Abstract
Avian leukosis virus subgroup J (ALV-J) was first isolated from meat-type chickens in 1988. No field cases of ALV-J infection or tumors in layer chickens were observed worldwide until 2004. However, layer flocks in China have experienced outbreaks of this virus in recent years. The molecular epidemiology of ALV-J strains isolated from layer flocks was investigated. The env genes of 77.8% (21/27) of the ALV-J layer isolates with a high degree of genetic variation were significantly different from the env genes of the prototype strain of ALV-J (HPRS-103) and American and Chinese strains from meat-type chickens (designated ALV-J broiler isolates). A total of 205 nucleotides were deleted from the 3′ untranslated region of 89.5% (17/19) of the ALV-J layer isolates. Approximately 94.7% (16/17) of the layer isolates contained a complete E element of 146 to 149 residues. The U3 sequences of 84.2% (16/19) of the ALV-J layer isolates displayed less than 92.5% sequence homology to those of the ALV-J broiler isolates, although the transcriptional regulatory elements that are typical of avian retroviruses were highly conserved. Several unique nucleotide substitutions in the env gene, the U3 region, and the E element of most of the ALV-J layer isolates were detected. These results suggested that the env gene, E element, and U3 region in the ALV-J layer isolates have evolved rapidly and were significantly different from those of the ALV-J broiler isolates. These findings will contribute to a better understanding of the pathogenic mechanism of layer tumor diseases induced by ALV-J.
INTRODUCTION
Avian leukosis viruses (ALVs), which belong to the genus Alpharetrovirus of the Retroviridae family, cause neoplastic diseases and other reproduction problems in the poultry industry worldwide. The ALVs can be classified as endogenous (ALV-E) or exogenous viruses according to their mode of transmission. Exogenous ALVs from chickens have been further divided into different subgroups (A, B, C, D, and J) on the basis of their host range, viral envelope interference, and cross-neutralization patterns (22). Avian leukosis virus subgroup J (ALV-J) was isolated from meat-type chickens in the late 1980s in the United Kingdom (21). ALV-J has primarily been associated with myeloid leukosis (ML) in broiler breeders and has caused more serious damage than any other subgroup. Although egg-type chickens have been experimentally infected with ALV-J to induce tumors (22), field cases of ALV-J infection and tumors in commercial layer chickens were not observed worldwide until 2004 (37).
In China, the ALV-J infection of broilers was first detected and reported in 1999 (12), followed by scattered reports of infection of broiler and local chickens in some areas of China (5, 7, 28). However, field cases of ALV-J infection and tumors in commercial layer chickens emerged in 2004 (37). ALV-J has been found to induce various tumors and cause significant economic losses in parent and commercial layer flocks in recent years, especially from 2008 to 2010 (6, 14, 18, 36). Many field cases of ALV-J infection and tumors occurred in 15- to 29-week-old egg-type chickens in several provinces. Dead or sick birds had hemangiomas of various sizes that were distributed on the surface of the head, claws, and wings; some birds had gray-white nodules in the liver, spleen, or kidneys, and the liver and spleen were enlarged to several times their normal size. Some affected flocks had dramatically reduced egg production and an increased rate of mortality (14).
Although several field cases and complete proviral genomic sequences for ALV-J strains isolated from layer chickens have been reported (6, 18, 36), little is known about the molecular epidemiology of ALV-J layer isolates in China. The first study conducted by our laboratory between 2007 and 2009 showed that ALV-J is predominantly responsible for layer avian leukosis in China (14). The present study completes the previous survey by focusing on the molecular epidemiology of ALV-J layer strains and the molecular characterization of ALV-J isolates that are circulating in China. A total of 16 ALV-J strains were isolated from clinical samples of different layer flocks from 7 provinces between 2008 and 2011. The env gene, 3′ untranslated region (UTR), and long terminal repeat (LTR) of ALV-J layer isolates (including 16 isolates in this study and other ALV-J layer isolates in China deposited in GenBank between 2007 and 2010) were sequenced and phylogenetically analyzed.
MATERIALS AND METHODS
Clinical samples.
Between April 2008 and May 2011, a total of 320 clinical samples (including tumor, whole-blood, spleen, kidney, liver, and other tissue samples) were collected from 6- to 36-week-old diseased egg-type chickens from 7 provinces in China (Heilongjiang, Henan, Hubei, Jilin, Liaoning, Shandong, and Jiangsu). The chickens originated from 48 different parents or commercial layer flocks. Most of the field cases of tumors occurred in 15- to 29-week-old egg-type chickens. The levels of egg production were dramatically reduced in the affected flocks. The clinical symptoms included hemorrhages in the skin of the phalanges and feather follicles. Some birds had gray-white nodules in the liver, spleen, or kidneys, and the liver and spleen were enlarged to several times the normal size.
Virus isolation and proviral DNA extraction.
All virus isolations were performed in DF-1 cells, which are known to be susceptible only to exogenous ALVs (19). The procedures for the isolation and identification of ALV in cell cultures were preformed according to previously described studies (1). Briefly, filtered tumor homogenates were inoculated into DF-1 cells, which were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, CA) supplemented with 10% fetal bovine serum (FBS) for two serial passages (5 days for each passage) at 37°C in a 5% CO2 incubator with daily monitoring. After the incubation, the infected DF-1 cells were tested for the ALV group-specific antigen (p27) by an antigen-capture enzyme-linked immunosorbent assay (AC-ELISA) with anti-p27 antibody-coated plates (IDEXX Inc., MA) and tested for the ALV-J-specific antigen by immunofluorescence assay (IFA) with ALV-J-specific monoclonal antibody FE9 (32). The positive samples detected by ELISA were harvested for DNA extraction and PCR amplification. The DNA was directly extracted from the positive cultured DF-1 cells or tumors using an established method. Briefly, the cultured cells were lysed in tissue lysis buffer (4 M guanidine hydrochloride, 25 mM sodium citrate, and 1% Triton X-100) and extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1). The DNA was precipitated with absolute isopropanol, washed with 70% isopropanol, and dried at room temperature. Subsequently, the DNA was resuspended in nuclease-free water and stored at −80°C.
PCR cloning and sequencing.
PCR was used to test genomic DNA from the cultured DF-1 cells or tumors for the presence of envelope sequences that are specific for ALV-J as previously described (27). The primer set H5 (5′-GGATGAGGTGACTAAGA-3′) and H7 (5′-CGAACCAAAGGTAACACACG-3′) was used for the specific detection of ALV-J proviral DNA, which generates a 544-bp PCR product (27). The primer set H5 and AD1 (5′-GGGAGGTGGCTGACTGTGT-3′) was used for the detection of subgroup A to E ALVs, which generate a 295- to 326-bp PCR product (27). According to the sequence of the ALV-J prototype strain, HPRS-103 (GenBank accession number AF097731), the primer pair EF (5′-CGACACTGATAAGGTTATTTGGGT-3′) and ER (5′-TCGGAACCTACAGCTGCTC-3′) was designed for the amplification of the ALV-J env gene. The PCR conditions included an initial denaturation cycle of 4 min at 94°C, followed by 30 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 54°C, and an extension for 2 min at 72°C, with a final extension of 7 min at 72°C. Another primer pair (UF [5′-CCGCGAAAGGTGTTAAGACG-3′] and UR [5′-TTCCCCCTCCCTATGCAAA-3′]) was designed according to the HPRS-103 sequence to amplify a 700-bp fragment that encompasses a partial region of gp37, the entire 3′ UTR (including a nonfunctional redundant transmembrane region [rTM], the direct repeat element [DR-1], and an E element) and the entire 3′ LTR. The PCR amplification scheme was the same as that for the env gene, but the annealing temperature was 56°C and the extension time was 1 min at 72°C. All PCRs were carried out with PrimerSTAR HS DNA high-fidelity polymerase (TaKaRa, Dalian, China).
The PCR products were excised from a 1.0% agarose gel, purified using an AxyPrep DNA gel extraction kit (Axygen Scientific, Inc., CA), and cloned into the TA vector pMD18-T (TaKaRa). Three independent clones of each ALV-J isolate were sequenced by the Beijing Genomics Institute (Beijing, China).
DNA alignments and phylogenetic analysis.
The nucleotide sequences were aligned using the Clustal W program, version 1.8 (17). A neighbor-joining tree was drawn using the MEGA program, version 3.1 (29), with confidence levels assessed using 1,000 bootstrap replications. The GenBank sequences of the ALV-J strains that were isolated from meat-type and layer chickens were included in the multiple-sequence alignment, and they are summarized in Table 1.
Table 1.
ALV-J strains used in the construction of phylogenetic trees
| No.a | Isolate | Origin | Yr | Accession no. | Hostb | Tumor typec | Reference or sourced |
|---|---|---|---|---|---|---|---|
| 1 | LN08SY10 | Liaoning, China | 2008 | HQ634802 | CL | ML | VI |
| 2 | JL08CH3-1 | Jilin, China | 2008 | HQ634809 | CL | He + ML | VI |
| 3 | LN08SY31 | Liaoning, China | 2009 | HQ634803 | CL | ML | VI |
| 4 | SD09DP04 | Shandong, China | 2009 | HQ634808 | CL | He + ML | VI |
| 5 | HLJ09SH01 | Heilongjiang, China | 2009 | HQ634806 | CL | He + ML | VI |
| 6 | HLJ09SH02 | Heilongjiang, China | 2009 | HQ634807 | CL | He | VI |
| 7 | HuB09JY03 | Hubei, China | 2009 | HQ634811 | CL | ML | VI |
| 8 | HuB09WH02 | Hubei, China | 2009 | HQ634804 | CL | He + ML | VI |
| 9 | HuB09WH03 | Hubei, China | 2009 | HQ634805 | CL | He + ML | VI |
| 10 | HLJ10SH03 | Heilongjiang, China | 2010 | HQ634813 | CL | He + ML | VI |
| 11 | HLJ10SH04 | Heilongjiang, China | 2010 | HQ634814 | CL | He + ML | VI |
| 12 | HN10PY01 | Henan, China | 2010 | CL | He | VI | |
| 13 | JL10HW01 | Jilin, China | 2010 | HQ634800 | CL | He + ML | VI |
| 14 | JL10HW02 | Jilin, China | 2010 | HQ634801 | CL | He + ML | VI |
| 15 | JS11HA94 | Jiangsu, China | 2011 | CL | He | VI | |
| 16 | HLJ08MDJ01 | Heilongjiang, China | 2008 | HQ634807 | CL | He | VI |
| 17 | JS09GY2 | Jiangsu, China | 2009 | GU982307 | CL | He | 36 |
| 18 | JS09GY3 | Jiangsu, China | 2009 | GU982308 | CL | He | 36 |
| 19 | JS09GY5 | Jiangsu, China | 2009 | GU982309 | CL | He | 36 |
| 20 | JS09GY6 | Jiangsu, China | 2009 | GU982310 | CL | He | 36 |
| 21 | NHH | China | 2009 | HM235668 | He | NF | |
| 22 | SD07LK1 | Shandong, China | 2007 | FJ216405 | PL | ML | 34 |
| 23 | SX090912J | Shanxi, China | 2009 | HQ386988 | CL | — | NF |
| 24 | SX090915J | Shanxi, China | 2009 | HQ386989 | CL | — | NF |
| 25 | HA08 | China | 2009 | HM235664 | ML | NF | |
| 26 | HN1001-1 | Henan, China | 2010 | HQ260974 | He | NF | |
| 27 | HN1001-2 | Henan, China | 2010 | HQ260975 | He | NF | |
| 28 | HPRS-103 | UK | 1988 | Z46390 | M | ML | 21 |
| 29 | ADOL-Hc1 | USA | 1993 | AF097731 | BB | ML | 13 |
| 30 | ADOL-7501 | USA | 1997 | AY027920 | CB | ML | 13 |
| 31 | UD5 | USA | 2000 | AF307952 | M | ML | NF |
| 32 | 10022-2 | USA | 2006 | GU222396 | — | 20 | |
| 33 | JS-nt | China | 2003 | HM235667 | ML | NF | |
| 34 | NX0101 | China | 2001 | AY897227 | BB | ML | 9 |
| 35 | YZ9901 | China | 1999 | AY897222 | CB | — | 9 |
| 36 | SD0101 | China | 2001 | AY897225 | BB | ML | 9 |
| 37 | NM2002-1 | China | 2002 | HM235669 | BB | ML | NF |
| 38 | YZ9902 | China | 1999 | HM235670 | CB | — | 12 |
| 39 | SD9901 | China | 1999 | AY897220 | BB | ML | 9 |
| 40 | SD9902 | China | 1999 | AY897221 | BB | ML | 9 |
| 41 | SD0001 | China | 2000 | AY897223 | BB | ML | 9 |
| 42 | SD0002 | China | 2000 | AY897224 | CB | ML | 9 |
| 43 | SD0102 | China | AY897224 | — | NF | ||
| 44 | HN0001 | China | 2000 | AY897219 | BB | ML | 9 |
| 45 | SDC2000 | China | 2000 | AY234052 | BB | ML | 38 |
| 46 | HB0301 | China | 2003 | AY897229 | BB | ML | 35 |
| 47 | BJ0303 | China | 2003 | AY897230 | BB | ML | 35 |
Isolates 1 to 16 are layer strains identified in this study. Isolates 17 to 27 are layer strains obtained from GenBank. Isolates 28 to 32 are the prototype and American strains. Isolates 33 to 47 are Chinese broiler isolates.
The host of the ALV-J isolates. CL, commercial layer chicken; PL, parental layer; M, meat-type chicken; BB, broiler breeder; CB, commercial broiler.
Type of tumor induced by ALV-J. ML, myeloma leukosis; He, hemangioma; —, unknown background of the isolate.
VI, viruses isolated and identified in this study; NF, no reference.
Nucleotide sequence accession numbers.
The sequences obtained in this study have been submitted to GenBank, and the accession numbers are provided in Table 1.
RESULTS
Virus isolation and identification of ALV-J.
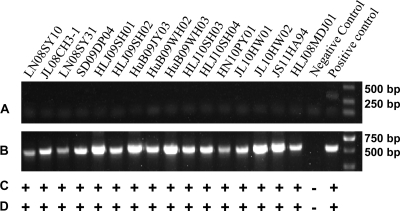
A total of 16 ALV-J strains (detailed information is summarized in Table 1) were isolated from clinical samples from different layer flocks from 2008 to 2011 in Heilongjiang, Henan, Hubei, Jilin, Liaoning, Shandong, and Jiangsu Provinces in China. The PCR of DNA extracted from infected DF-1 cells with 16 ALV-J isolates produced an ALV-J-specific 545-bp fragment with primers H5 and H7 (Fig. 1A); however, no specific fragments were produced with primers H5 and AD1 (Fig. 1B). Further evidence of ALV-J in the samples was demonstrated by the positive result in the IFA with ALV-J-specific monoclonal antibody FE9 (Fig. 1C) and in the AC-ELISA with anti-p27 antibody-coated plates (Fig. 1D).
Fig 1.
Identification of 16 ALV-J isolates with PCR (A, primer pair H5/AD1; B, primer pair H5/H7), IFA (C) and AC-ELISA (D). Uninfected DF-1 cells served as the negative control. DF-1 cells infected with Rous-associated virus type 1 (subgroup A) served as the positive control for PCR with primer pair H5/AD1. DF-1 cells infected with HPRS-103 served as the positive control for PCR with primer pair H5/AD1, IFA, and AC-ELISA.
Molecular characterization of env gene of layer isolates.
A total of 27 ALV-J layer isolates (including 16 isolates in this study and 11 isolates from GenBank) were analyzed (Table 1). The prototype ALV-J strain HPRS-103 (21), American ALV-J isolates (e.g., ADOL-Hc1 and ADOL-7501), and Chinese strains from meat-type chickens (designated ALV-J broiler isolates) were used as references for comparisons in the molecular studies. All reference viruses are summarized in Table 1.
The env genes of layer isolates were 1,512 to 1,518 nucleotides long. The nucleotide changes that occurred throughout the env gene showed a maximum divergence of 7.4%, with nucleotide sequence identities ranging from 92.6 to 100%. The analysis of the deduced amino acid sequences showed that the maximum divergence in the amino acid sequence was 11.2%, with sequence identities ranging from 88.8 to 100%.
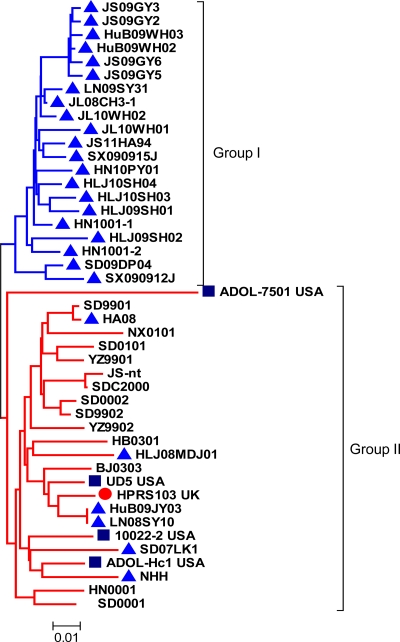
The phylogenetic analysis indicated that 77.8% (21/27) of the ALV-J layer isolates showed close homology (91.3 to 99.0%) with each other and belonged to one branch, which was designated group I (Fig. 2). These isolates were 88.7 to 94.3% identical to prototype strain HPRS-103, 88.7 to 92.5% identical to American strains, and 88.1 to 94.3% identical to Chinese broiler isolates at the amino acid level. However, 22.2% (6/27, including iso-lates HA08, NHH, HLJ08MDJ1, HuB09JY03, LN08SY10, and SD07LK1) of the layer isolates showed relatively close homology (88.7 to 99.0%) with the ALV-J broiler isolates (excluding the ADOL-7501 isolate), which belonged to the other branch and which was designated group II (Fig. 2).
Fig 2.
Phylogenetic analysis of the nucleotide sequences of env genes of ALV-J layer and broiler isolates. The tree was constructed on the basis of the minimum-evolution method using MEGA 4 software. Bootstrap values were calculated with 1,000 replicates of the alignment. The two groups are marked. Triangles represent the ALV-J layer isolates. The circle represents the prototype strain of ALV-J, HPRS-103. Squares represent the American ALV-J strains from meat-type chickens.
Compared with the ALV-J broiler isolates, the amino acid substitutions of all layer isolates were distributed throughout the envelope glycoprotein, and there was a clustering of sequence variations near the hr1, hr2, and 48- to 76-amino-acid variable domains of the gp85 gene. Thirteen amino acids (48S, 49A, 61Q, 63D, 76T, 117G, 123I, 143T/E, 146G, 150H, 189K, 203N, and 218Q) of gp85 were relatively conserved in 21 layer isolates.
Molecular characterization of 3′UTR of layer isolates.
The 3′ UTR and LTR sequences from a total of 19 ALV-J layer isolates (including 15 isolates in this study and 4 isolates from GenBank) were analyzed (Table 1). A comparison of the nucleotide sequences of 3′ UTRs with the ALV-J broiler isolates revealed that 205 bp were deleted in the rTM and DR-1 regions of 89.5% (17/19) of the ALV-J layer isolates (excluding HLJ08MDJ01 and SD07LK1) (Fig. 3). A total of 175 bp were deleted from the 3′ end of the rTM region, and 30 nucleotides were deleted from the 5′ end of the DR-1 region. This deletion was similar to the mutation in the 3′ UTRs of American ALV-J isolates (ADOL-7501 and UD5). Approximately 210 bp were deleted from the 3′ end of the rTM region, and 8 bp of nucleotides were deleted from the 5′ end of the DR-1 region in the HLJ08MDJ01 isolate. A total of 113 bp of nucleotides were deleted from the 3′ end of the rTM region, which contained the entire DR-1 region in the SD07LK1 isolate.
Fig 3.
Comparison of the deletions at the 3′ UTR in the genomic proviral DNA of ALV-J layer and broiler isolates. The top two lines represent the base numbers and elements in the genomic proviral DNA of HPRS-103. The deletions are indicated by empty spaces between the thick black lines.
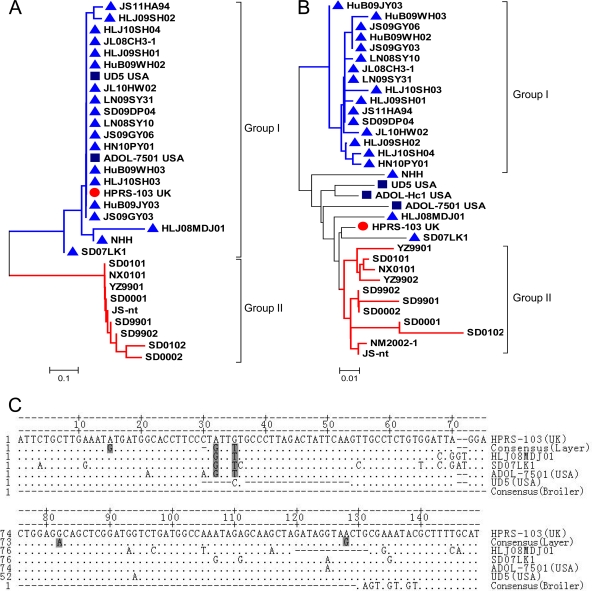
Approximately 94.7% (16/17) of the ALV-J layer isolates contained an intact E element (146 to 149 nucleotides) with a high degree of identity to the E element of the prototype HPRS-103 and American strains (ADOL-7501 and UD5) (Fig. 4A). Only a single layer isolate (HLJ08MDJ01) contained a 12-bp deletion in the E element. In contrast, nine Chinese broiler strains displayed E elements with substantial deletions (127-nucleotide deletions) (Fig. 4C) (9).
Fig 4.
Phylogeny and alignment of the E elements and U3 region of ALV-J layer and broiler isolates. (A and B) Phylogenetic analysis of E elements and U3 regions. Triangles represent the ALV-J layer isolates. Squares represent the American ALV-J strains from meat-type chickens. The circle represents the prototype strain of ALV-J, HPRS-103. (C) Comparison of nucleotide sequences of the E elements. Consensus (layer) sequences were derived from 16 layer isolates (JS11HA94, HLJ09SH02, HLJ10SH04, JL08CH3-1, HLJ09SH01, HuB09WH02, JL10HW02, LN09SY31, SD09DP04, LN08SY10, JS09GY06, HN10PY01, HuB09WH03, HLJ10SH03, HuB09JY03, JS09GY03). The consensus (broiler) sequences were derived from the nine Chinese ALV-J strains (YZ9901, SD9901, SD9902, SD0001, SD0002, HN0001, SD0101, NX0101, and JS-nt). The dots indicate identical residues, while the letters indicate base substitutions. The dashes indicate gaps produced in the alignment. The shading indicates unique nucleotide substitutions of the ALV-J layer isolates.
The nucleotide sequences of the E element of the ALV-J layer isolates were aligned with those of the ALV-J reference strains. The consensus sequence from layer chickens is derived from 15 isolates and termed “consensus (layer).” The consensus sequence from broiler chickens is derived from eight Chinese isolates and termed “consensus (broiler)” (9), as presented in Fig. 4C. The sequence alignment revealed that most of the sequence of the E element was conserved in all of the ALV-J layer isolates, and only minor mutations were observed. There was a single nucleotide deletion (bases 29 and 31) and 3 unique nucleotide substitutions at positions 15 (A to G), 79 (G to A), and 125 (A to G) in the E elements of 15 layer isolates and 2 unique nucleotide substitutions at positions 32 (A to G) and 35 (G to T) in the E elements of all layer isolates (Fig. 4C).
Molecular characterization of 3′LTR of layer isolates.
The sequence analysis of 3′ LTRs of all layer isolates revealed the same 325-bp sequence, except for the 3′ LTR of the NHH isolate (314 bp). The R and U5 regions of all layer isolates were well conserved compared with those of the ALV-J broiler isolates. The phylogenetic analysis indicated that the U3 sequences of 84.2% (16/19) of the layer isolates belonged to a new branch (Fig. 4B) and have 96.5 to 100% DNA sequence identity with each other, which is less than the 92.5% sequence homology to the ALV-J broiler isolates. The U3 sequences of NHH, HLJ08MDJ01, and SD07LK1 layer isolates displayed relatively greater homology to the ALV-J broiler isolates (Fig. 4B).
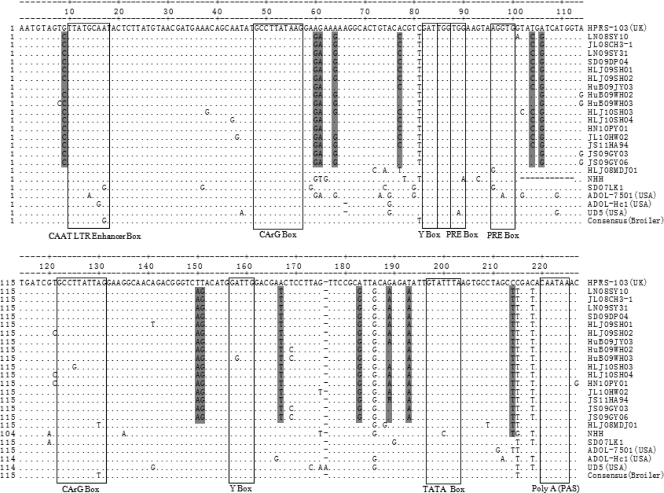
Transcriptional regulation elements were identified in the 3′ U3 region of the LTR in all of the ALV-J layer isolates, including two CArG, Y, and PRE boxes and one C/EBP, CAAT, and TATA box (Fig. 5). Two CArG boxes were located at positions 49 to 58 and 123 to 132, respectively. Similarly, two highly conserved inverse Y boxes with the sequence 5′-ATTGG-3′ were located at positions 83 to 87 and 158 to 162, respectively. Two copies of the PRE motif GGTGG were located at positions 86 to 90 and 98 to 102, flanking the nucleotide sequence AAGTAA. The CCAAT/enhancer (5′-TTATGCAAT-3′) was located at positions 10 to 18. A TATA box with a consensus sequence TATTTAA was located at positions 197 to 203. A key polyadenylation signal with the sequence AATAAA was located at the end of the 3′ U3 region. Almost all of the putative transcription regulatory elements identified in the U3 region were conserved and homologous to those of ALV-J broiler isolates (Fig. 5), which further indicated the importance of these regulatory elements during viral replication (24).
Fig 5.
Comparison of the nucleotide sequences in the U3 region of the ALV-J layer isolates, prototype strain HPRS-103, American ALV-J strains, and the consensus (broiler) sequence of the nine Chinese ALV-J strains. The dots indicate identical residues, while the letters indicate base substitutions. The dashes indicate gaps produced in the alignment. The locations of putative transcription regulatory elements are indicated in boxes and marked. The shading indicates unique nucleotide substitutions in the ALV-J layer isolates.
As shown in Fig. 5, compared with the ALV-J broiler isolates, 14 unique nucleotide substitutions were detected in the U3 region of most of the ALV-J layer isolates.
DISCUSSION
Since ALV-J emerged in the 1980s, it has become highly prevalent in meat-type chickens and has caused serious problems in meat-type birds (13). However, in China, the host range of ALV-J is changing. ALV-J has become a major problem in layer chickens since 2008 (6, 14). In the present study, the analysis of sequences of ALV-J layer isolates showed that (i) the env genes of 77.8% (21/27) of the layer isolates with a high degree of genetic variation were significantly different from the env genes of the ALV-J broiler isolates, (ii) a 205-bp deletion in the 3′ UTR and unique nucleotide mutations in the 3′ UTR and 3′ LTR were detected, and (iii) almost all layer isolates contained a complete E element of 147 to 149 residues. This is the first comprehensive report concerning the molecular characterization of ALV-J layer isolates.
Genetic and antigenic variations with sequence changes in the variable regions of the env gene of ALV-J have been observed (26, 33). To characterize the molecular changes of the env gene of these layer isolates, the env genes were amplified and compared with those of the ALV-J broiler isolates (the prototype strain HPRS-103 and American and Chinese strains from meat-type chickens). A total of 21 layer isolates showed significant differences in the nucleotide sequences compared with the ALV-J broiler isolates, forming a separate group (Fig. 2). The envelope glycoprotein of ALV primarily functions as a ligand for receptor binding for viral entry into the susceptible cell and determines the host range (7). Studies with other ALV subgroups have shown that the central region of the gp85 subunit contains regions of sequence variability in five clusters designated hr1, hr2, vr1, vr2, and vr3 (4). Analyses of ALV env genes have identified the hr1 and hr2 domains to be the principal receptor interaction determinants (11); vr3 also plays a role in determining the specificity of receptor recognition (10, 31). In the present study, an alignment of the deduced amino acid sequences of 27 layer isolates and ALV-J broiler isolates revealed 13 amino acid substitutions distributed within the central region of the gp85 subunit. These results suggested that these amino acid substitutions might be associated with changes in the host range and pathogenicity of ALV-J.
Previous studies have demonstrated that ALV-J is prone to significant molecular variation because the envelope gene of this virus displays multiple mutations, resulting in antigenic variants, possibly as a result of immune pressure (26, 33). Among the variable domains within gp85, the hr1, hr2, and vr3 regions are the main targets of selection pressure (30). In this study, 7 of the 13 amino acid substitutions were distributed within the hr1 and hr2 regions. The pattern of amino acid substitutions presented here suggested that the antigenic variation might have resulted from selection pressure.
Because the 3′ UTR of ALV, which contains potent regulatory sequences that influence chromosomal and viral gene expression, is important in viral pathogenesis and replication (23, 24), the rTM, DR-1, and E elements of layer ALV-J isolates were analyzed. The entire rTM region consists of 210 nucleotides. In the present study, 15 out of 17 layer isolates displayed a 175-bp deletion at the 3′ end of the rTM region. Although the number and region of the deletion in the rTM region varied between ALV-J layer and broiler isolates, the deletion mutation in the rTM region was common in most of these ALV-J strains, suggesting that this region is not necessary for viral replication or spreading (3). A previous study indicated that DR-1 elements are exclusively found in sarcoma viruses and ALV-J (2), and they seem to contribute to the fitness of the viruses because these elements have been associated with the efficient accumulation of unspliced RNA in the cytoplasm and the selective increase of spliced src mRNA in avian sarcoma virus (ASV). However, a 30-bp deletion at the 5′ end of the DR-1 region in most of ALV-J layer isolates suggested that at least the 5′ end might be dispensable for viral fitness.
Recent reports have demonstrated substantial deletions (>50%) in the E element of ALV-J proviruses that were isolated from breeding stocks and broiler chickens between 1988 and 2007 (39), and the E element had an almost identical deletion of 127 bp in earlier Chinese strains (8) and the recently isolated strain JS-nt from meat-type chickens. In contrast, almost all layer isolates in this study contained an intact E element. An analysis of the E-element sequence revealed unique nucleotide substitutions (A15G, G79A, and A125G) and deletions (bases 29 and 31) in this region in most of the ALV-J layer isolates. Previous studies indicated that the E element might exert a tissue-specific effect on the transcription of cellular oncogenes in the vicinity of the integrated provirus and might also be involved in determining the oncogenic spectrum of avian retrovirus-induced disease (15, 31). Although an intact E element was not a critical requirement for the ALV-J induction of tumors, the presence of the E element could be associated with a higher frequency of tumors in susceptible chickens (8). The ALV-Js isolated from layer flocks induced a high incidence of hemangioma in China (6, 36). However, previous studies indicated that the dominant tumor induced by ALV-J infection in meat-type chickens was a myelocytoma (ML) (21). Whether these properties of the E element of ALV-J layer isolates are related to the change in tumor spectrum in the layer flocks in China is unknown and should be further investigated.
The U3 sequences of the ALV-J broiler strains have been previously compared (16, 36). In the present study, we analyzed the U3 sequences of ALV-J layer isolates. The phylogenetic analysis indicated that the U3 sequences of 16 layer isolates were highly homologous with each other and showed relatively low homology (less than 92.5%) with those of the ALV-J broiler isolates. Fourteen unique nucleotide substitutions in the U3 region were found in most of the layer isolates. These results indicated that the U3 region of the ALV-J layer isolates has evolved rapidly. The U3 region of the LTR contained a transcriptional promoter and enhancer elements and determined both the level of viral transcription and the oncogenic potential of ALVs (10, 25). These nucleotide mutations in the U3 region were likely associated with the tumor disease of layers that can be induced by ALV-J.
ALV-Js have evolved rapidly in pathogenicity, which results in a change in host range and tumor spectrum. The genetic diversity of ALV-J might not be restricted to the env gene; genetic rearrangements and high mutation rates were also found in noncoding genomic regions (16, 39). The two noncoding regions of the ALV-J genome that might activate oncogenes and be involved in oncogenesis are the U3 sequences of the LTR and the E element located in the 3′ UTR (3). Our results indicated significant genetic variation in these genes. The env gene, U3 region, and E element of most of the ALV-J layer isolates formed a separate branch from the prototype meat-type strain HPRS-103 and other ALV-J broiler strains, and several unique nucleotide substitutions were detected in these regions. Although the present study did not provide information concerning the functional characteristics of the genes described, it provided some interesting information on the molecular characterization of ALV-J layer isolates. These findings will contribute to a better understanding of the pathogenic mechanism of layer tumor diseases induced by ALV-J.
ACKNOWLEDGMENTS
The study was supported by the National Natural Science Foundation of China (31072146), the earmarked fund for Modern Agro-Industry Technology Research System (no. nycytx-42-G3-01), and Harbin Programs for Science and Technology Development (no. 2010AA6AN034).
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Bagust TJ, Fenton SP, Reddy MR. 2004. Detection of subgroup J avian leukosis virus infection in Australian meat-type chickens. Aust. Vet. J. 82:701–706 [DOI] [PubMed] [Google Scholar]
- 2. Bai J, Howes K, Payne LN, Skinner MA. 1998. Sequence analysis of an infectious proviral clone of HPRS-103 shows that it represents a new avian retrovirus envelope subgroup (designated J). Avian Pathol. 27(Suppl 1):S92–S93 [Google Scholar]
- 3. Bai J, Payne LN, Skinner MA. 1995. HPRS-103 (exogenous avian leukosis virus, subgroup J) has an env gene related to those of endogenous elements EAV-0 and E51 and an E element found previously only in sarcoma viruses. J. Virol. 69:779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bova CA, Manfredi JP, Swanstrom R. 1986. Env genes of avian retroviruses: nucleotide sequence and molecular recombinants define host range determinants. Virology 152:343–354 [DOI] [PubMed] [Google Scholar]
- 5. Cheng ZQ, Zhang L, Liu SD, Zhang LJ, Cui ZZ. 2005. Emerging of avian leukosis virus subgroup J in a flock of Chinese local breed. Acta Microbiol. Sinica 45:584–587 (In Chinese.) [PubMed] [Google Scholar]
- 6. Cheng Z, Liu J, Cui Z, Zhang L. 2010. Tumors associated with avian leukosis virus subgroup J in layer hens during 2007 to 2009 in China. J. Vet. Med. Sci. 72:1027–1033 [DOI] [PubMed] [Google Scholar]
- 7. Chesters PM, et al. 2002. The viral envelope is a major determinant for the induction of lymphoid and myeloid tumours by avian leukosis virus subgroups A and J, respectively. J. Gen. Virol. 83:2553–2561 [DOI] [PubMed] [Google Scholar]
- 8. Chesters PM, Smith LP, Nair V. 2006. The E (XSR) element contributes to the oncogenicity of avian leukosis virus (subgroup J). J. Gen. Virol. 87:2685–2692 [DOI] [PubMed] [Google Scholar]
- 9. Cui Z, Du Y, Zhang Z, Silva RF. 2003. Comparison of Chinese field strains of avian leukosis subgroup J viruses with prototype strain HPRS-103 and United States strains. Avian Dis. 47:1321–1330 [DOI] [PubMed] [Google Scholar]
- 10. Cullen BR, Raymond K, Ju G. 1985. Functional analysis of the transcription control region located within the avian retroviral long terminal repeat. Mol. Cell. Biol. 5:438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dorner AJ, Coffin JM. 1986. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell 45:365–374 [DOI] [PubMed] [Google Scholar]
- 12. Du Y, Cui Z, Qin A. 1999. Subgroup J avian leukosis viruses in China. China Poult. Sci. 3:1–4 (In Chinese.) [Google Scholar]
- 13. Fadly AM, Smith EJ. 1999. Isolation and some characteristics of a subgroup J-like avian leukosis virus associated with myeloid leukosis in meat-type chickens in the United States. Avian Dis. 43:391–400 [PubMed] [Google Scholar]
- 14. Gao YL, et al. 2010. Avian leukosis virus subgroup J in layer chickens, China. Emerg. Infect. Dis. 16:1637–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hayward WS, Neel BG, Astrin SM. 1981. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 290:475–480 [DOI] [PubMed] [Google Scholar]
- 16. Hue D, et al. 2006. Major rearrangements in the E element and minor variations in the U3 sequences of the avian leukosis subgroup J provirus isolated from field myelocytomatosis. Arch. Virol. 151:2431–2446 [DOI] [PubMed] [Google Scholar]
- 17. Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150–163 [DOI] [PubMed] [Google Scholar]
- 18. Lai H, et al. 2011. Isolation and characterization of emerging subgroup J avian leukosis virus associated with hemangioma in egg-type chickens. Vet. Microbiol. 151:275–283 [DOI] [PubMed] [Google Scholar]
- 19. Maas R, van Zoelen D, Oei H, Claassen I. 2006. Replacement of primary chicken embryonic fibroblasts (CEF) by the DF-1 cell line for detection of avian leukosis viruses. Biologicals 34:177–181 [DOI] [PubMed] [Google Scholar]
- 20. Pandiri AR, et al. 2010. Subgroup J avian leukosis virus neutralizing antibody escape variants contribute to viral persistence in meat-type chickens. Avian Dis. 54:848–856 [DOI] [PubMed] [Google Scholar]
- 21. Payne LN, et al. 1991. A novel subgroup of exogenous avian leukosis virus in chickens. J. Gen. Virol. 72:801–807 [DOI] [PubMed] [Google Scholar]
- 22. Payne LN, Howes K, Gillespie AM, Smith LM. 1992. Host range of Rous sarcoma virus pseudotype RSV(HPRS-103) in 12 avian species: support for a new avian retrovirus envelope subgroup, designated J. J. Gen. Virol. 73:2995–2997 [DOI] [PubMed] [Google Scholar]
- 23. Robinson HL, Blais BM, Tsichlis PN, Coffin JM. 1982. At least two regions of the viral genome determine the oncogenic potential of avian leukosis viruses. Proc. Natl. Acad. Sci. U. S. A. 79:1225–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ruddell A. 1995. Transcription regulatory elements of the avian retroviral long terminal repeat. Virology 206:1–7 [DOI] [PubMed] [Google Scholar]
- 25. Ryden TA, Mde Mars Beemon K. 1993. Mutation of the C/EBP binding sites in the Rous sarcoma virus long terminal repeat and gag enhancers. J. Virol. 67:2862–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Silva RF, Fadly AM, Hunt HD. 2000. Hypervariability in the envelope genes of subgroup J avian leukosis viruses obtained from different farms in the U.S. Virology 272:106–111 [DOI] [PubMed] [Google Scholar]
- 27. Smith LM, et al. 1998. Development and application of polymerase chain reaction (PCR) tests for the detection of subgroup J avian leukosis virus. Virus Res. 54:87–98 [DOI] [PubMed] [Google Scholar]
- 28. Sun S, Cui Z. 2007. Epidemiological and pathological studies of subgroup J avian leukosis virus infections in Chinese local “yellow” chickens. Avian Pathol. 36:221–226 [DOI] [PubMed] [Google Scholar]
- 29. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thu WL, Wang CH. 2003. Phylogenetic analysis of subgroup J avian leucosis virus from broiler and native chickens in Taiwan during 2000-2002. J. Vet. Med. Sci. 65:325–328 [DOI] [PubMed] [Google Scholar]
- 31. Tsichlis PN, Coffin JM. 1980. Recombinants between endogenous and exogenous avian tumor viruses: role of the C region and other portions of the genome in the control of replication and transformation. J. Virol. 33:238–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Venugopal K, Howes K, Barron GS, Payne LN. 1997. Recombinant env-gp85 of HPRS-103 (subgroup J) avian leukosis virus: antigenic characteristics and usefulness as a diagnostic reagent. Avian Dis. 41:283–288 [PubMed] [Google Scholar]
- 33. Venugopal K, Smith LM, Howes K, Payne LN. 1998. Antigenic variants of J subgroup avian leukosis virus: sequence analysis reveals multiple changes in the env gene. J. Gen. Virol. 79:757–766 [DOI] [PubMed] [Google Scholar]
- 34. Wang H, Cui Z. 2008. The identification and sequence analysis of ALV-J isolated from layers. Chin. J. Virol. 24:369–370 (In Chinese.) [PubMed] [Google Scholar]
- 35. Wang Z, Cui Z, Zang Z, Wu Y. 2005. The mutation tendency of the gp85 gene of Chinese field strains of ALV-J from 1999 to 2003. Vitologica Siniga 20:393–398 [Google Scholar]
- 36. Wu X, et al. 2010. Recombinant avian leukosis viruses of subgroup J isolated from field infected commercial layer chickens with hemangioma and myeloid leukosis possess an insertion in the E element. Vet. Res. Commun. 34:619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu B, et al. 2004. Occurrence of avian leukosis virus subgroup J in commercial layer flocks in China. Avian Pathol. 33:13–17 [DOI] [PubMed] [Google Scholar]
- 38. Yang Y, Yie J, Zhao Z, Qin A, Gu Y. 2003. Isolation and identification of inner Mongolia Strain of ALV subgroup J. Vitologica Siniga 18:454–458 (In Chinese.) [Google Scholar]
- 39. Zavala G, Cheng S, Jackwood MW. 2007. Molecular epidemiology of avian leukosis virus subgroup J and evolutionary history of its 3′ untranslated region. Avian Dis. 51:942–953 [DOI] [PubMed] [Google Scholar]