Abstract
Several apoptotic regulators, including Bcl-x, are alternatively spliced to produce isoforms with opposite functions. We have used an RNA interference strategy to map the regulatory landscape controlling the expression of the Bcl-x splice variants in human cells. Depleting proteins known as core (Y14 and eIF4A3) or auxiliary (RNPS1, Acinus, and SAP18) components of the exon junction complex (EJC) improved the production of the proapoptotic Bcl-xS splice variant. This effect was not seen when we depleted EJC proteins that typically participate in mRNA export (UAP56, Aly/Ref, and TAP) or that associate with the EJC to enforce nonsense-mediated RNA decay (MNL51, Upf1, Upf2, and Upf3b). Core and auxiliary EJC components modulated Bcl-x splicing through different cis-acting elements, further suggesting that this activity is distinct from the established EJC function. In support of a direct role in splicing control, recombinant eIF4A3, Y14, and Magoh proteins associated preferentially with the endogenous Bcl-x pre-mRNA, interacted with a model Bcl-x pre-mRNA in early splicing complexes, and specifically shifted Bcl-x alternative splicing in nuclear extracts. Finally, the depletion of Y14, eIF4A3, RNPS1, SAP18, and Acinus also encouraged the production of other proapoptotic splice variants, suggesting that EJC-associated components are important regulators of apoptosis acting at the alternative splicing level.
INTRODUCTION
Nearly all multiexon genes in the human genome are subjected to alternative splicing (51, 78). In addition to producing proteins with distinct activities (47, 68), alternative variants with frameshifts or stop codons can also have important regulatory functions (79). For example, to establish homeostatic levels of alternative splicing regulators, many genes produce splice variants that become degraded through nonsense-mediated RNA decay (NMD) (31, 46, 59). This tight control is important because perturbations created by the aberrant expression of splicing regulators are associated with human diseases such as muscular dystrophy and cancer (19, 27, 76).
Apoptotic genes are often regulated through alternative splicing (61), and many examples occur in the Bcl-2 family of apoptotic regulators (14, 26). Antiapoptotic (e.g., Bcl-2, Mcl1, and Bcl-xL) and proapoptotic (e.g., Bax, Bim, and Bcl-xS) Bcl-2 members usually differ in the number and variety of BH domains that they contain (81). Alternative usage of two competing 5′ splice sites (5′ss) produces the antiapoptotic Bcl-xL regulator and the shorter proapoptotic Bcl-xS variant lacking one BH domain (5) (Fig. 1A). Bcl-xL is highly expressed in cancer tissues; its overexpression confers resistance to apoptotic stimuli and favors metastasis (6, 20, 49). In contrast, Bcl-xS can induce apoptosis and alleviate multidrug resistance (12, 41). Because Bcl-x splicing likely integrates the opposing actions of a multitude of pathways that affect survival and apoptosis, the mechanisms governing this important splicing decision must be subjected to tight control. For this reason, the regulatory aspects of Bcl-x splicing have received more attention recently.
Fig 1.
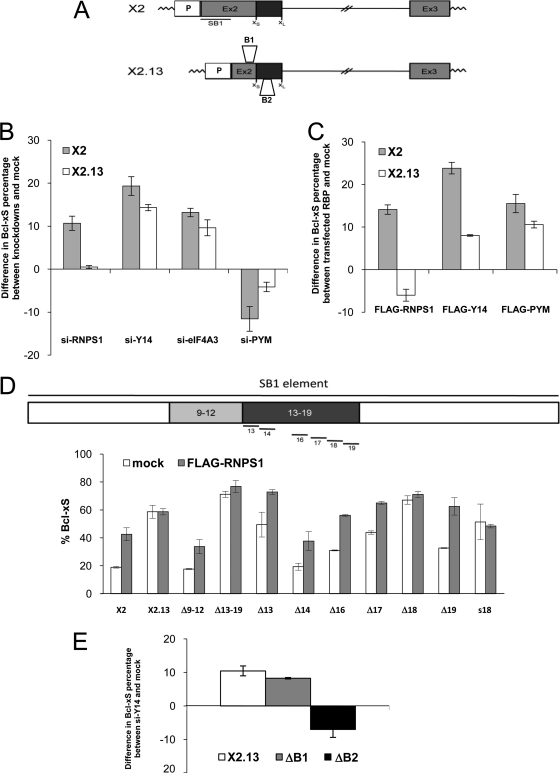
EJC components modulate the ratio of Bcl-x splice forms in different cell lines in a NMD-independent manner. (A) Diagram representing the structure of the Bcl-x gene and its splice variants. (B) Impact of depleting Y14 and RNPS1 in MDA-MB-231, PC-3, 293, and HeLa cells on the relative abundance of Bcl-x mRNA splice forms. The percentage of Bcl-xS is indicated below each lane. (C) RNAi assays were extended to other core and auxiliary EJC components. Two different siRNAs per gene (black and white bars) were transfected in 293 and HeLa cells. Seventy-two hours later, the impact of these depletions on the Bcl-x splicing profile was investigated. Experiments were done in triplicates, and standard deviations are shown. Western blot analyses are shown on the right. (D) The samples generated for panel C were investigated for changes in the expression of TIMM8B and ILK splice forms. Isoforms containing a premature stop codon (PTC) are indicated. (E) HeLa cells transfected with siRNAs targeting Upf1, Upf2, and Upf3b were investigated for changes in the expression of Bcl-x, TIMMB8, ILK, and CPNE2 splice variants.
The production of Bcl-xS is stimulated by ceramide, a regulator of stress response (37). One sequence element mediating this regulation is bound by SAP155, and activation of protein phosphatase 1 by ceramide has been proposed to dephosphorylate and inactivate SAP155 (11, 36). While hnRNP A1 can cooperate with Sam68 to improve the production of Bcl-xS, the phosphorylation of Sam68 by the Fyn kinase decreases the expression of Bcl-xS (53). An intronic region downstream of the Bcl-xL 5′ss is required to modulate the expression of Bcl-x splice variants when cells are treated with growth factors (34). We have identified four regions in Bcl-x exon 2 that contribute to splicing control (see Fig. S1 in the supplemental material). The B2 element is located downstream of the Bcl-xS 5′ss, where it interacts with the hnRNP F/H proteins to enforce splice site usage (18, 23). The B3 element enhances the use of the Bcl-xL 5′ss through SRp30c but also contains two pseudo-5′ss that antagonize splicing to the authentic site (13). B1 is located directly upstream of the proapoptotic Bcl-xS 5′ss and is a composite element made up of adjacent and overlapping enhancers and silencers. hnRNP K binds to B1 to repress the production of Bcl-xS (55). A fourth element (SB1) located further upstream in exon 2 also represses the production of Bcl-xS (23). In 293 cells, the activity of SB1 requires protein kinase C (PKC) activity (56), but the identities of proteins that bind to SB1 are currently unknown.
Here we report that many factors commonly associated with the exon junction complex (EJC) control Bcl-x splicing decisions. Their splicing-regulatory activity appears to be distinct from their usual EJC function because (i) core and auxiliary components regulate splicing through different cis-acting elements, (ii) proteins that normally associate with these EJC components to elicit mRNA export and nonsense-mediated mRNA decay (NMD) do not affect the production of Bcl-x isoforms, and (iii) individual EJC components preferentially associate with the Bcl-x pre-mRNA rather than the spliced product and can alter its alternative splicing in nuclear splicing extracts. Since lowering the levels of EJC components also favors the expression of proapoptotic splice variants in other genes, our results uncover an important new class of regulators of apoptosis acting at the splicing level.
MATERIALS AND METHODS
Cell lines and small interfering RNAs (siRNAs).
293 and HeLa S3 cells were grown in Dulbecco's modified Eagle's medium (DMEM). MDA-MB-231 were grown in DMEM with l-glutamine and 15% fetal bovine serum (FBS). The prostate cancer cell line PC-3 was cultivated in Ham's medium-nutrient mixture F12 with l-glutamine and 10% FBS.
The siRNAs used to knock down the expression of RNA binding proteins (RBPs) and EJC components were purchased from IDT (Coralville, IA), and their sequences are listed in Table S2 in the supplemental material. siRNAs were transfected into cells at a concentration that was never greater than to 100 nM using Lipofectamine 2000 (Invitrogen). Proteins and RNA were extracted from mock-transfected and siRNA-transfected cells at different time posttransfection, as indicated.
Plasmid transfections.
Bcl-x inserts of plasmids X2 and X2.13 were produced by PCR amplification using plasmids CMV-X2 and CMV-X2.13 (23) as templates, the Pfu-Turbo polymerase, and primers AscI-X-Fwd (GGCGCGCCTCACTATAGGGAGACCCAAGC-TGGCTAG) and X-Age-Rev (CTTACCGGTGGATCCCCCGGGCTGCAGGAATTCG-AT). The derived PCR products were cleaved with AscI and AgeI and ligated into SVEDA-HIV-2 vector (a kind gift of Alberto Kornblihtt, Buenos Aires, Argentina) cut with the same enzymes. Overlap PCR mutagenesis was used to generate deletions in the SB1 element (Δ9-12, Δ13-19, Δ13, Δ14, Δ16, Δ17, Δ18, and Δ19). For s18, substitution was performed by transversion mutation of each single nucleotide of region 18, replacing G with C and A with T. Plasmid transfections were carried out with polyethyleneimide (Polysciences Inc.). Conditions for transfection were as described previously (56), and transfection was performed 24 h after the application of siRNAs or involved cotransfection with plasmids allowing expression of FLAG-RNPS1, FLAG-Y14, and FLAG-PYM (kindly provided by Matthias Hentze, Heidelberg, Germany).
qPCR and Western blot analysis.
To assess the efficiency of the knockdown in the first RBP screen, we carried out quantitative real-time reverse transcription-PCR (RT-PCR) assays using SYBR green (2× Power SYBR green master mix; ABI 4367660) to assess transcript levels. The aldolase A gene (RTPrimerDB no. 915) was used as the housekeeping gene with the same samples. A total of 200 ng of RNA, measured for integrity (using the Agilent Lab-on-Chip station) and quantification (using the Thermo Scientific NanoDrop instrument), was reverse transcribed using random hexamers (Roche 11034731001) with Transcriptor reverse transcriptase (Roche 03531317) in a final volume of 10 μl. Ten nanograms of cDNA was used for the quantification in the presence of the specific primers at 0.2 μM in a 10-μl reaction mixture in triplicates. Reactions were carried out in the ABI 7500 quantitative PCR (qPCR) (Applied Biosystems) or Eppendorf Realplex instrument. A first cycle of 10 min at 95°C was followed by 40 cycles of 15 s at 94°C, 20 s at 55°C, and 20 s at 68°C. For later analysis of knockdowns and the testing of other EJC components, we used antibodies. The same blot was decorated with an antiactin antibody (Sigma A2066) to verify total protein contents in different lanes. Anti-RNPS1 antibodies were kindly provided by Akila Mayeda. Anti-eIF4A3 was kindly provided by Adrian Krainer. Antibodies against Magoh, Aly/Ref, and TAP were generous gifts from Elisa Izaurralde. Catherine Tomasetto kindly provided the antibody against MLN51. Antibodies against UAP56 and Y14 were purchased from Abnova and Abcam, respectively. Western blotting against poly(ADP-ribose) polymerase (PARP) was done using anti-PARP antibodies (Biosource AHF0262).
RT-PCR assays.
A set of 96 alternative splicing units from human apoptotic and cancer-related genes was selected from the AceView database. Sets of primers mapping in the exons flanking the sites of the alternative splicing events (ASEs) were designed using Primer3 with default parameters. For the global analysis of splice isoforms from apoptotic genes, total RNA was extracted using TRIzol and quantitated using the Lab-on-Chip station (Agilent Inc.). A total of 2 μg of RNA was reverse transcribed using a mix of random hexamers and oligo(dT) and the Omniscript reverse transcriptase (Qiagen, Germantown, MD) in a final volume of 20 μl. Twenty nanograms of cDNA was amplified with 0.2 U/10 μl of HotStar Taq DNA polymerase (Qiagen) in the buffer provided by the manufacturer, without extra MgCl2 and in the presence of the specific primers (IDT) for each splicing unit (at concentrations ranging from 0.3 to 0.6 μM) and deoxynucleoside triphosphates (dNTPs). The units, oligonucleotides, and expected sizes of RT-PCR products have been described previously (76, 77). Reactions were carried out in the GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA). A first cycle of 15 min at 95°C was followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C. The reaction was ended with an extension step of 10 min at 72°C. Visualization and analysis of amplified products were done using the LabChip HT DNA assay on an automated microfluidic station (Caliper, Hopkinton, MA) (30). Changes in the percentage of Bcl-xS were calculated relative to control transfections performed on the same day. All raw data can be consulted at http://palace.lgfus.ca/pcrreactiongroup/list/165 (user name, michelle_reviewers; password, Lae2010).
The splicing profile of Bcl-x was also assessed by RT-PCR. Reverse transcription was performed using the OmniScript RT kit (Qiagen) with random hexamers for endogenously derived Bcl-x mRNAs, while oligonucleotide RT-Sveda-Rev (GGGAAGCTAGAGTAAGTAG) was used for the plasmid-derived mRNAs. One-eighth of the cDNA material was used as a template for the PCR. Primers X3 (ATGGCAGCAGTAAAGCAAGCG) and X2 (TCATTTCCGACTGAAGAGTGA) were used to amplify fragments of splicing isoforms derived from endogenous Bcl-x, while primers X34 (AGGGAGGCAGGCGACGAGTTT) and X-Age-Rev (CTTACCGGTGGATCCCCCGGGCTGCAGGAATTCGAT) were used for plasmid-derived transcripts. For the conventional PCR, [α-32P]dCTP (Perkin-Elmer Canada Inc.) was added to PCR mixtures, and amplification products were fractionated onto 5% native polyacrylamide gels. Bands were revealed and quantified after scanning on a Storm PhosphorImager 860 (GE).
Amplification of TIMM8B, ILK, and CPNE2 was performed with primers TIMM8B-F (AAGCCGATGAAGCGGAGTT) and TIMM8B-R (CTACTGCCCTCCTTTCTGTAC), ILK-F (TTTTCACTCAGTGCCGGG) and ILK-R (CTCTGCCACTTGATCTTGG), and CPNE2-F (GTCTCCCATGAGTTTGCCAT) and CPNE2-R (CAGGAACTCCATGGCAGCG), respectively.
RNA immunoprecipitation assays.
Sepharose-protein A beads (GE) were incubated in radioimmunoprecipitation assay (RIPA) buffer (containing 0.5% Triton X-100) for 1 h at room temperature, washed three times in the same buffer, and then resuspended for a final volume of 50% beads. Total cell extract was prepared by lysing cells in 1 ml of RIPA buffer containing 100 U of RNase inhibitor (Invitrogen). After centrifugation at 13,000 rpm for 20 min at 4°C, the supernatants were collected and cleared by incubation with 100 μl of Sepharose-protein A beads (GE) for 1 h. This step was repeated under the same conditions but for 30 min. After centrifugation, the cleared supernatants were incubated with the antibodies overnight at 4°C. One hundred microliters of protein A-Sepharose beads was then added along with 0.5 mg of yeast tRNA and 100 U of RNase inhibitor, and the incubation was continued for 4 h at 4°C. The beads were washed two times with RIPA buffer containing 1 M urea and then three times with RIPA buffer and finally resuspended in 100 μl of RIPA buffer. After proteinase K treatment, RNA was extracted with phenol-chloroform-isoamyl alcohol and precipitated with ethanol. RNA precipitates were resuspended in 15 μl of water and treated with DNase I for 15 min at 37°C, and the material was used for RT-PCR analysis. One-third of the recovered RNA was reverse transcribed with primers Bcl-x-Ex3-R (5′-TCATTTCCGACTGAAGAGTGAGC-3′) and ILK-Ex4-R (5′-GTTACAGATGCTGACAAGGGC-3′). For the subsequent PCR amplification, two different reverse primers were used for both Bcl-x and ILK to discriminate the pre-mRNA from the mRNA. The oligonucleotide sequences were Bcl-x-Ex2-end-F (GTAAACTGGGGTCGCATTGTG) and Bcl-x-Int2-320-R (ACTTCCTACCCTCACAGGTTT) for Bcl-x pre-mRNA or Bcl-x-Ex3-R for Bcl-x mRNA and ILK-Ex2-F (TGAGATGTTGATCATGCGGG) and ILK-Int2-R (AGGCAGAAAGTACAGTCAGTTC) for ILK pre-mRNA or ILK-R (CTCTGCCACTTGATCTTGG) for ILK mRNA. PCR products were separated on a 2% agarose gel and visualized under UV light.
For immunoprecipitation of exogenous RNAs from nuclear extracts, equivalent quantities of radiolabeled transcripts were incubated 30 min at 30°C in 12.5 μl of splicing mix depleted of ATP by incubating nuclear extracts for 20 min at room temperature (15). Forty microliters of a 50% slurry of antibody-bound protein A-Sepharose beads was added and incubated for 30 min with the splicing mixture at 4°C. The beads were washed three or four times with NET-2 buffer containing 0.05% NP-40. The cpm at input and recovered after the washes were measured and normalized to cpm remaining on control IgG coated-beads.
In vitro splicing assays.
Plasmid S2.13 was cut with XbaI and plasmid 45 was cut with ScaI and transcribed with T3 RNA polymerase in the presence of a cap analog and [α-32P]UTP (Perkin Elmer Life Sciences). The pre-mRNA was purified as described previously (23). A HeLa nuclear extract (17) was preincubated for 10 min at 30°C with recombinant His-tagged eIF4A3 or Magoh-Y14ΔN (3) produced using standard protocols (66). Two femtomoles of transcript was then added to the mixture under standard splicing conditions (13) and incubated for 2 h at 30°C. Reverse transcription for Bcl-x was carried out using the X2b primer (CGCTCTAGAACTAGTGGATC) and Omniscript (Qiagen). PCRs were then performed in the presence of [α-32P]dCTP using Taq DNA polymerase (NEB) and primers X2b and X3 (ATGGCAGCAGTAAAGCAAGCG). Amplification products were separated on a 5% acrylamide-bisacrylamide (29:1) native gel and quantitated on a PhosphorImager (Molecular Dynamics). Reverse transcription for plasmid 45 was performed as described previously (35).
UV cross-linking.
To produce labeled transcripts, DNA templates for transcription were produced by PCR using a forward primer containing the T7 promoter sequence and sequence upstream from the Bcl-xS 5′ splice site, a reverse primer complementary to the first nucleotides of intron 2 of Bcl-x, and plasmids corresponding to wild-type (WT) Bcl-x and the ΔB2 derivative. Transcription with T7 RNA polymerase was performed in the presence of [α-32P]UTP (3000 Ci/mmol; Perkin-Elmer). Reaction mixtures containing 400,000 cpm of radiolabeled wild-type or ΔB2 transcripts, 2.5 μg of recombinant eIF4A3 or 10 μl of HeLa nuclear extract in the presence of MgCl2 (final concentration of 4 mM), dithiothreitol (DTT) (2.5 mM), and buffer D were incubated on ice for 30 min. Samples were transferred on an ice-cold surface and immediately UV irradiated (350 mJ) in a Stratalinker apparatus (Stratagene). UV-cross-linked samples were treated with RNase A (final concentration of 2 mg/ml) for 30 min at 37°C. One-quarter of the reaction mixture was used as input, while the rest was immunoprecipitated using a monoclonal anti-eIF4A3 antibody (a generous gift of A. Krainer), as described above. Both input and immunoprecipitated samples were boiled for 3 min in 2× SDS-PAGE loading buffer and run on a 10% SDS-polyacrylamide gel. Cross-linked profiles were revealed on a Storm PhosphorImager 860.
Gel shift assay.
Two femtomoles of radiolabeled RNAs (6,000 cpm) was incubated for 25 min at 30°C in 6 μl of a binding mixture containing 25 mM MES (morpholineethanesulfonic acid)-KOH (pH 6.0), 150 mM potassium acetate, 2 mM MgCl2, 4 mM DTT, 2 U of RNase inhibitor, and 25 μM ATP-γ-S (Sigma-Aldrich), supplemented or not with 1 μg of His-tagged eIF4A3, prior to the addition of 2.5 μl of dye containing 50 μg of heparin. The reactions were run on a 4.5% native acrylamide gel (29:1 acrylamide-bisacrylamide, 5% glycerol, 50 mM Tris [pH 8.8], 50 mM glycine) in Tris-glycine running buffer (50 mM Tris [pH 8.8], 50 mM glycine) for 3 h at 150 V and visualized by phosphorimaging after gel drying.
Apoptotic assays.
Cells were stained at 96 h posttransfection with 1:350 annexin V-Alexa Fluor 647 (Invitrogen) and 800 ng/ml of Hoechst 33342 (Invitrogen) in an annexin binding buffer (pH 7.4) composed of 10 mM HEPES, 140 mM NaCl, and 2.5 mM CaCl2. High-content endpoint fluorescence images were acquired on a BD Pathway 855 high-content BioImager (BD Biosciences) using a 20× objective with a laser-based autofocus and excitation and emission filters appropriate for each fluorescent dye. Thresholds were applied manually to identify annexin V-positive cells.
RESULTS
Components more prevalently associated with EJC function affect the expression of Bcl-x splice variants.
To identify proteins that regulate the production of the Bcl-xS and Bcl-xL mRNA splice forms, we used RNA interference (RNAi) to knock down 61 RNA binding proteins (RBPs) selected from different groups of proteins (hnRNPs, SR and other splicing regulators, constitutive splicing factors, and a variety of RRM-containing proteins implicated in other aspects of RNA processing) (see Table S1 in the supplemental material). To minimize potential off-target effects, we used two distinct and nonoverlapping siRNAs per RBP (see Table S2 in the supplemental material). Depletion efficiencies were determined using real-time quantitative RT-PCR (qRT-PCR), and only depletions of at least 50% were considered. We selected the prostate cancer PC-3 and breast cancer MDA-MB-231 cell lines for this assay because we used them previously for the successful knockdown of Fox2 and several hnRNP proteins (76, 77). We define a hit as a change of greater than 10% in the level of the Bcl-xS product obtained with the two siRNAs in two cell lines. The 10% change in splicing provides a Z score above 1.5, which represents 1.5 standard deviations above or below the average variation observed for thousands of controls performed for all splicing units in a variety of cell lines (77). This 10% threshold makes the assay more apt to detect increases rather than decreases in the level of Bcl-xS, since endogenous levels of the Bcl-xS isoform usually range between 5 and 15%.
As shown in Table S1 in the supplemental material, our analysis confirmed a role for hnRNP K in the production of Bcl-x splice variants (55). hnRNPs F and H are known regulators of Bcl-x splicing (23) but did not score as hits here because their knockdowns promoted a reduction in Bcl-xS that was below the 10% cutoff. In contrast, Sam68 and hnRNP A1, which have been described as upregulators of Bcl-xS, had no impact, but this may reflect cell line specificity since the original study was done using HEK293 cells (53). RNPS1 and Y14 were the only other RBPs whose depletion increased the level of Bcl-xS by more than 10% in the two cell lines (Fig. 1B). Similar shifts were observed by depleting RNPS1 and Y14 in 293 and HeLa cells (Fig. 1B). Because our previous work on Bcl-x was performed mostly with 293 and HeLa cells, we used these cells for subsequent analyses.
Y14 and RNPS1 are components of the exon junction complex (EJC); Y14 is part of the core EJC with eIF4A3 and Magoh, while RNPS1 is an auxiliary factor (32). We asked if other core EJC components also affected Bcl-x splicing. Knocking down eIF4A3 increased the relative expression of Bcl-xS in HeLa and 293 cells (Fig. 1C; see Fig. S2 in the supplemental material). We attempted the depletion of Magoh with at least three different siRNAs but obtained only marginal (less than 50%) knockdown efficiencies. Nevertheless, under these conditions we noted consistent increases in Bcl-xS levels, which varied between 5 and 10% (not shown). Likewise, depleting SAP18 and Acinus, which like RNPS1 are auxiliary components of the EJC (70), increased the level of Bcl-xS (Fig. 1C; see Fig. S2 in the supplemental material).
The change in Bcl-x mRNA splice forms mediated by the depletion of EJC components could be due to different mechanisms. Since mRNA export factors (Aly/Ref, UAP56, p15, and TAP) are recruited by the EJC (32, 82), an export defect may unbalance the production of Bcl-x isoforms. To address this possibility, we knocked down TAP, Aly/Ref, and UAP56 in HeLa cells. Although we have not verified if the efficient depletion that was achieved in all cases was sufficient to compromise mRNA export, the impact on the production of Bcl-x variants was minimal (no effect or less than 5% in most cases) (Fig. 1C), suggesting that defective mRNA export was unlikely to be the major cause for the increase in Bcl-xS when eIF4A3, Y14, RNPS1, SAP18, and Acinus are depleted.
Although Bcl-xS and Bcl-xL share the same stop codon, EJC components may recruit NMD effectors specifically on the Bcl-xS mRNA to reduce its level. Alternatively, NMD may affect the relative levels of Bcl-x splice forms indirectly by changing the levels of alternative splicing regulators that are controlled by NMD (8, 31, 46, 50, 59, 79). We considered these possibilities unlikely because the depletion of EJC components was not sufficient to alter the level of the splice variants that contained a premature termination codon (PTC) from TIMM8B and ILK (Fig. 1D). In addition, the knockdown of eIF4A3, Y14, and RNPS1 did not promote major changes in the expression of known splicing regulators of Bcl-x (hnRNP A1, K, F/H, and SRp30c) based on Western blot analyses performed with HeLa and 293 cells (see Fig. S3 in the supplemental material). To conclusively assess the contribution of the NMD pathway, we tested the impact of depleting Upf1, Upf2, and Upf3b in HeLa cells. Upf1 is also required for the Staufen 1-mediated and replication-dependent histone mRNA turnover pathways (reviewed in reference 29). For this assay we obtained HeLa RNA samples used previously to report the global effects of Upf1, Upf2, and/or Upf3b on a variety of alternative splice isoforms (59) (kindly provided by B. Blencowe and L. Maquat). Less than 5% of Upf proteins remain in these treated cells (59). Using these samples, we confirmed that the PTC-containing mRNA isoforms from the TIMM8B, ILK, and CPNE2 genes were stabilized (Fig. 1E). Notably, the relative level of the Bcl-x splice forms remained unchanged in the same samples (Fig. 1E), a strong indication that NMD does not directly or indirectly alter the relative level of the Bcl-x mRNA isoforms expressed within this experimental time frame. Also, the knockdown of the predominantly cytoplasmic protein MLN51 (also called CASC3) (confirmed to be superior to 50% by Western blot analysis and qRT-PCR) did not alter Bcl-x splicing (Fig. 1C; see Fig. S2 in the supplemental material). Since MNL51 has been proposed to bridge the EJC to Upf1 (24), this result supports the view that the change in Bcl-x splice variants elicited by many EJC components is NMD independent.
A change in Bcl-x splice forms may also result from a difference in cytoplasmic stability that is unrelated to NMD. To address this possibility, we monitored the Bcl-x ratio of splice variants after blocking transcription with actinomycin D and 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (see Fig. S4 in the supplemental material). These treatments antagonized the shift toward Bcl-xS induced by depleting RNPS1 and Y14, indicating that a change in the relative levels of Bcl-x splice variants requires active transcription. Thus, differential cytoplasmic stability does not appear to make a major contribution in changing the levels of the Bcl-x mRNA splice variants when RNPS1 and Y14 are depleted.
Overall, our results suggest that several proteins associated with EJC function may also regulate Bcl-x splicing. RNPS1, SAP18, and Acinus have already been implicated in a few cases of splicing modulation in vivo or in vitro (38, 58, 60, 67). Notably, however, the auxiliary EJC components UAP56 and SRm160, which have been functionally associated with splicing before being implicated in EJC function (4, 22, 62), did not alter the Bcl-x splicing profile when depleted (Fig. 1C and data not shown). More recently, two studies reported that core components of the EJC could regulate the generic splicing of mitogen-activated protein kinase (MAPK) and other long-intron-containing transcripts in Drosophila (1, 57). Thus, EJC factors may accomplish multiple functions to coordinate different processes. It is unlikely, however, that the EJC-mediated regulation of Bcl-x is caused by the large size of the intron downstream of exon 2 (greater than 55 kb in humans) because, as will be shown below, a minigene carrying a shortened intron of 1.3 kb can mimic the response of endogenous Bcl-x when the levels of EJC components are altered.
RNPS1 and core EJC proteins control Bcl-x splicing through distinct cis-acting elements.
The depletion of EJC components stimulates the production of Bcl-xS, suggesting that these factors normally repress the use of the Bcl-xS 5′ splice site. The 361-nucleotide (nt)-long cis-acting repressor sequence SB1 is located approximately 200 nt upstream of the Bcl-xS 5′ splice site (Fig. 2A), but the identity of the factor(s) that mediates this repression is unknown (56). To determine whether RNPS1, Y14, and eIF4A3 act through the SB1 element, we tested the impact of their depletions using Bcl-x minigenes containing or lacking SB1 (X2 and X2.13, respectively). As shown in Fig. 2B and in Fig. S5 in the supplemental material, depleting RNPS1 in 293 cells shifted splicing toward the production of Bcl-xS on X2 but not on X2.13 transcripts, indicating that RNPS1 requires the SB1 portion for activity. This SB1 requirement was confirmed by expressing recombinant FLAG-RNPS1 (kindly provided by M. Hentze), which shifted Bcl-x splicing on X2 but had a slight opposite effect on X2.13 (Fig. 2C; see Fig. S5 in the supplemental material). Notably, the impact of expressing FLAG-RNPS1 was similar to that of a depletion of RNPS1. This dominant negative effect is unlikely to be caused by the FLAG tag, since a similarly tagged RNPS1 displayed wild-type activity (73). On the other hand, overexpression, by changing the relative stoichiometry of a component that is part of a complex, can titrate or sequester components of the complex. Overexpression can also alter the mechanism of assembly of a complex. In all cases, this can lead to dominant negative effects (9, 74).
Fig 2.
EJC components require distinct cis-acting elements to modulate Bcl-x pre-mRNA splicing. (A) Structures of the Bcl-x minigenes. X2 differs from X2.13 by the presence of the SB1 element (underlined, 361 nt) located at the 5′ end of exon 2 (Ex2) (56). Derivatives of X2.13 lacking the 50-nucleotide-long B1 or the 77-nucleotide-long B2 element are also shown (23). P, promoter. (B) RT-PCR analysis of minigene expression was carried out in 293 cells treated with siRNAs targeting RNPS1, Y14, eIF4A3, and PYM, followed by transfection of the Bcl-x minigenes. (C) RT-PCR analysis of Bcl-x expression in 293 cells was carried out after cotransfecting FLAG-RNPS1, FLAG-Y14, and FLAG-PYM constructs with the Bcl-x minigenes. (D) Diagram of the SB1 element and of the various 40-bp (Δ9-12), 70-bp (Δ13-19), and 10-nt (Δ13 to Δ19) deletion mutants tested. Plasmid transfection was performed in 293 cells in the absence or presence of the FLAG-RNPS1 plasmid. To illustrate the impact of the mutant and that of RNPS1 coexpression, Bcl-x splicing is expressed as the percentage of the Bcl-xS isoform. s18 represents a substitution mutation with the sequence 5′-CTTCTCTTGT-3′. (E) RT-PCR analysis of Bcl-x alternative splicing in 293 cells transfected with an siRNA targeting Y14 and then with minigene X2.13 and versions lacking the B1 or B2 element. RT-PCR was performed to amplify the Bcl-x isoforms produced from the minigenes. Bar graphs in panels B, C, and E show the positive or negative difference in the production of Bcl-xS in cells where the expression of specific EJC components was increased or decreased relative to that in mock-treated cells.
To better define the portion of SB1 that confers regulation by RNPS1, we tested a variety of deletion and substitution mutants. We attempted to perform these experiments in the context of an RNPS1 depletion. However, since our analysis with 293 cells produced a 10% differential between knockdown and mock for X2 and X2.13 (Fig. 2B), it was difficult to extract statistically significant results from intermediate values (data not shown). Instead, we relied on overexpressing RNPS1, which provides a more robust differential (20%) (Fig. 2C). Mutants Δ9-12 and Δ13-19 lack adjacent 40- and 70-nt portions in SB1 (Fig. 2D). While the production of Bcl-xS was improved with Δ13-19, FLAG-RNPS1 increased Bcl-xS usage on the Δ9-12 mutant but had little impact on the Δ13-19 mutant (Fig. 2D). Six 10-nt deletion mutants were next produced from the 13-19 region of SB1; Δ16, Δ17, Δ18, and Δ19 all individually stimulated the production of Bcl-xS, and only Δ18 was refractory to a further increase when FLAG-RNPS1 was coexpressed (Fig. 2D). Because an RNPS1 effect may be obscured by the strong impact of the Δ18 mutation, we tested a substitution mutation for the same region (s18), whose effect on Bcl-x splicing was not as strong as Δ18. Coexpressing FLAG-RNPS1 did not improve the production of Bcl-xS on s18, suggesting that region 18 plays an important role in mediating the activity of RNPS1.
In contrast, the depletion of Y14 and eIF4A3 in 293 cells modulated the splicing of both X2 and X2.13 (Fig. 2B; see Fig. S5 in the supplemental material). As a further indication that SB1 is not essential for the activity of EJC core components, we observed that knocking down PYM, a protein that sequesters Y14 (25), increased the relative level of endogenous Bcl-xL (not shown), and this increase was also seen when the knockdown of PYM was tested on the Bcl-x minigenes X2 and X2.13 (Fig. 2B; see Fig. S5 in the supplemental material). Expressing FLAG-PYM (a kind gift of M. Hentze and N. Gehring) stimulated the expression of Bcl-xS on both X2 and X2.13 (Fig. 2C; see Fig. S5 in the supplemental material). Although the impact of expressing FLAG-Y14 (provided by N. Gehring and M. Hentze) was stronger on X2, FLAG-Y14 was clearly active in the absence of SB1 (Fig. 2C; see Fig. S5 in the supplemental material). Similar to the case for RNPS1, the FLAG-Y14-induced splicing shift occurred in the same direction as the knockdown, suggesting a titration effect and its activity as part of a complex. To identify a region on the Bcl-x pre-mRNA required for the activity of Y14, we tested deletion mutants (Fig. 2A) (23). Removing the B1 element did not prevent the Y14-associated increase in Bcl-xS, but deleting the 77-nt-long B2 element abrogated this increase and favored the expression of Bcl-xL (Fig. 2E).
Thus, different sequences mediate the activity of EJC components; RNPS1 requires region 18 in the SB1 element, while Y14 requires the B2 element located nearly 300 nt downstream of region 18. Notably, region 18 and B2 are not directly upstream of a 5′ splice site, at the position where EJCs would normally be deposited. Although the exact mechanism of regulation remains to be investigated, our results suggest that splicing activity requires distinct complexes whose positions are significantly different from the established mode of assembly of the EJC.
EJC components associate with the Bcl-x pre-mRNA and modulate splicing.
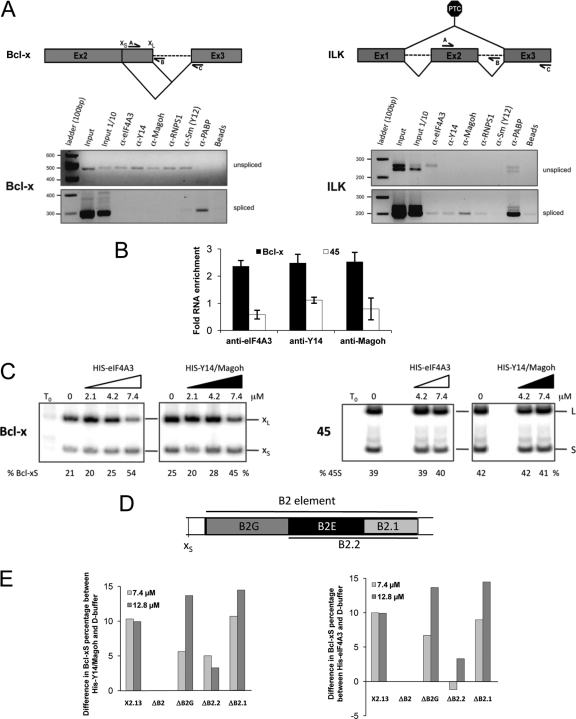
Our results suggest a direct role for EJC components in splicing regulation. If so, they should interact with the Bcl-x pre-mRNA. To determine if we could detect their association with the endogenous Bcl-x pre-mRNA, we performed immunoprecipitation assays under stringent conditions (incubation in the presence of 0.5 mg of tRNA and washing buffer containing 1 M urea) using a 293 whole-cell extract and antibodies against various EJC components. RT-PCR assays on the recovered material were then performed to identify intron-containing and spliced portions of endogenous Bcl-x and the PTC-containing ILK transcripts. In the case of ILK, except for eIF4A3, the unspliced product was not recovered with antibodies against EJC components. In contrast, the association of eIF4A3, Y14, Magoh, and RNPS1 was detected on spliced ILK (Fig. 3A). The situation was reversed for Bcl-x, since in the same recovered samples, antibodies directed at the eIF4A3, Y14, Magoh, RNPS1, and Sm proteins of the snRNP proteins, but not against cytoplasmic poly(A) binding protein (PABP), recovered the intron-containing transcript but not the spliced product (Fig. 3A). Overall, these results indicate that EJC components associate with the Bcl-x pre-mRNA, consistent with their proposed roles as regulators of Bcl-x splicing.
Fig 3.
EJC components associate with the Bcl-x pre-mRNA and modulate its alternative splicing. (A) The structures of the Bcl-x and ILK pre-mRNA are diagrammed, and the positions of the primers used to amplify intron-containing (A-B pairs) or intron-lacking (A-C pairs) transcripts are shown. RT-PCR analysis was carried out after immunoprecipitating the RNA using a variety of antibodies in 293 total cell extracts. Sepharose-protein A beads linked to antibodies against the eIF4A3, Y14, Magoh, RNPS1, and Sm proteins (Y12) and PABP were added to extracts. Bound RNA was recovered after extensive washing and treatment with proteinase K and was analyzed by RT-PCR using primers specific to Bcl-x or the PTC-containing ILK splicing unit. (B) Following incubation of Bcl-x and control hnRNP A1-derived (45) in vitro-transcribed RNA in a HeLa nuclear extract, labeled RNA was immunoprecipitated with anti-eIF4A3, anti-Y14, and anti-Magoh antibodies. Incubation was performed in splicing mixtures depleted of ATP. The amount of immunoprecipitated RNA was quantitated and plotted to show the fold difference of precipitated material relative to control beads, with a value of 1 indicating no difference. (C) In vitro splicing assay in HeLa nuclear extracts. Splicing assays were performed with 2 fmol of the Bcl-x pre-mRNA S2.13 (13) or RNA 45 (derived from hnRNP A1 [45]) in the presence of increasing amounts of His-tagged recombinant eIF4A3 or Magoh-Y14ΔN (3). (D) Diagram of the B2 element and the subregions that define the ΔB2G, ΔB2.2, and ΔB2.1 deletion mutants. (E) In vitro splicing assays performed as for panel C using the recombinant Y14-Magoh mixture (left panel) or the recombinant eIF4A3 (right panel). The Bcl-x pre-mRNAs tested are indicated, as well as the final concentrations of recombinant proteins. Bar graphs show difference in the production of Bcl-xS in Y14-Magoh- or eIF4A3-supplemented extracts relative to extracts receiving only buffer D.
The core proteins eIF4A3, Y14, and Magoh are components of late spliceosomal C and activated B complexes but not the earlier A complexes (42). However, on the Bcl-x pre-mRNA, these interactions may occur earlier to regulate splice site choice. To address this possibility, we performed immunoprecipitation assays with HeLa nuclear extracts supplemented with our model Bcl-x pre-mRNA S2.13 (13). S2.13 is identical to X2.13 (Fig. 2A) except that it is synthesized in vitro using T3 RNA polymerase. We performed the immunoprecipitation using an extract depleted of ATP (15). Under these conditions, we detected the association of EJC components with the Bcl-x pre-mRNA, while the recovery of a control pre-mRNA (45 RNA [45]) was similar to the level obtained with no antibodies (Fig. 3B). The same results were reproduced when ATP was depleted from the nuclear extract by hexokinase treatment as recommended (43) (data not shown). Thus, EJC components can interact specifically with a model Bcl-x pre-mRNA under conditions where spliceosome assembly is compromised.
To test if EJC components can modulate the splicing of Bcl-x, we incubated S2.13 in HeLa nuclear extracts supplemented with purified recombinant eIF4A3 or a mixture of recombinant heterodimer Magoh-Y14. The relative abundance of the Bcl-x splice forms was determined by RT-PCR following 2 h of incubation (13). The addition of eIF4A3 and Magoh-Y14 shifted splicing in favor of Bcl-xS, thereby reproducing the impact of FLAG-Y14 in vivo (Fig. 3C). These shifts were consistently observed in several independent experiments and were specific since no shift occurred when the recombinant proteins were tested on the unrelated control pre-mRNA 45 harboring competing 5′ss (Fig. 3C). While the amount of recombinant proteins added was elevated, micromolar levels correspond to the concentration of splicing regulators such as SRp30c and PTB in our splicing extracts (52). These results therefore show that core components of the EJC can specifically affect the alternative splicing of Bcl-x in vitro.
Our ability to reproduce in vitro the impact of components of the EJC core offered an opportunity to better delineate the regulatory sequence on the Bcl-x pre-mRNA using a set of mutant transcripts used previously in splicing extracts (23). First, we observed that the impact of recombinant eIF4A3 on Bcl-x splicing in vitro required the B2 element, because the shift toward Bcl-xS was lost completely when B2 was deleted (Fig. 3E; see Fig. S6A in the supplemental material), mimicking the in vivo results with Y14 (Fig. 2E). Second, we tested three deletion mutants (ΔB2G, ΔB2.2, and ΔB2.1) (Fig. 3D) used previously to show that hnRNP F modulates Bcl-x splicing through the B2G region (23). Here, B2G was not essential for the Y14-Magoh-induced shift, but the flanking 26-nt B2E region appeared to be critical (Fig. 3E, left panel; see Fig. S6A in the supplemental material). Identical results were obtained with eIF4A3 (Fig. 3E, right panel; see Fig. S6A in the supplemental material), suggesting that eIF4A3, Y14, and Magoh might form a complex on B2E to regulate Bcl-x splicing.
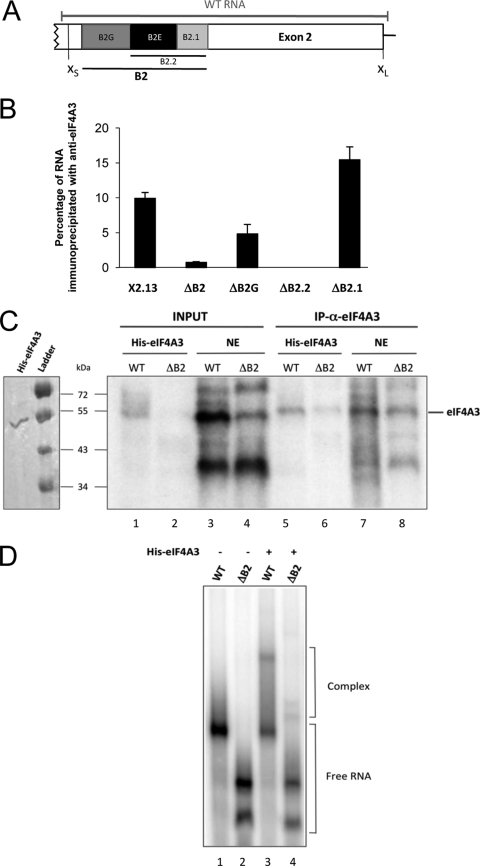
We tested the eIF4A3/B2E interaction by performing an anti-eIF4A3 immunoprecipitation with splicing extract using mutated transcripts (Fig. 4B). The assay shows that RNA recovery was most severely compromised when B2E was lacking. The same conclusion was reached when using an anti-Y14 antibody (see Fig. S6B in the supplemental material). The fact that eIF4A3 and Y14 require the same element for activity and interaction suggests that they act as part of a complex that may also include Magoh. In contrast to the case for Y14 and eIF4A3, which act through the B2E element, RNPS1 activity required the SB1 region, located several hundreds of nucleotides upstream. We took advantage of the RNA immunoprecipitation assay to ask whether we could detect in a nuclear extract an interaction between RNPS1 and this region of the Bcl-x pre-mRNA. As shown in Fig. S6C in the supplemental material, recovery of the transcript required SB1. To test if the eIF4A3 interaction with the Bcl-x RNA might be direct, we carried out a UV cross-linking assay using recombinant His-tagged eIF4A3 incubated with a 32P-labeled transcript containing the B2 element (WT) (Fig. 4A) and a derivative lacking it (ΔB2). The assay with WT RNA revealed a protein of the size of eIF4A3 whose intensity was reduced considerably when the B2 element was absent (Fig. 4C, lanes 1 and 2). As expected, these products were recovered with the anti-eIF4A3 antibody (lanes 5 and 6). Likewise, a band comigrating with recombinant eIF4A3 was detected when WT RNA was used for cross-linking in a HeLa nuclear extract, and the intensity of this band was strongly reduced when ΔB2 RNA was used (Fig. 4C, lanes 3 and 4). These proteins were recovered with the anti-eIF4A3 antibody (lanes 7 and 8). These results indicate that eIF4A3 interacts directly with the Bcl-x transcript through the B2 element. Gel shift assays carried out with recombinant proteins confirmed that complexes were formed when a B2-containing WT RNA was incubated with recombinant eIF4A3 (Fig. 4D, lanes 1 and 3). In contrast, the efficiency of complex formation was reduced when ΔB2 RNA was used (lanes 2 and 4). Attempts to detect an RNPS1 cross-link product in a HeLa extract remained negative, and our inability to produce recombinant RNPS1 prevented us from carrying out gel shift assays.
Fig 4.
eIF4A3 interacts directly with the Bcl-x pre-mRNA via the B2 element. (A) Diagram of the alternative exonic region located between the alternative Bcl-x 5′ splice sites. The B2 element is depicted with its subregions B2G, B2.2, and B2.1. The segment designated WT RNA represents the in vitro-transcribed RNA used for panels C and D. (B) Following the incubation of the various Bcl-x transcripts in a HeLa nuclear extract, labeled RNA was immunoprecipitated with anti-eIF4A3. The percentage of immunoprecipitated labeled RNA (relative to input and subtracted from that for a mock immunoprecipitation performed with each pre-mRNA) is plotted. (C) Cross-linking assays were performed in the presence of labeled WT or ΔB2 transcripts incubated for 30 min on ice with recombinant His-eIF4A3 or a HeLa nuclear extract, followed by UV irradiation and immunoprecipitation with anti-eIF4A3. Samples were fractionated on a 10% polyacrylamide-SDS gel. The left panel displays a Coomassie blue-stained gel of our purified His-eIF4A3 protein. (D) Labeled WT or ΔB2 transcripts were incubated for 25 min at 30°C in the presence (+) or the absence (−) of 1 μg recombinant protein His-eIF4A3. Complexes were separated on a native 4.5% acrylamide gel.
Other apoptotic splicing events regulated by EJC components.
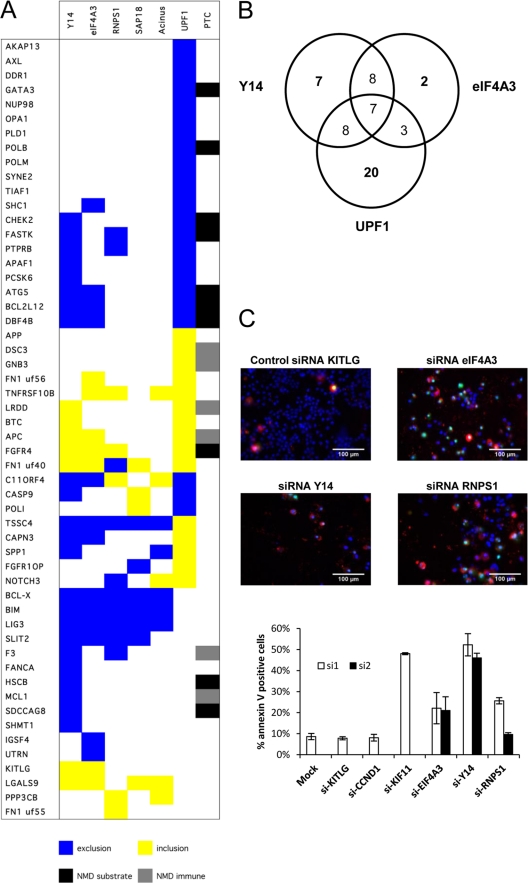
To identify other genes similarly regulated by EJC components, we monitored 96 alternative splicing events (ASEs) in apoptotic and cancer-related genes using a high-throughput RT-PCR platform (30, 64, 75–77). We looked for events that, like for Bcl-x, would be affected by depleting EJC components but not Upf1. Of the 54 ASEs that reacted to at least one knockdown, 16 did not react to a depletion of Upf1, including 5 ASEs (in Bcl-x, Bim, Lig3, Slit2, and Lgals9) that shifted with 4 or all EJC knockdowns (Fig. 5A). In the case of Bim (a Bcl-2 family member also known as Bcl2l11), the splicing shift produces an isoform with more potent apoptotic activity (48). The depletion of Y14 also affected Mcl1, another Bcl-2 family member. The Mcl1 splicing shift encourages the production of the shorter proapoptotic isoform Mcl1S, whereas the longer Mcl1L variant is antiapoptotic (2). Thus, the depletion of several EJC components coregulates the alternative splicing of at least three Bcl-2 family members by promoting the synthesis of proapoptotic splice variants.
Fig 5.
EJC components control a network of splicing events linked to apoptosis. (A) RT-PCR analysis monitoring the alternative splicing of 54 units following the knockdown of EJC components and Upf1 in HeLa cells. Total RNA was extracted from HeLa cells at 72 h after transfection with siRNAs. The colored boxes represent significant shifts (minimum 10%) in the percent splicing index (PSI). The “PTC” column indicates the presence of a premature termination codon introduced by alternative splicing by either inclusion or exclusion. If the PTC is located more than 50 nucleotides upstream of an exon-exon junction, it is predicted to be a target for NMD and hence the event is indicated as a black box (NMD substrate). If the PTC is located within the 50 nucleotides upstream of the exon-exon junction or downstream of the last exonic junction, the transcript is assumed to escape NMD (NMD immune) and is indicated as a gray box. (B) Venn diagram showing the numbers of transcripts regulated by Y14, eIF4A3, and UPF1. Transcripts are considered coregulated by two or three of these proteins only if the splicing ratio shifts in the same direction upon depletion of the proteins. (C) Annexin V assay in HeLa cells transfected with siRNAs targeting EJC components. Cells were immunostained for annexin V (red), and nuclei were labeled with Hoechst stain (blue) (69). The percentage of apoptotic cells was quantitated (bottom graph) upon transfection of siRNAs targeting eIF4A3, Y14, and RNPS1 (2 siRNAs each) and compared to that for various controls (Lipofectamine alone, negative-control siRNAs targeting KITLG and cyclin D1, and a positive-control siRNA targeting KIF11 [71]).
Depleting EJC components also elicited isoform shifts in the direction opposite to the one observed when Upf1 was depleted, also suggesting splicing regulation. For a few units in this category (Tssc4, Capn3, and Spp1), at least two EJC components produced this effect. Finally, several ASEs responded similarly to a depletion in Upf1, Y14, and eIF4A3, likely representing direct or indirect NMD effects. Transcript-specific differences may arise because the depletions were partial and the threshold concentration for optimal assembly of a functional EJC may vary for different transcripts.
Depleting EJC components in 293 cells confirmed the behavior of ASEs in Mcl1, Lig3, Capn3, Tssc4, Slit2, Fanca, Kitlg, and Lgals9 (data not shown).
The splicing changes imposed by depleting individual EJC components favored the production of at least three proapoptotic isoforms. The splicing shifts that occurred are likely to be biologically relevant because (i) we could detect corresponding shifts in protein variants (see Fig. S7 in the supplemental material), (ii) the 10 to 30% increases in Bcl-xS that we have obtained by knocking down EJC components are within the range of changes that have associated Bcl-xS with the promotion of apoptosis or the sensitization of cells to chemotherapeutic agents (41), (iii) the depletion of Y14, RNPS1, and eIF4A3 stimulated the caspase-mediated cleavage of PARP in 293 and HeLa cells (see Fig. S8A and S8B in the supplemental material), and (iv) the knockdown of eIF4A3, Y14, and RNPS1 promoted apoptosis, as judged by staining for the apoptotic marker annexin V in HeLa and PC-3 cells (Fig. 5C; see Fig. S9 in the supplemental material). This apoptotic signature is consistent with the observation that the depletion of eIF4A3 and Y14, respectively, decreases cell growth (63) and promotes apoptosis in mouse mesothelioma cells (69). Depleting other RNA binding proteins that did not modulate Bcl-x splicing did not elicit apoptosis (see Table S1 and Fig. S9 in the supplemental material).
In a separate study, we have assessed the impact of specifically depleting the large antiapoptotic isoform of Mcl1 compared to both the long and the short proapoptotic variants. Interestingly, the siRNA-mediated depletion of the long Mcl1 isoform promoted PARP cleavage and apoptosis, whereas this phenotype was reduced considerably when both isoforms were depleted (54). These results suggest that the splicing shifts that favor the production of proapoptotic variants have an important phenotypic impact.
Since the activation of caspases can alter the integrity of splicing-regulatory components, which in turn can affect alternative splicing (10, 16, 21), we assessed whether the Bcl-x splicing shift was caspases dependent. 293 and HeLa cells were pretreated with z-VAD-fmk, a pan-caspase inhibitor that compromises PARP cleavage. z-VAD-fmk had no basal effect on Bcl-x splicing and, importantly, did not antagonize the switch in Bcl-x splicing induced by the depletion of RNPS1 and Y14 (see Fig. S8C in the supplemental material). This result indicates that the Bcl-x splicing shift is independent of the activation of caspases, consistent with the notion that individual EJC components or subcomplexes directly affect the alternative splicing of Bcl-x.
DISCUSSION
Splicing decisions affecting Bcl-x have a profound impact on cell fate, since Bcl-x splice variants are antagonistic and can either promote or inhibit cell death. An RNAi screen aimed at further exploring Bcl-x splicing regulation identified eIF4A3, Y14, RNPS1, SAP18, and Acinus as controlling the relative proportions of Bcl-x splice variants. Although these proteins are all components of the exon junction complex (EJC), which is deposited on the mRNA concomitantly with splicing to coordinate mRNA export and surveillance, the depletion of other EJC-associated proteins implicated in mRNA export (UAP56, Aly/Ref, and TAP) and mRNA surveillance (MLN51 and the Upf proteins) did not alter the relative levels of the Bcl-x splice variants.
A more direct participation of these proteins in splicing control is also supported by the observations that RNPS1, eIF4A3, Y14, and Magoh associated more prevalently with the endogenous Bcl-x pre-mRNA than with spliced products and that recombinant versions of these proteins specifically altered Bcl-x splice site selection in splicing extracts.
One intriguing question is whether the splicing-modulatory activity of these proteins is in any way associated with their function as components of the EJC. While the EJC is deposited on the mRNA at the time of splicing, the core proteins eIF4A3, Y14, and Magoh are found in late spliceosomal C and activated B complexes but not in earlier A complexes (42). A pre-EJC complex made up of eIF4A3, Magoh, and Y14 is most likely assembled before exon ligation, providing a binding platform for auxiliary components (24). Although it is not known if low levels of EJC components can reprogram splicing decisions within the spliceosome, our results indicate that in the case of Bcl-x, the interaction of these factors occurs in a manner that is distinct from the conventional EJC assembly mode. First, core EJC components associate with the pre-mRNA when ATP is depleted, suggesting an interaction before assembly of complex A. Second, splicing regulation by core and auxiliary EJC components acts through distinct cis-acting elements. While RNPS1 activity required a region in SB1 located more than 250 nt upstream of the Bcl-xS 5′ss with which it could interact, Y14-Magoh and eIF4A3 acted through and interacted with an element in B2 located downstream of the Bcl-xS 5′ss. Thus, the requirement for regions separated by approximately 300 nt that did not correspond to the expected deposition sites for EJCs (i.e., 20 to 24 nt upstream of a splice junction) suggests that these components are assembled on the Bcl-x pre-mRNA distinctly from the typical mode of assembly of EJCs.
It will be interesting to explore the molecular mechanisms by which regulation of splice site selection is enforced by these factors. One model is that subcomplexes made up of core and auxiliary components would assemble independently at B2 and SB1. eIF4A3, Y14, and Magoh activity required the same region in B2, suggesting the existence of a bound complex. As for RNPS1, which acts through the SB1 region, it has been described as a component of ASAP, a complex that contains Acinus and SAP18 and that can modulate splicing in vitro (60). The RNA binding ability of eIF4A3 and RNPS1 may therefore trigger the assembly of distinct complexes that may individually impact the use of the Bcl-xS 5′ss. Another interesting model is that the individual subcomplexes at SB1 and B2 would interact to form an EJC-like complex that would now impose modulation of splice site selection by looping out the 5′ ss of Bcl-xS, in a manner that is reminiscent of the model for repression of 5′ss usage by hnRNP A1 (35).
EJC components regulate a group of alternative splicing events in apoptotic genes.
All five EJC components that we tested, but not Upf1, also affected the proportion of Bim splice variants, whereas the depletion of Y14 affected that of Mcl1. Bim and Mcl1 encode proteins that, like Bcl-x, belong to the Bcl-2 family of apoptotic regulators. The shift occurred in the direction of the most potent proapoptotic form of Bim and switched splicing from the antiapoptotic MCL1L to the proapoptotic MCL1S variant. The depletion of EJC components also favored the production of a splice variant for the DNA repair gene Lig3, which lacks the initiation codon. Since Lig3 is degraded by calpain during cell death induced by DNA-damaging agents (7), preventing its synthesis by producing a noncoding variant links the control of splicing by EJC components with genomic instability and apoptosis. Moreover, the depletion of core EJC components promoted exon 6 skipping on the intracellular cysteine protease calpain 3, whereas the depletion of Upf1 encouraged the expression of the exon 6-included form. Although the exon 6-lacking variant has distinct substrate specificity (28), it is not known if this calpain variant cleaves LIG3 more efficiently. Overall, decreasing the levels of EJC components therefore shifts splicing in favor of proapoptotic splice forms, a situation that may explain why the depletion of EJC components induced apoptosis in all the cell lines that we have studied.
What could be the advantage of using components of the EJC to control the alternative splicing of apoptotic regulators? Approximately 10% of the human transcriptome is regulated by NMD (40). The NMD pathway eliminates mRNAs containing premature stop codons (PTCs) (33, 39). This task requires that components of NMD machinery be produced in sufficient amounts. Thus, the regulation of alternative splicing by EJC components may function as a checkpoint to ensure that NMD is fully operational; a decrease in the level of these components would alter the alternative splicing of key apoptotic regulators and trigger apoptosis. Interestingly, casein kinase 2, an enzyme that activates RNPS1, is overexpressed in many types of cancer (72, 73), and depleting casein kinase 2 promotes apoptosis (80). In addition to apoptosis, EJC components may similarly control other pathways related to cell growth. A likely candidate would be cell division, since the knockdown of core EJC components cause spindle defects in neural stem cells (65). In addition, Moore et al. recently observed coordinated apoptotic splicing switches in Bcl-x and Mcl1 upon inhibition of key cell cycle factors connected to aurora kinase A and other regulators of cell division and mitotic spindle assembly (44). This study also identified Y14 as a regulator of Bcl-x and Mcl1 alternative splicing. Finally, two recent studies with Drosophila indicate that defective expression of core EJC components elicits a splicing defect that reduces the level of MAPK, a signaling protein important for cell proliferation, differentiation, and survival (1, 57). Notably, the splicing of large introns was also more frequently compromised in Drosophila cells depleted of EJC core components. Although the regulated 5′ splice sites in the human Bcl-x gene are bordering a large 50-kb intron, this does not appear to be an essential characteristic in the mammalian system since we could not detect a bias for long introns in the human target units, and regulation was reproduced with a Bcl-x minigene carrying a small intron.
Our results highlight the importance of maintaining appropriate levels of specific EJC components. Failure to do so alters the splicing regulation of several apoptotic genes and promotes cell death. Targeting EJC components may therefore represent an interesting anticancer strategy because in addition to favoring the accumulation of harmful products, it should encourage the production of proapoptotic effectors.
Supplementary Material
ACKNOWLEDGMENTS
We thank Benjamin Blencowe, Arneet Salzmann, Lynne Maquat, Elisa Izaurralde, Nahum Sonenberg, Alberto Kornblihtt, Akila Mayeda, Adrian Krainer, Jens Lykke-Andersen, Catherine Tomasetto, Niels Gehring, and Matthias Hentze for generously providing materials. We thank Marco Blanchette for critical comments on the manuscript.
This work was supported by a grant from the Canadian Institute of Health Research to B.C. B.C. is the Canada Research Chair in Functional Genomics.
Footnotes
Published ahead of print 27 December 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Ashton-Beaucage D, et al. 2010. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell 143:251–262 [DOI] [PubMed] [Google Scholar]
- 2. Bae J, Leo CP, Hsu SY, Hsueh AJ. 2000. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J. Biol. Chem. 275:25255–25261 [DOI] [PubMed] [Google Scholar]
- 3. Ballut L, et al. 2005. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 12:861–869 [DOI] [PubMed] [Google Scholar]
- 4. Blencowe BJ, Issner R, Nickerson JA, Sharp PA. 1998. A coactivator of pre-mRNA splicing. Genes Dev. 12:996–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boise LH, et al. 1993. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74:597–608 [DOI] [PubMed] [Google Scholar]
- 6. Boise LH, Thompson CB. 1997. Bcl-x(L) can inhibit apoptosis in cells that have undergone Fas-induced protease activation. Proc. Natl. Acad. Sci. U. S. A. 94:3759–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bordone L, Campbell C. 2002. DNA ligase III is degraded by calpain during cell death induced by DNA-damaging agents. J. Biol. Chem. 277:26673–26680 [DOI] [PubMed] [Google Scholar]
- 8. Boutz PL, et al. 2007. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 21:1636–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cardarelli L, et al. 2010. Phages have adapted the same protein fold to fulfill multiple functions in virion assembly. Proc. Natl. Acad. Sci. U. S. A. 107:14384–14389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Casciola-Rosen LA, Miller DK, Anhalt GJ, Rosen A. 1994. Specific cleavage of the 70-kDa protein component of the U1 small nuclear ribonucleoprotein is a characteristic biochemical feature of apoptotic cell death. J. Biol. Chem. 269:30757–30760 [PubMed] [Google Scholar]
- 11. Chalfant CE, et al. 2002. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. J. Biol. Chem. 277:12587–12595 [DOI] [PubMed] [Google Scholar]
- 12. Clarke MF, et al. 1995. A recombinant bcl-x s adenovirus selectively induces apoptosis in cancer cells but not in normal bone marrow cells. Proc. Natl. Acad. Sci. U. S. A. 92:11024–11028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cloutier P, et al. 2008. Antagonistic effects of the SRp30c protein and cryptic 5′ splice sites on the alternative splicing of the apoptotic regulator Bcl-x. J. Biol. Chem. 283:21315–21324 [DOI] [PubMed] [Google Scholar]
- 14. Cory S, Huang DC, Adams JM. 2003. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene 22:8590–8607 [DOI] [PubMed] [Google Scholar]
- 15. Das R, Reed R. 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 5:1504–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Degen WG, Aarssen Y, Pruijn GJ, Utz PJ, van Venrooij WJ. 2000. The fate of U1 snRNP during anti-Fas induced apoptosis: specific cleavage of the U1 snRNA molecule. Cell Death Differ. 7:70–79 [DOI] [PubMed] [Google Scholar]
- 17. Dignam JD. 1990. Preparation of extracts from higher eukaryotes. Methods Enzymol. 182:194–203 [DOI] [PubMed] [Google Scholar]
- 18. Dominguez C, Fisette JF, Chabot B, Allain FH. 2010. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat. Struct. Mol. Biol. 17:853–861 [DOI] [PubMed] [Google Scholar]
- 19. Du H, et al. 2010. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 17:187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du YC, Lewis BC, Hanahan D, Varmus H. 2007. Assessing tumor progression factors by somatic gene transfer into a mouse model: Bcl-xL promotes islet tumor cell invasion. PLoS Biol. 5:e276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fischer U, Janicke RU, Schulze-Osthoff K. 2003. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 10:76–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fleckner J, Zhang M, Valcarcel J, Green MR. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11:1864–1872 [DOI] [PubMed] [Google Scholar]
- 23. Garneau D, Revil T, Fisette JF, Chabot B. 2005. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 280:22641–22650 [DOI] [PubMed] [Google Scholar]
- 24. Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. 2009. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 7:e1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. 2009. Disassembly of exon junction complexes by PYM. Cell 137:536–548 [DOI] [PubMed] [Google Scholar]
- 26. Green DR, Kroemer G. 2004. The pathophysiology of mitochondrial cell death. Science 305:626–629 [DOI] [PubMed] [Google Scholar]
- 27. Grosso AR, Martins S, Carmo-Fonseca M. 2008. The emerging role of splicing factors in cancer. EMBO Rep. 9:1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Herasse M, et al. 1999. Expression and functional characteristics of calpain 3 isoforms generated through tissue-specific transcriptional and posttranscriptional events. Mol. Cell. Biol. 19:4047–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Isken O, Maquat LE. 2008. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 9:699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klinck R, et al. 2008. Multiple alternative splicing markers for ovarian cancer. Cancer Res. 68:657–663 [DOI] [PubMed] [Google Scholar]
- 31. Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. 2007. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446:926–929 [DOI] [PubMed] [Google Scholar]
- 32. Le Hir H, Izaurralde E, Maquat LE, Moore MJ. 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lejeune F, Maquat LE. 2005. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 17:309–315 [DOI] [PubMed] [Google Scholar]
- 34. Li CY, et al. 2004. Regulation of alternative splicing of Bcl-x by IL-6, GM-CSF and TPA. Cell Res. 14:473–479 [DOI] [PubMed] [Google Scholar]
- 35. Martinez-Contreras R, et al. 2006. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 4:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Massiello A, Roesser JR, Chalfant CE. 2006. SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 20:1680–1682 [DOI] [PubMed] [Google Scholar]
- 37. Massiello A, et al. 2004. Identification of two RNA cis-elements that function to regulate the 5′ splice site selection of Bcl-x pre-mRNA in response to ceramide. J. Biol. Chem. 279:15799–15804 [DOI] [PubMed] [Google Scholar]
- 38. Mayeda A, et al. 1999. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J. 18:4560–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McGlincy NJ, Smith CW. 2008. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 33:385–393 [DOI] [PubMed] [Google Scholar]
- 40. Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36:1073–1078 [DOI] [PubMed] [Google Scholar]
- 41. Mercatante DR, Mohler JL, Kole R. 2002. Cellular response to an antisense-mediated shift of Bcl-x pre-mRNA splicing and antineoplastic agents. J. Biol. Chem. 277:49374–49382 [DOI] [PubMed] [Google Scholar]
- 42. Merz C, Urlaub H, Will CL, Luhrmann R. 2007. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA 13:116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Michaud S, Reed R. 1991. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 5:2534–2546 [DOI] [PubMed] [Google Scholar]
- 44. Moore MJ, Wang Q, Kennedy CJ, Silver PA. 2010. An alternative splicing network links cell-cycle control to apoptosis. Cell 142:625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nasim FU, Hutchison S, Cordeau M, Chabot B. 2002. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA 8:1078–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ni JZ, et al. 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 21:708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nilsen TW, Graveley BR. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463:457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. O'Connor L, et al. 1998. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 17:384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Olopade OI, et al. 1997. Overexpression of BCL-x protein in primary breast cancer is associated with high tumor grade and nodal metastases. Cancer J. Sci. Am. 3:230–237 [PubMed] [Google Scholar]
- 50. Pan Q, et al. 2006. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 20:153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40:1413–1415 [DOI] [PubMed] [Google Scholar]
- 52. Paradis C, et al. 2007. hnRNP I/PTB can antagonize the splicing repressor activity of SRp30c. RNA 13:1287–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. 2007. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell Biol. 176:929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Prinos P, et al. 2011. Alternative splicing of SYK regulates mitosis and cell survival. Nat. Struct. Mol. Biol. [DOI] [PubMed] [Google Scholar]
- 55. Revil T, Pelletier J, Toutant J, Cloutier A, Chabot B. 2009. Heterogeneous nuclear ribonucleoprotein K represses the production of pro-apoptotic Bcl-xS splice isoform. J. Biol. Chem. 284:21458–21467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Revil T, et al. 2007. Protein kinase C-dependent control of Bcl-x alternative splicing. Mol. Cell. Biol. 27:8431–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roignant JY, Treisman JE. 2010. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell 143:238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sakashita E, Tatsumi S, Werner D, Endo H, Mayeda A. 2004. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol. Cell. Biol. 24:1174–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Saltzman AL, et al. 2008. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 28:4320–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schwerk C, et al. 2003. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol. Cell. Biol. 23:2981–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schwerk C, Schulze-Osthoff K. 2005. Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell 19:1–13 [DOI] [PubMed] [Google Scholar]
- 62. Shen H, et al. 2008. Distinct activities of the DExD/H-box splicing factor hUAP56 facilitate stepwise assembly of the spliceosome. Genes Dev. 22:1796–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shibuya T, Tange TO, Sonenberg N, Moore MJ. 2004. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 11:346–351 [DOI] [PubMed] [Google Scholar]
- 64. Shkreta L, et al. 2008. Anticancer drugs affect the alternative splicing of Bcl-x and other human apoptotic genes. Mol. Cancer Ther. 7:1398–1409 [DOI] [PubMed] [Google Scholar]
- 65. Silver DL, et al. 2010. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat. Neurosci. 13:551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Simard MJ, Chabot B. 2002. SRp30c is a repressor of 3′ splice site utilization. Mol. Cell. Biol. 22:4001–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Singh KK, et al. 2010. Human SAP18 mediates assembly of a splicing regulatory multiprotein complex via its ubiquitin-like fold. RNA 16:2442–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stamm S, et al. 2005. Function of alternative splicing. Gene 344:1–20 [DOI] [PubMed] [Google Scholar]
- 69. Sudo H, et al. 2010. Knockdown of COPA, identified by loss-of-function screen, induces apoptosis and suppresses tumor growth in mesothelioma mouse model. Genomics 95:210–216 [DOI] [PubMed] [Google Scholar]
- 70. Tange TO, Shibuya T, Jurica MS, Moore MJ. 2005. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 11:1869–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tao W, et al. 2005. Induction of apoptosis by an inhibitor of the mitotic kinesin KSP requires both activation of the spindle assembly checkpoint and mitotic slippage. Cancer Cell 8:49–59 [DOI] [PubMed] [Google Scholar]
- 72. Tawfic S, et al. 2001. Protein kinase CK2 signal in neoplasia. Histol. Histopathol. 16:573–582 [DOI] [PubMed] [Google Scholar]
- 73. Trembley JH, et al. 2005. Activation of pre-mRNA splicing by human RNPS1 is regulated by CK2 phosphorylation. Mol. Cell. Biol. 25:1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Veitia RA, Bottani S, Birchler JA. 2008. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 24:390–397 [DOI] [PubMed] [Google Scholar]
- 75. Venables JP, et al. 2008. Identification of alternative splicing markers for breast cancer. Cancer Res. 68:9525–9531 [DOI] [PubMed] [Google Scholar]
- 76. Venables JP, et al. 2009. Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 16:670–676 [DOI] [PubMed] [Google Scholar]
- 77. Venables JP, et al. 2008. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol. Cell. Biol. 28:6033–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang ET, et al. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13:91–100 [DOI] [PubMed] [Google Scholar]
- 80. Yamane K, Kinsella TJ. 2005. Casein kinase 2 regulates both apoptosis and the cell cycle following DNA damage induced by 6-thioguanine. Clin. Cancer Res. 11:2355–2363 [DOI] [PubMed] [Google Scholar]
- 81. Youle RJ, Strasser A. 2008. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9:47–59 [DOI] [PubMed] [Google Scholar]
- 82. Zhou Z, et al. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407:401–405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.