Abstract
Infection by a novel canine astrovirus was associated with gastroenteritis in two dogs. The virus displayed 70.3 to 73.9% amino acid identity to other canine astroviruses in the full-length capsid. Specific antibodies were detected in the convalescent-phase sera of the dogs, indicating seroconversion. Also, the virus appeared weakly related antigenically to the prototype canine astrovirus isolate ITA/2008/Bari.
TEXT
Astroviruses (AstVs) (family Astroviridae) are small, rounded viruses (SRVs) with a peculiar starlike shape when observed by electron microscopy (EM), although this conformation is not always readily recognizable. The AstV genome is composed of a plus-sense single-stranded RNA of 6.4 to 7.3 kb in size, containing 3 open reading frames (ORFs) and with a 3′ poly(A) tail (13). Two ORFs, located at the 5′ end of the genome (ORF1a and ORF1b), encode nonstructural proteins, while ORF2, located at the 3′ end, encodes the capsid protein (13). AstV infection is associated with gastroenteritis in most animal species, and human AstVs are regarded as the second or third most common cause of viral diarrhea in children (13). AstVs have also been associated with extraintestinal diseases, such as nephritis in chicken (10), hepatitis in ducks (16), and shaking syndrome in minks (1).
AstV-like particles have been detected only occasionally in dogs by EM, either alone or in coinfection with other enteric viruses (11, 19, 21). More recently, AstVs have been identified in dogs with enteric signs and characterized molecularly, suggesting that the detected viruses may represent a distinct AstV species (12, 17, 22). Also, a canine AstV, strain ITA/08/Bari, was successfully adapted to replicate in vitro, and AstV-specific antibodies were detected in canine sera (12).
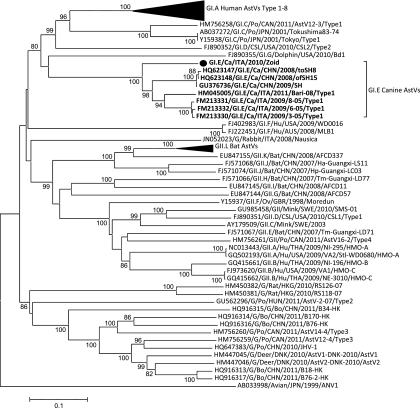
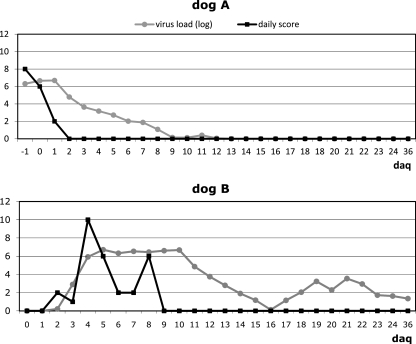
Here, we report an outbreak of AstV infection affecting two household dogs housed together, in Bari, Italy, in 2010. One of the dogs (dog A), a 2-year-old mixed-breed animal, developed anorexia and diarrhea, while the other animal (dog B), a 60-day-old mixed-breed pup, was overtly healthy. The owner of the animals was requested to separate the animals and adopt preventive measures to avoid exposure of the pup (dog B) to potential infectious agents. Stools and sera of the two dogs were collected and subjected to virological and serological investigations. The samples tested negative for canine parvovirus (CPV-2), enteric coronaviruses (CCoVs), rotavirus, norovirus, adenovirus (CAV), and canine distemper (CDV) (4, 5, 6, 7, 9, 18). Dog A had been subjected to regular vaccination for major canine pathogens (CPV-2, CDV, CAV, Leptospira), while the pup (dog B) had started the immunization schedule with a modified live CPV-2 vaccine at 45 days of age. The feces of dog A tested positive for astrovirus by using primer pairs panAstVFor1 (GARTTYGATTGGRCKCGKTAYGA), panAstVFor2 (GARTTYGATTGGRCKAGGTAYGA), and panAstVRev (GGYTTKACCCACATNCCRAA) (2). These primers target conserved nucleotide (nt) stretches in the RdRp region (ORF1b) and amplify a 420-bp fragment (2). The stools of both the animals were collected daily, the animals were examined clinically, and the clinical signs were scored as described elsewhere (3), but not including the white blood cells count. Bacterial and parasite pathogens were not identified in the stools of both the animals. On day 2 of quarantining (daq 2), dog A recovered completely from the disease. Despite the prophylaxis measures adopted, on daq 4 dog B developed acute severe gastroenteritis with fluid diarrhea and vomiting. Astrovirus infection in dog B was confirmed by reverse transcription (RT)-PCR and EM observation of the stools. Appropriate therapy (rehydration, antiemetic, antibiotic, and probiotics) was immediately started in the pup, although clinical signs were observed until daq 8. The 3′ end of ORF1b, the full-length ORF2, and the 3′ noncoding region (approximately 3.5 kb) of the virus from dog B (strain ITA/2010/Zoid) was amplified with a 3′ rapid amplification of cDNA ends (RACE) protocol, as previously described (20), and cloned and sequenced (GenBank accession number JN193534). The virus from dog A was also sequenced from the 3′ end of ORF1b to the poly(A) tail and was found to be identical (100% nt identity) to the virus identified from dog B. In the full-length capsid protein (ORF2), the virus displayed 70.3 to 73.9% amino acid (aa) identity to other canine AstV strains, while amino acid identity to other mammalian AstVs ranged form 18.3 to 33.2% (Table 1 and Fig. 1). According to a new taxonomy proposal for mammalian AstVs, based on analysis of the full-length capsid protein, recently submitted to the International Committee for Taxonomy of Viruses (http://talk.ictvonline.org/files/proposals/taxonomy_proposals_vertebrate1/m/vert01/2287.aspx), AstV strains within the same species display a p-distance lower than 0.312. Therefore, all the canine AstV strains, including ITA/2010/Zoid, can be classified as a unique species, GI.E (Table 1). The sequences of the novel AstV were used to optimize a specific real-time quantitative RT-PCR (qRT-PCR) (data not shown), designing novel primers CaAstV-F2 (ATTACCACGATGTTGYTCTGTR) and CaAstV-R2 (CATGATTGGTATGTTGAAAAYYTG) and probe CaAstVPb2 (FAM-ATATGTACTTTTGCCATCAGGAGAG-BHQ1, where FAM is 6-carboxyfluorescein and BHQ1 is Black Hole Quencher 1). The stools of dogs A and B were retested by the qRT-PCR in order to quantify the virus load. In dog A, AstV RNA was detected from daq −1 to 12, with the highest virus loads being detected between daq −1 and 1. Dog B shed AstV RNA from daq 3 to 36, when the quarantine and the study were ended, with the highest virus loads from daq 4 to 10 (Fig. 2). By immunofluorescence using the canine isolate AstV ITA/2008/Bari as the antigen (12), antibodies specific for canine AstV were not detected in the two dogs at daq 0 or at daq 14 and daq 28. In immune electron microscopy (IEM), antibodies aggregating AstV particles were observed in convalescent (daq 28)- but not in acute (daq 0)-phase sera, indicating seroconversion. Also, weak antigenic cross-reactivity was observed between strain ITA/2008/Bari and ITA/2010/Zoid virus particles and sera in IEM. This could be accounted for by the genetic heterogeneity observed between the virus ITA/2010/Zoid and the prototype canine AstV isolate ITA/2008/Bari used as the antigen. Attempts to adapt the virus ITA/2010/Zoid to growth in vitro on primary canine cells and on canine cell lines (MDCK and A72) were not successful, thus hampering a precise evaluation of the antigenic relationships between the canine AstVs.
Table 1.
Comparison in the full-length capsid protein (ORF2) of the strain GI.E/Zoid/ITA/2010 with other canine strains
| Strain | Dog/ITA/2010/Zoida |
|
|---|---|---|
| % identity | p-distance | |
| FM213332/GI.E/Ca/ITA/2009/6-05 | 73.2 | 0.226 |
| FM213331/GI.E/Ca/ITA/2009/8-05 | 73.6 | 0.223 |
| FM213330/GI.E/Ca/ITA/2009/3-05 | 73.9 | 0.22 |
| HM045005/GI.E/Ca/ITA/2008/Bari | 70.3 | 0.257 |
| HQ623147/GI.E/Ca/CHN/2008/toSH8 | 71.4 | 0.247 |
| HQ623148/GI.E/Ca/CHN/2008/ofSH15 | 71.6 | 0.244 |
| GU376736/GI.E/Ca/CHN/2009/SH | 72.2 | 0.238 |
| Noncanine AstVs | 33.2–18.3 | 0.775–0.627 |
The amino acid identity was calculated using a selection of sequences of 68 mammalian AstVs, without removing the gaps and with no distance correction, using the software Bioedit version 7.0.9.0 (8). The p-distance was calculated using the same sequence alignment, with MEGA package version 4.0 (15).
Fig 1.
Phylogenetic tree displaying the relationships among mammalian AstVs. The tree was calculated on the full-length amino acid capsid alignment without removing the gaps, using the neighbor-joining method with the p-distance correction, with 1,000-replicate bootstrapping using the MEGA package version 4.0 (15). Bootstrap values lower than 75% are not shown. The canine AstV strain ITA/2010/Zoid is indicated with a solid circle. AstV species with assigned designation are indicated with the letter G followed by the roman number and a capital letter. Strains with no assigned designation are indicated with a G. Abbreviations: ca, canine; fe, feline; hu, human; po, porcine; bo, bovine.
Fig 2.
Virus shedding and clinical scores in the two animals (dog A and dog B). Virus load is expressed as the logarithmic (log) value of the RNA copies/g of feces. The clinical scores were calculated by following the outlines of Decaro et al. (3), with minor modifications.
The findings of this study indicate that a unique canine AstV infected two dogs in a short time span. It is not clear whether dog B was already infected at daq 0 or it got infected after quarantining and which was the source of infection for the two animals.
It is also of relevance that we could correlate peaks of AstV shedding with the clinical signs, at least in pup B, which was followed almost entirely during the course of infection. Gastroenteric signs (daq 4 to 8) in the pup were associated with the highest viral loads in the feces (107 to 108 copies of genome equivalents/g of feces). Such a severe clinical picture could be easily attributed to other canine pathogens (CPV-2 or CCoV) by veterinarians. Surveys in Italy and China have revealed differences in AstV distribution between symptomatic (12.0 to 24.5%) and asymptomatic (0 to 9.3%) animals (12, 22). Overall, these findings suggest that AstVs may play a role as enteric pathogens of dogs.
Finally, there is evidence that canine AstVs are genetically and antigenically heterogeneous, with ranges of variation similar to those defined for human GI.A AstV types 1 to 8 (14). Whether some strains can also vary in their pathobiological features is not clear. For instance, some mink AstVs are able to invade and replicate in the central nervous system, causing disease, while others are not (1). Including canine AstVs in the algorithms of gastroenteric diseases of dogs will be pivotal in order to portray a more complete picture of these viruses.
ACKNOWLEDGMENTS
This work was funded by a grant from the Italian Ministry of Health, Ricerca corrente 2009, project IZS VE 21/09 RC, “Definizione di una procedura validata per la selezione di cani per programmi di Interventi Assistiti dagli Animali (IAA).”
Footnotes
Published ahead of print 21 December 2011
REFERENCES
- 1. Blomström AL, Widén F, Hammer AS, Belák S, Berg M. 2010. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J. Clin. Microbiol. 48:4392–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chu DKW, Poon LLM, Guan Y, Peiris JSM. 2008. Novel astroviruses in insectivorous bats. J. Virol. 82:9107–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Decaro N, et al. 2008. Experimental infection of dogs with a novel strain of canine coronavirus causing systemic disease and lymphopenia. Vet. Microbiol. 128:253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Decaro N, et al. 2005. A real-time PCR assay for rapid detection and quantitation of canine parvovirus type 2 in the feces of dogs. Vet. Microbiol. 105:19–28 [DOI] [PubMed] [Google Scholar]
- 5. Decaro N, et al. 2004. Quantitation of canine coronavirus RNA in the faeces of dogs by TaqMan RT-PCR. J. Virol. Methods 119:145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Elia G, et al. 2006. Detection of canine distemper virus in dogs by real-time RT-PCR. J. Virol. Methods 136:171–176 [DOI] [PubMed] [Google Scholar]
- 7. Gentsch JR, et al. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 9. Hu RL, et al. 2001. Detection and differentiation of CAV-1 and CAV-2 by polymerase chain reaction. Vet. Res. Commun. 25:77–84 [DOI] [PubMed] [Google Scholar]
- 10. Imada T, et al. 2000. Avian nephritis virus (ANV) as a new member of the family Astroviridae and construction of infectious ANV cDNA. J. Virol. 74:8487–8493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marshall JA, et al. 1984. Viruses and virus-like particles in the faeces of dogs with and without diarrhoea. Aust. Vet. J. 61:33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martella V, et al. 2011. Detection and characterization of astroviruses in dogs. J. Gen. Virol. 92:1880–1887 [DOI] [PubMed] [Google Scholar]
- 13. Mendez E, Arias CF. 2007. Astroviruses, p 981–1000 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed Lippincott Willliams & Wilkins, Philadelphia, PA [Google Scholar]
- 14. Méndez-Toss M, Romero-Guido P, Munguía ME, Méndez E, Arias CF. 2000. Molecular analysis of a serotype 8 human astrovirus genome. J. Gen. Virol. 81:2891–2897 [DOI] [PubMed] [Google Scholar]
- 15. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 16. Todd D, et al. 2009. Identification of chicken enterovirus-like viruses, duck hepatitis virus type 2 and duck hepatitis virus type 3 as astroviruses. Avian Pathol. 38:21–30 [DOI] [PubMed] [Google Scholar]
- 17. Toffan A, et al. 2009. Genetic characterization of a new astrovirus detected in dogs suffering from diarrhoea. Vet. Microbiol. 139:147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vennema H, de Bruin E, Koopmans M. 2002. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J. Clin. Virol. 25:233–235 [DOI] [PubMed] [Google Scholar]
- 19. Vieler E, Herbst W. 1995. Electron microscopic demonstration of viruses in feces of dogs with diarrhea. Tierarztl. Prax. 23:66–69 (In German.) [PubMed] [Google Scholar]
- 20. Wang Q-H, et al. 2005. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 11:1874–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Williams FP., Jr 1980. Astrovirus-like, coronavirus-like, and parvovirus-like particles detected in the diarrheal stools of beagle pups. Arch. Virol. 66:215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu AL, et al. 2011. Isolation and characterization of canine astrovirus in China. Arch. Virol. 156:1671–1675 [DOI] [PubMed] [Google Scholar]