (23) | ||
| P2 | R4 | A+, B+ | 1 (1) | |
| P3 | R4 | A+, B+ | 11d,f (8) | |
| P4 | R4 | A+, B+ | 1c (1) | |
| P5 | R5 | A−, B− | 3 (1) | |
| P6 | R6 | A+, B+ | 1 (1) | |
| P7 | R7 | A+, B+ | 1 (1) | |
| P8 | R8 | A−, B+ | 1 (1) | |
| P9 | R9 | A+, B+ | 1c (1) | |
| P10 | R8 | A−, B+ | 1e (1) | |
| P11 | R10 | A−, B− | 1 (1) | |
| P12 | R11 | A+, B+ | 2 (1) | |
TMG, Tokyo Metropolitan Geriatric Hospital.
KY, Kyorin University Hospital.
Three different TMG strains (no. 6, type P1c/R2; no. 7, type P4/R4; and no. 8, type P9/R9) were isolated from a patient.
Three different KY strains (no. 37, type P3/R4; no. 38, type P1f/R2; and no. 39, type P1f/R2) were isolated from a patient.
Two different KY strains (no. 66, type P10/R8; and no. 69, type P1c/R1) were isolated from a patient.
Two different KY strains (no. 123, type P1c/R1; and no. 12, type P3/R4) were isolated from a patient.
Fig 2.
Dendrogram and PFGE patterns of C. difficile isolates. Lanes P1a to P12, representative pattern of each PFGE type; lanes M, bacteriophage lambda ladder size marker. The dendrogram was constructed with Fingerprinting II software (Bio-Rad Laboratories, Inc.).
Among the 20 recurrent cases, 16 patients were regarded to be relapse cases caused by the same strain (80.0%), and the remaining 4 patients were regarded to be reinfection cases caused by different strains (20.0%) (Table 2). In the relapse cases, the major PFGE type and DNA ribotype were P1 (12 strains, 75.0%; Table 2) and R2 (8 strains, 50.0%; data not shown). There was no significant association between DNA typing group (PFGE type or PCR ribotype) and incidence of recurrence.
Table 2.
Major PFGE type and number of patients with single infection, relapse, and reinfection
| Major PFGE type | No. of patients |
|||
|---|---|---|---|---|
| Single infection | Relapse | Reinfectiona | Total | |
| P1 | 11 | 12 | 4 | 27 |
| P2 | 1 | 0 | 0 | 1 |
| P3 | 4 | 2 | 2 | 8 |
| P4 | 0 | 0 | 1 | 1 |
| P5 | 0 | 1 | 0 | 1 |
| P6 | 1 | 0 | 0 | 1 |
| P7 | 1 | 0 | 0 | 1 |
| P8 | 1 | 0 | 0 | 1 |
| P9 | 0 | 0 | 1 | 1 |
| P10 | 0 | 0 | 1 | 1 |
| P11 | 1 | 0 | 0 | 1 |
| P12 | 0 | 1 | 0 | 1 |
| Total | 20 (20)b | 16 (16) | 9 (4) | 45 (40) |
Cumulative number of patients.
Data in parentheses are the actual number of patients.
Toxin type and cytotoxicity.
Among 73 strains used in the study, 67 strains were toxin A+, B+ (91.8%), 2 were toxin A−, B+ (2.7%), and 4 were toxin A−, B− (5.4%) (Table 3). Toxin A+, B+ strains were mainly categorized to types P1/R2, P3/R4, and P1/R1. In contrast, toxin A−, B+ and A−, B− strains were categorized to type P8/R8 or P10/R8 and P5/R5 or P11/R10, respectively. There was no correlation between toxin type and infection status (Table 3). Cytotoxicity assay showed that 13 strains (17.8%) were highly toxigenic (toxin titer, >28), and 5 strains (6.8%) had a low toxicity with a titer of 22 (data not shown). There was no significant relationship between the cytotoxicity of C. difficile isolates and DNA typing group (PFGE type and PCR ribotype), except that toxin A−, B− strains (type P5/R5 and P11/R10) showed no cytotoxic activities. Similarly, no correlation between the cytotoxicity and the incidence of recurrence was seen (data not shown).
Table 3.
Toxin type of C. difficile clinical isolates compared to infection status
| Toxin type | No. (%) of C. difficile isolates |
|||
|---|---|---|---|---|
| Single infection | Relapse | Reinfection | Total | |
| A+, B+ | 18 (90.0) | 40 (93.0) | 9 (90.0) | 67 (91.8) |
| A−, B+ | 1 (5.0) | 0 (0.0) | 1 (10.0) | 2 (2.7) |
| A−, B− | 1 (5.0) | 3 (7.0) | 0 (0.0) | 4 (5.5) |
| Total | 20 (100) | 43 (100) | 10 (100) | 73 (100) |
Antibiotic susceptibility.
MIC50s, MIC90s, and the range of MICs of vancomycin, metronidazole, clindamycin, ceftriaxone, erythromycin, and ciprofloxacin against the 73 C. difficile isolates are all shown in Table 4. All isolates were susceptible to vancomycin and metronidazole. Resistance against clindamycin, ceftriaxone, erythromycin, and ciprofloxacin were found in 87.7%, 93.2%, 87.7%, and 100% of the isolates tested, respectively.
Table 4.
Antibiotic susceptibility of C. difficile clinical isolates
| Antibiotic | MIC (mg/líter) |
No. (%) of resistant isolates (n = 73) | |||
|---|---|---|---|---|---|
| 50% | 90% | Range | Breakpoint | ||
| Vancomycin | 2 | 4 | 1–8 | ≥32 | 0 (0) |
| Metronidazole | 0.19 | 0.25 | 0.094-0.25 | ≥32 | 0 (0) |
| Clindamycin | >256 | >256 | 1–>256 | ≥8 | 64 (87.7) |
| Ceftriaxone | >256 | >256 | 32–>256 | ≥64 | 68 (93.2) |
| Erythromycin | >256 | >256 | 0.38–>256 | ≥8 | 64 (87.7) |
| Ciprofloxacin | >32 | >32 | 6–>32 | ≥4 | 73 (100) |
C. difficile isolates with PFGE types P1, P3, P4, P8, and P10 showed high-level resistance against clindamycin (MIC50s > 256 mg/liter), ceftriaxone (MIC50s ≥ 128 mg/liter), erythromycin (MIC50s > 256 mg/liter), and ciprofloxacin (MIC50s > 32 mg/liter) (Table 5), as did isolates with ribotypes R1, R2, R3, R4, and R8 (data not shown). On the other hand, the susceptibility of C. difficile isolates to vancomycin and/or metronidazole was not related to either the PFGE type (Table 5) or the PCR ribotype (data not shown).
Table 5.
Comparison of antibiotic susceptibility with PFGE subtype
| PFGE type | MIC50 (mg/liter) |
|||||
|---|---|---|---|---|---|---|
| Vancomycin | Metronidazole | Clindamycin | Ceftriaxone | Erythromycin | Ciprofloxacin | |
| P1a | 1.5 | 0.19 | >256 | >256 | >256 | >32 |
| P1b | 3 | 0.19 | >256 | >256 | >256 | >32 |
| P1c | 3 | 0.125 | >256 | >256 | >256 | >32 |
| P1d | 2 | 0.125 | >256 | >256 | >256 | >32 |
| P1e | 2 | 0.19 | >256 | >256 | >256 | >32 |
| P1f | 1.5 | 0.19 | >256 | >256 | >256 | >32 |
| P1g | 2 | 0.19 | >256 | >256 | >256 | >32 |
| P2 | 1.5 | 0.19 | 2 | 48 | 0.5 | 6 |
| P3 | 1.5 | 0.19 | >256 | >256 | >256 | >32 |
| P4 | 2 | 0.19 | >256 | 128 | >256 | >32 |
| P5 | 2 | 0.25 | 1.5 | 96 | 0.5 | 12 |
| P6 | 3 | 0.125 | 4 | 64 | 0.5 | 8 |
| P7 | 1.5 | 0.19 | 2 | 32 | 0.38 | 6 |
| P8 | 1.5 | 0.25 | >256 | 192 | >256 | >32 |
| P9 | 1.5 | 0.19 | 3 | 32 | 0.38 | 8 |
| P10 | 1.5 | 0.25 | >256 | >256 | >256 | >32 |
| P11 | 3 | 0.19 | >256 | 64 | >256 | 8 |
| P12 | 1.5 | 0.125 | 1 | 32 | 0.38 | 6 |
There was no correlation observed between the antibiotic susceptibility of the C. difficile isolates to the above-mentioned antimicrobial drugs and recurrence (data not shown).
Germination and sporulation.
From preliminary experiments testing a variety of media and agar types, we chose the GAM agar plate as the testing medium for spore formation rate, as it yielded the highest number of spores (data not shown). To determine the germination rate, GAM agar plates with or without the C. difficile germination stimulant sodium taurocholate were used.
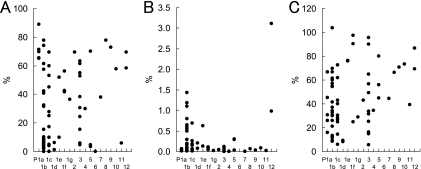
The mean spore formation rate of the 73 C. difficile isolates after 5 days was 32% (range, <1.0 to 89%), and sporulation ability was not related to PFGE type (Fig. 3A). Similarly, there was no significant correlation between sporulation ability and PCR ribotype (data not shown) or recurrence (Fig. 4A).
Fig 3.
Spore formation rate (A), germination rate on GAM agar without sodium taurocholate (B), and germination rate on GAM agar supplemented with 0.1% sodium taurocholate (C) of C. difficile isolates compared to PFGE subtype.
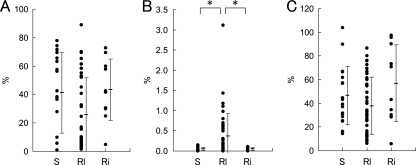
Fig 4.
Spore formation rate (A), germination rate on GAM agar without sodium taurocholate (B), and germination rate on GAM agar supplemented with 0.1% sodium taurocholate (C) of C. difficile isolates compared to infection status. S (n = 20), Rl (n = 43), and Ri (n = 10) indicate isolates from single, relapse, and reinfection cases, respectively. Bars represent mean ± SDs. Asterisks denote statistical significance (P < 0.05).
The mean germination rate of the 73 isolates on GAM agar was 0.24% (range, 0.00094 to 3.1%), and the germination rate was elevated about 10- to 50,000-fold when GAM agar was supplemented with sodium taurocholate (mean, 42.7%; range, 5.7 to 103.7%) (Fig. 3B and C). The germination rate of the isolates either in the presence or in the absence of sodium taurocholate was not related to PFGE type (Fig. 3B and C) or PCR ribotype (data not shown). However, the germination rate of strains isolated from relapse cases was significantly higher than the germination rates of strains isolated from single and reinfection cases when sodium taurocholate was not added (Fig. 4B), though this was not observed with sodium taurocholate (Fig. 4C). This observation on germination rate was reproducible, and a similar result was obtained when GAM agar was replaced by BHI agar (see Fig. S1 in the supplemental material).
DISCUSSION
In this study, 73 clinical isolates of C. difficile were analyzed by PFGE and PCR ribotyping to determine infection status and to relate this to microbiological characteristics. PFGE and PCR ribotyping are popular methods of genetic characterization worldwide and have been found to correlate well (7, 14, 35). We categorized the 73 strains into 12 major PFGE types (type P1 was further categorized into 7 subtypes) and 11 PCR ribotypes. A good correlation between PFGE and PCR ribotyping was observed, and as previously reported, PFGE was able to discriminate strains more accurately than PCR ribotyping (7).
PFGE and PCR ribotyping showed that 16 out of 20 recurrent CDAD patients (80.0%) were relapse cases caused by the same strains. This relapse rate was high compared with the rates of 25 to 87.5% previously reported (with the remaining cases suffering reinfection or a combination of relapse plus reinfection) (2, 4, 9, 32, 41, 45). Although some relapse cases may include reinfection from the original strain in the patient's environment, some may occur through persistence of the original C. difficile isolate in the intestinal tract despite antibiotic treatment (2, 9, 41). In such cases, recurrence may occur more frequently with strains that have phenotypic characteristics advantageous for intestinal survival.
Most pathogenic C. difficile strains produce the two large clostridial glucosylating toxins A and B, and these toxins are the main virulence factors in CDAD (26, 35). Previously, it was thought that toxin A was the only toxin essential for the pathogenesis of CDAD, and early experiments in animal models showed that sole administration of toxin A caused symptoms of CDAD but administration of toxin B without toxin A had no effect (10, 27, 35). However, recent studies report outbreaks caused by A−, B+ strains and therefore conclude that toxin B also plays an important role in the virulence of C. difficile (10, 26, 27). Toxin A−, B+ strains have been isolated in many countries with a prevalence of 0 to 97.9% and recently seem to be on the increase (10, 23). In this study, only 2 out of 73 isolates (2.7%) were A−, B+ strains, and their DNA genotypes were P8/R8 and P10/R8. Although it is not clear why the prevalence of A−, B+ strains was relatively low, it may be associated with the fact that C. difficile strains isolated from recurrent CDAD cases made up the majority of the isolates used in this study.
It is known that certain DNA typing groups are hypervirulent, such as the C. difficile BI/NAP1/027 strain (28, 35). However, no correlation between the cytotoxicity and PFGE or PCR ribotype was observed in this study. Therefore, virulence determinants may be independent of DNA typing group.
It is important to determine the current resistance status of C. difficile and to find out the correlation between the DNA type and antimicrobial susceptibilities. We found that all of the isolates were susceptible to vancomycin and metronidazole, and high rates of resistance against other antibiotics were also observed, such as clindamycin (87.7%), ceftriaxone (93.2%), erythromycin (87.7%), and ciprofloxacin (100%). The rates of resistance of C. difficile isolates against these antibiotics in Europe and South Korea have been shown to be similar to the rates obtained in this study (23, 30, 33), and high susceptibility of C. difficile isolates to vancomycin and metronidazole has been reported by many researchers (1, 23, 30, 33). Isolates with PFGE types P1, P3, P4, P8, and P10 and PCR ribotypes R1, R2, R3, R4, and R8 showed high-level resistance against clindamycin, ceftriaxone, erythromycin, and ciprofloxacin. Although our DNA typing data cannot be compared with those from other reports, these results indicate that antibiotic selection pressure may cause the selection of certain dominant DNA typing groups, as reported by Taori et al. (42).
From the results of epidemiological studies on the antimicrobial susceptibilities of C. difficile isolates, it is well-known that oral antibiotics, such as the commonly used vancomycin and metronidazole, are the most efficacious for the treatment of CDAD. However, approximately 20% of patients suffer a recurrence of CDAD following cessation of antibiotic treatment. Recurrences usually occur within 5 to 8 days after withdrawal of antibiotics, but their onset can be delayed for several weeks. Recurrence of CDAD is a serious and difficult clinical problem in terms of prolongation of the duration of hospitalization and increasing treatment costs (4, 36, 40).
PFGE type P1 and PCR ribotype R2 strains were most frequently found in single and relapse cases. However, there was no correlation between DNA typing groups and infection status, indicating that the incidence of recurrence does not depend on the DNA type of isolates in cases treated with vancomycin or metronidazole. No correlation between the susceptibility of isolates against these antibiotics and DNA typing group was observed.
We postulated that the sporulation and germination abilities of C. difficile are likely to be related to the incidence of relapse after treatment with antibiotics due to increased persistence in the intestinal tract and rapid growth prior to restoration of the normal intestinal flora. Indeed, it has been reported that epidemic strains of C. difficile produced more spores than the nonprevalent strains and the sporulation rate was increased by exposure to a cleaning agent or germicide (11, 44).
We found no correlation between either the sporulation or germination rate with PFGE type, PCR ribotype, or infection status. However, the isolates recovered from relapse cases showed a significantly higher germination rate when incubated on GAM agar plates without sodium taurocholate, though this difference was not observed when taurocholate was present. Sodium taurocholate is known to stimulate germination of C. difficile spores (38, 46, 47), and the presence of a putative receptor for taurocholate has recently been reported (34). Compared to the isolates recovered from single or reinfection cases, relapse strains may be more sensitive to cogerminants other than taurocholate in the test medium or may have an atypical spore coat structure which facilitates germination.
According to Ramirez et al. (34), it is thought that the concentration of active taurocholate in the lower intestine is negligible due to deconjugation into taurin and cholate by the intestinal flora and that depletion of flora by strong antibiotic therapy may increase the concentration of active taurocholate. Therefore, a high germination ability in the absence of cogerminants such as taurocholate may contribute to the increased reinfection rates observed in these CDAD cases. Further work may be required to investigate the effects of other cogerminants and intestinal concentrations of these chemicals during the first infective episode and recurrence, but our results suggest that the germination ability of C. difficile strains may be a potential risk factor for the recurrence of CDAD.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. W. Njoroge and C. C. Bii, Kenya Medical Research Institute, Nairobi, Kenya, for technical assistance.
Footnotes
Published ahead of print 28 December 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Ackermann G, Degner A, Cohen H, Silva J, Jr, Rodloff AC. 2003. Prevalence and association of macrolide-lincosamide-streptogramin B (MLSB) resistance with resistance to moxifloxacin in Clostridium difficile. J. Antimicrob. Chemother. 51:599–603 [DOI] [PubMed] [Google Scholar]
- 2. Alonso R, et al. 2001. Molecular analysis of relapse vs re-infection in HIV-positive patients suffering from recurrent Clostridium difficile associated diarrhoea. J. Hosp. Infect. 48:86–92 [DOI] [PubMed] [Google Scholar]
- 3. Baines SD, O'Connor R, Saxton K, Freeman J, Wilcox MH. 2009. Activity of vancomycin against epidemic Clostridium difficile strains in a human gut model. J. Antimicrob. Chemother. 63:520–525 [DOI] [PubMed] [Google Scholar]
- 4. Barbut F, et al. 2000. Epidemiology of recurrences or reinfections of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2386–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartlett JG, Chang TW, Gurwith M, Gorbach SL, Onderdonk AB. 1978. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N. Engl. J. Med. 298:531–534 [DOI] [PubMed] [Google Scholar]
- 6. Bidet P, et al. 2000. Comparison of PCR-ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for typing Clostridium difficile. J. Clin. Microbiol. 38:2484–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brazier JS. 2001. Typing of Clostridium difficile. Clin. Microbiol. Infect. 7:428–431 [DOI] [PubMed] [Google Scholar]
- 8. Choi HK, Kim KH, Lee SH, Lee SJ. 2011. Risk factors for recurrence of Clostridium difficile infection: effect of vancomycin-resistant enterococci colonization. J. Korean Med. Sci. 26:859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Do AN, et al. 1998. Risk factors for early recurrent Clostridium difficile-associated diarrhea. Clin. Infect. Dis. 26:954–959 [DOI] [PubMed] [Google Scholar]
- 10. Drudy D, Fanning S, Kyne L. 2007. Toxin A-negative, toxin B-positive Clostridium difficile. Int. J. Infect. Dis. 11:5–10 [DOI] [PubMed] [Google Scholar]
- 11. Fawley WN, et al. 2007. Efficacy of hospital cleaning agents and germicides against epidemic Clostridium difficile strains. Infect. Control Hosp. Epidemiol. 28:920–925 [DOI] [PubMed] [Google Scholar]
- 12. Garey KW, Sethi S, Yadav Y, DuPont HL. 2008. Meta-analysis to assess risk factors for recurrent Clostridium difficile infection. J. Hosp. Infect. 70:298–304 [DOI] [PubMed] [Google Scholar]
- 13. Gumerlock PH, Tang YJ, Meyers FJ, Silva J., Jr 1991. Use of the polymerase chain reaction for the specific and direct detection of Clostridium difficile in human feces. Rev. Infect. Dis. 13:1053–1060 [DOI] [PubMed] [Google Scholar]
- 14. Janezic S, Rupnik M. 2010. Molecular typing methods for Clostridium difficile: pulsed-field gel electrophoresis and PCR ribotyping. Methods Mol. Biol. 646:55–65 [DOI] [PubMed] [Google Scholar]
- 15. Kamiya S, Borriello SP. 1992. A non-haemagglutinating form of Clostridium difficile toxin A. J. Med. Microbiol. 36:190–197 [DOI] [PubMed] [Google Scholar]
- 16. Kamiya S, Ogura H, Meng XQ, Nakamura S. 1992. Correlation between cytotoxin production and sporulation in Clostridium difficile. J. Med. Microbiol. 37:206–210 [DOI] [PubMed] [Google Scholar]
- 17. Kamiya S, Yamakawa K, Ogura H, Nakamura S. 1987. Effect of various sodium taurocholate preparations on the recovery of Clostridium difficile spores. Microbiol. Immunol. 31:1117–1120 [DOI] [PubMed] [Google Scholar]
- 18. Kato H, et al. 2001. Colonisation and transmission of Clostridium difficile in healthy individuals examined by PCR ribotyping and pulsed-field gel electrophoresis. J. Med. Microbiol. 50:720–727 [DOI] [PubMed] [Google Scholar]
- 19. Kato H, et al. 1994. Application of typing by pulsed-field gel electrophoresis to the study of Clostridium difficile in a neonatal intensive care unit. J. Clin. Microbiol. 32:2067–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kato H, et al. 1998. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J. Clin. Microbiol. 36:2178–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kato N, et al. 1991. Identification of toxigenic Clostridium difficile by the polymerase chain reaction. J. Clin. Microbiol. 29:33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kelly CP, Pothoulakis C, LaMont JT. 1994. Clostridium difficile colitis. N. Engl. J. Med. 330:257–262 [DOI] [PubMed] [Google Scholar]
- 23. Kim H, et al. 2010. Investigation of toxin gene diversity, molecular epidemiology, and antimicrobial resistance of Clostridium difficile isolated from 12 hospitals in South Korea. Korean J. Lab. Med. 30:491–497 [DOI] [PubMed] [Google Scholar]
- 24. Kim JW, et al. 2010. Proton pump inhibitors as a risk factor for recurrence of Clostridium-difficile-associated diarrhea. World J. Gastroenterol. 16:3573–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klaassen CH, van Haren HA, Horrevorts AM. 2002. Molecular fingerprinting of Clostridium difficile isolates: pulsed-field gel electrophoresis versus amplified fragment length polymorphism. J. Clin. Microbiol. 40:101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuehne SA, et al. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713 [DOI] [PubMed] [Google Scholar]
- 27. Lyras D, et al. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McDonald LC, et al. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433–2441 [DOI] [PubMed] [Google Scholar]
- 29. McFarland LV, Elmer GW, Surawicz CM. 2002. Breaking the cycle: treatment strategies for 163 cases of recurrent Clostridium difficile disease. Am. J. Gastroenterol. 97:1769–1775 [DOI] [PubMed] [Google Scholar]
- 30. Mutlu E, Wroe AJ, Sanchez-Hurtado K, Brazier JS, Poxton IR. 2007. Molecular characterization and antimicrobial susceptibility patterns of Clostridium difficile strains isolated from hospitals in south-east Scotland. J. Med. Microbiol. 56:921–929 [DOI] [PubMed] [Google Scholar]
- 31. Nakamura S, Yamakawa K, Izumi J, Nakashio S, Nishida S. 1985. Germinability and heat resistance of spores of Clostridium difficile strains. Microbiol. Immunol. 29:113–118 [DOI] [PubMed] [Google Scholar]
- 32. O'Neill GL, Beaman MH, Riley TV. 1991. Relapse versus reinfection with Clostridium difficile. Epidemiol. Infect. 107:627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pituch H, et al. 2011. Characterization and antimicrobial susceptibility of Clostridium difficile strains isolated from adult patients with diarrhoea hospitalized in two university hospitals in Poland, 2004-2006. J. Med. Microbiol. 60:1200–1205 [DOI] [PubMed] [Google Scholar]
- 34. Ramirez N, Liggins M, Abel-Santos E. 2010. Kinetic evidence for the presence of putative germination receptors in Clostridium difficile spores. J. Bacteriol. 192:4215–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 36. Shah D, et al. 2010. Clostridium difficile infection: update on emerging antibiotic treatment options and antibiotic resistance. Expert Rev. Anti Infect. Ther. 8:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shakov R, Salazar RS, Kagunye SK, Baddoura WJ, DeBari VA. 2011. Diabetes mellitus as a risk factor for recurrence of Clostridium difficile infection in the acute care hospital setting. Am. J. Infect. Control 39:194–198 [DOI] [PubMed] [Google Scholar]
- 38. Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190:2505–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stubbs SL, Brazier JS, O'Neill GL, Duerden BI. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Surawicz CM. 2004. Treatment of recurrent Clostridium difficile-associated disease. Nat. Clin. Pract. Gastroenterol. Hepatol. 1:32–38 [DOI] [PubMed] [Google Scholar]
- 41. Tang-Feldman Y, Mayo S, Silva J, Jr, Cohen SH. 2003. Molecular analysis of Clostridium difficile strains isolated from 18 cases of recurrent Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 41:3413–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taori SK, Hall V, Poxton IR. 2010. Changes in antibiotic susceptibility and ribotypes in Clostridium difficile isolates from southern Scotland, 1979-2004. J. Med. Microbiol. 59:338–344 [DOI] [PubMed] [Google Scholar]
- 43. Tenover FC, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilcox MH, Fawley WN. 2000. Hospital disinfectants and spore formation by Clostridium difficile. Lancet 356:1324. [DOI] [PubMed] [Google Scholar]
- 45. Wilcox MH, Fawley WN, Settle CD, Davidson A. 1998. Recurrence of symptoms in Clostridium difficile infection—relapse or reinfection? J. Hosp. Infect. 38:93–100 [DOI] [PubMed] [Google Scholar]
- 46. Wilson KH. 1983. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J. Clin. Microbiol. 18:1017–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilson KH, Kennedy MJ, Fekety FR. 1982. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 15:443–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wultańska D, et al. 2010. Clostridium difficile infection in Polish pediatric outpatients with inflammatory bowel disease. Eur. J. Clin. Microbiol. Infect. Dis. 29:1265–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.