Abstract
Genetic and morphological studies have revealed that the radial spokes regulate ciliary and flagellar bending. Functional and biochemical analysis and the discovery of calmodulin in the radial spokes suggest that the regulatory mechanism involves control of axonemal protein phosphorylation and calcium binding to spoke proteins. To identify potential regulatory proteins in the radial spoke, in-gel kinase assays were performed on isolated axonemes and radial spoke fractions. The results indicated that radial spoke protein 2 (RSP2) can bind ATP and transfer phosphate in vitro. RSP2 was cloned and mapped to the PF24 locus, a gene required for motility. Sequencing revealed that pf24 contains a point mutation converting the first ATG to ATA, resulting in only trace amounts of RSP2 and confirming the RSP2 mapping. Surprisingly, the sequence does not include signature domains for conventional kinases, indicating that RSP2 may not perform as a protein kinase in vivo. However, the predicted RSP2 protein sequence contains Ca2+-dependent calmodulin binding motifs and a GAF domain, a domain found in diverse signaling proteins for binding small ligands including cyclic nucleotides. As predicted from the sequence, recombinant RSP2 binds calmodulin in a calcium-dependent manner. We postulate that RSP2 is a regulatory subunit of the radial spoke involved in localization of calmodulin for control of motility.
The radial spokes play an important role in the control of ciliary and flagellar motility. For example, in Chlamydomonas reinhardtii or humans failure in assembly of the radial spokes results in ciliary and flagellar paralysis (51, 56). The spokes are complex T-shaped structures anchored on the A microtubule of each outer doublet, adjacent to the inner dynein arms, and project toward the central pair apparatus where the spoke heads interact with the central pair projections (Fig. 1) (reviewed in reference 12). In the long axis, radial spokes repeat in groups every 96 nm along each microtubule, in exact register with the inner dynein arms and the dynein regulatory complex (16, 18, 32, 42, 43). The radial spokes are composed of at least 22 proteins (40, 63), and five of them, including radial spoke protein 2 (RSP2), are phosphorylated in vivo (22, 40). Some of the spoke proteins have been characterized (12, 17, 38), including RSP3, believed to target and anchor the spoke to the doublet microtubule (13, 22, 55) and perform as an A-kinase anchor protein (15).
FIG. 1.
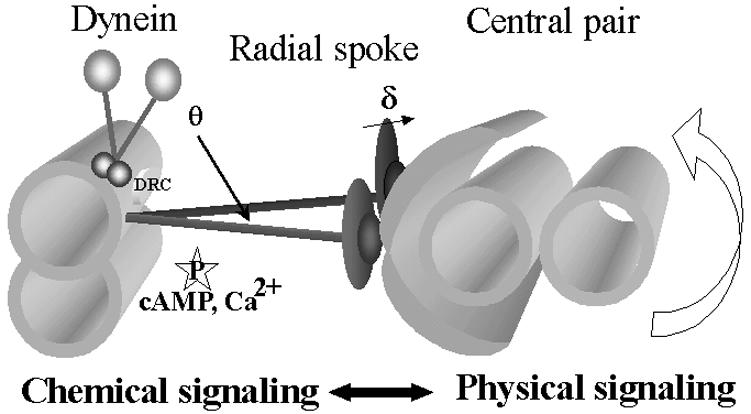
Hypothetical model depicting the radial spoke as a mechanochemical transducer extending between the central pair apparatus and outer doublet microtubules, anchored near the base of the inner arm dynein. In this model, the signal input includes calcium binding and/or mechanical strain induced by transient interaction of the spoke head with the central apparatus as microtubules slide (54) or as the central pair rotates (35). Transduction output, at the junction between the spoke stalk and doublet microtubule, is predicted to include localized control of axonemal protein phosphorylation and localized regulation of dynein-driven microtubule sliding. cAMP, cyclic AMP; DRC, dynein regulatory complex.
The radial spokes are thought to operate by local control of dynein-driven microtubule sliding and thus regulate the size and shape of the bend. The mechanism is not understood but likely involves both mechanical control of microtubule sliding and regulation of axonemal protein phosphorylation (Fig. 1). For example, structural analyses indicate that radial spokes, through transient interactions with the central apparatus, control axonemal bending (35, 54). Suppressor mutations in Chlamydomonas that rescue motility in radial spoke mutants have revealed a control system that regulates dynein activity (23, 42), and importantly, phenotypic analysis of suppressor mutants demonstrated that the radial spoke plays a key role in control of the shape of flagellar bends (6). Moreover, in vitro functional assays of dynein-driven microtubule sliding indicate that the radial spoke mechanism involves control of axonemal kinases and phosphorylation of dynein subunits (48-50, 62) and that the mechanism, in conjunction with the central apparatus, may mediate calcium control of bending (33, 49, 53).
Several experimental systems have demonstrated that changes in intracellular calcium regulate ciliary and flagellar motility (7, 8) and that in vitro reactivation of axonemes from several organisms, including Chlamydomonas, has revealed that calcium alters motility by direct interaction with structural components of the axoneme (3, 19, 25, 27, 34). Physiological studies indicate that flagellar dynein-driven microtubule sliding can be regulated by changes in calcium in a process which, in certain instances, involves the radial spokes, central apparatus, and possibly calmodulin (2, 5, 33, 48, 49; for calcium regulation independent of the radial spokes, see for example references 52 and 59). Furthermore, biochemical analysis has revealed that calmodulin is a component of the radial spoke stalk (63), presumably in a position to mediate calcium control of bending.
Based on these diverse data, we postulated that the radial spokes act as signal transducers that mediate structural and chemical signals—including calcium binding—for control of protein phosphorylation and motility (Fig. 1). To test this idea, we have taken advantage of a novel isolation of intact radial spokes and spoke stalks (63) to identify potential signaling proteins located in the structure of the spoke and to determine the mechanism for calmodulin localization. We cloned a novel Chlamydomonas gene encoding RSP2, mapped the gene near the PF24 locus in linkage group X, identified a point mutation in pf24, and determined that anti-RSP2 antibodies specifically recognize the recombinant RSP2 protein. We demonstrate that RSP2 is a calmodulin binding protein and contains a GAF domain (named for its presence in cGMP-regulated phosphodiesterases, certain adenylyl cyclases, and the bacterial transcription factor FhlA), a domain present in numerous signal transduction proteins thought to bind small molecules that influence catalytic activity (1, 21, 24). We conclude that RSP2 is a key component of the radial spoke, mediating calcium-calmodulin signaling and control of flagellar motility.
MATERIALS AND METHODS
Cell strains and growth conditions.
Chlamydomonas mutants defective in radial spoke assembly or phosphorylation, including pf14, pf17, pf24, and pf27, were previously defined (22) and provided by E. H. Harris (Duke University, Chlamydomonas Genetics Center). pf28pf30 was described previously (41). All cells were grown in liquid modified medium I with aeration over a 14-h-10-h light-dark cycle (57).
Biochemistry.
Preparation of flagella and axonemes, KI extraction of axonemes, and velocity sedimentation of radial spoke complexes on sucrose gradients were carried out as described before (63). For purifying the 20S radial spokes, axonemes were isolated from the double mutant pf28pf30, which lacks the 20S dynein complexes that could otherwise contaminate the fraction. Protein samples from axonemes and gradient fractions were prepared for and separated by standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by silver or Coomassie blue staining or Western blotting (63). For Western blots, a mouse monoclonal antibody (3G3) and polyclonal rabbit antibodies to RSP2 (63) were kindly provided by Dennis Diener and Joel Rosenbaum (Yale University).
For band purification of RSP2, the 15S radial spoke stalks were isolated from axonemes derived from pf17, a mutant lacking the radial spoke head and RSP1. This strategy was used to avoid the contamination of RSP1, which migrates near RSP2 in gels (see Results). The 15S stalk fractions were concentrated by centrifugal filtration (Ultrafree-CL filter unit; Millipore); proteins, separated in the SDS-6% polyacrylamide gel, were transferred to a polyvinylidene difluoride membrane; and bands were revealed by imido black staining as described before (61). The RSP2 band was identified based on RSP2 Western analysis and excised for microsequencing (performed by John Leszyk at the University of Massachusetts Proteomic Mass Spectrometry Laboratory). The resulting peptide sequences included APTQAGHDTAYLK, ETVGEALAR, NAEVEGNFYR, ALLAADTLFPEGSGQPLSADD, LFEDMAAYDATAER, and AAAEAEAAAAAEAAAEAAAEAAAAAAE.
In-gel kinase assays (26) were carried out with modifications as described by Yang and Sale (62). Briefly, dephosphorylated casein (Sigma, St. Louis, Mo.), 10 mg/ml dissolved in 1× resolution buffer, was added into a 6 or 9% acrylamide gel mixture to a final concentration of 1 mg/ml. Following electrophoresis, the gel was briefly rinsed, using several changes in 20% isopropanol in buffer B (50 mM HEPES [pH 7.4], 5 mM β-mercaptoethanol) to remove excess SDS-PAGE buffer. The gel was then immersed in the same isopropanol buffer for 60 min with two buffer changes and then with buffer B alone for 30 min. The gel was then immersed in 0.04% Tween 20 in buffer B at 4°C overnight for renaturation. This buffer was changed six times, and the renatured gel was immersed in 10 ml of reaction buffer (50 mM HEPES [pH 7.4], 100 μM sodium orthovanadate, 10 mM MnCl2, 5 mM β-mercaptoethanol) first at 30°C for 30 min and then for another 3 h after 7 μl of [γ-32P]ATP, 3,000 Ci/mM (3.3 μM), was added. (The experiment can also be performed in the presence of 50 μM ATP but with reduced labeling.) To control for ATP binding, [α-32P] ATP was used in place of [γ-32P]ATP at the equivalent specific activity. The reaction was terminated, and free nucleotides were removed by rinsing the gel in the fixative solution (10 mM sodium pyrophosphate and 5% trichloroacetic acid) for 20 min, five to seven times. The gel was stained with Coomassie blue and then dried for phosphorimaging with a 1- to 2-h exposure.
For calmodulin affinity purification of the recombinant RSP2 (see below), calmodulin-Sepharose was used according to the instructions of the manufacturer (Amersham Pharmacia, Piscataway, N.J.). Briefly, the bacterial pellet was resuspended in 60 mM NaCl-50 mM Tris, pH 7.5 (calmodulin binding buffer). Following sonication and ultracentrifugation, the extract was supplemented with aprotinin, phenylmethylsulfonyl fluoride, complete protease inhibitor cocktail (Roche, Indianapolis, Ind.), and either 2 mM Ca2+ or 0.5 mM EGTA. Following overnight incubation with calmodulin-Sepharose 4B and extensive washing with the same buffer with either Ca2+ or EGTA, elution was carried out either with or without 2 mM EGTA in the calmodulin binding buffer.
Molecular cloning.
Genomic DNA and RNA were prepared as described previously (61). For reverse transcription-PCR (RT-PCR), sense and antisense degenerate primers (ACTGAATTCGGNCAYGAYACIGCITACCT and AGTCTCGAGTCGTAIGCCATRTCYTC) were designed based on the peptide sequences GHDTAYL and EDMAAYD, respectively. (The first 9 nucleotides were added for cloning convenience.) For PCR, multiple primer pairs were used to amplify the entire gene. RT and PCR, using Superscript II reverse transcriptase (Invitrogen) and High-Fidelity Pfu polymerase (Stratagene), were performed as described previously (61). Thermocycling procedures were performed as described previously (61). For library screening, pScreen-1 and λgt10 cDNA libraries and the λfixII genomic library were provided by P. Lefebvre (University of Minnesota), G. Pazour (University of Massachusetts), and E. F. Smith (Dartmouth College). The libraries were screened using probes labeled with [α-32P]dCTP, and Northern blot analysis was carried out as described previously (61). For genomic mapping of RSP2, restriction fragment length polymorphism (RFLP) mapping was performed by Carolyn Silflow (University of Minnesota) as described previously (44), and results were previously reported (28).
The expression construct containing the entire coding sequence of RSP2 was derived from cDNA and genomic library clones. Briefly, PCR, using the 5′ cDNA clone as a template, was used to create an NcoI restriction site at the first ATG. The 5′ fragment was released by NcoI/NdeI digestion of the PCR clone. The middle fragment was released by NdeI/SacI digestion of the second, overlapping cDNA clones. The 3′ SacI/SalI fragment, which was not interrupted by introns and included the intrinsic stop codon, was released by digestion of the 3′ genomic subclone. The three fragments were ligated with T4 DNA ligase and cloned into the NcoI and SalI sites in the expression vector pET28a (Novagen). Based upon sequencing and Western blotting with antibodies to RSP2, the construct expressed the authentic RSP2 (see Results and Discussion).
A construct for expressing the six-His-tagged RSP2 fusion protein was engineered by PCR using a sense primer containing the six-His coding sequence after the first ATG. The strategy was to introduce minimal additional residues to the recombinant proteins for testing the kinase activity and phosphorylation. For expression, freshly prepared RSP2 constructs were transformed into Escherichia coli strain BL21(DE3) (Novagen). A single colony was inoculated into 100 ml of Luria-Bertani medium supplemented with kanamycin and allowed to grow at 37°C until cell densities reached optical densities at 600 nm of 0.4 to 0.6. The expression of recombinant RSP2 was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 26°C for 4 h.
Primary sequence and helical wheel analyses were performed using the software Protean in Lasergene from DNASTAR (http://www.dnastar.com/products/Protean.html). SMART (Simple Modular Architecture Research Tool, http://smart.embl-heidelberg.de), Pfam at Washington University (http://pfam.wustl.edu/index.html), and PSI Blast search at the National Center for Biotechnology Information website were used for searching the pattern and domains. Default parameters were used.
RESULTS
In-gel kinase and Western analyses reveal that RSP2 binds ATP in vitro.
As discussed above, biochemical and physiological evidence indicates that axonemal kinases, including PKA and CK1, regulate dynein-driven microtubule sliding and that the radial spokes, through an unknown mechanism, locally regulate the axonemal kinases. We postulated that the radial spokes contain additional kinases or signaling molecules to mediate such a function. To test this hypothesis, we employed an “in-gel” protein kinase assay that can reveal candidate protein kinases by phosphorylation of substrates embedded in the acrylamide gel (see Materials and Methods). Two strategies were employed. First, the assay was used to compare SDS-PAGE axonemal samples from Chlamydomonas wild-type cells and a panel of mutants with defects in spoke assembly. Predictably, if a candidate spoke protein kinase is present in wild-type axonemes, then the same candidate kinase may be absent or reduced in axonemes from selective mutant cells defective in radial spoke proteins or radial spoke assembly. Second, we also used the in-gel kinase assay to assess protein kinase activity in isolated 20S radial spokes and 15S radial spoke stalks (63). Predictably, the same candidate kinase bands identified in the isolated axonemes would be found in the spoke and spoke stalk fractions.
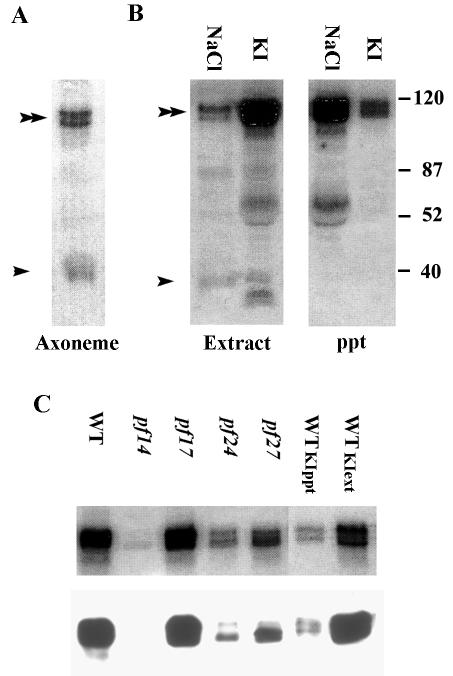
An in-gel kinase assay of axonemal proteins revealed prominent 32P-labeled bands with molecular masses of 39 kDa (arrowhead, Fig. 2A) and a doublet at 118 kDa (double arrowhead, Fig. 2A), indicating two candidate axonemal protein kinases. The 39-kDa candidate kinase is soluble in 0.6 M NaCl (arrowhead, Fig. 2B) and has previously been identified as the axonemal CK1 (62). In contrast, the 118-kDa candidate protein kinase, like the radial spokes, is not extractable with 0.6 M NaCl (double arrow, Fig. 2B) but is soluble in 0.6 M KI. The 118-kDa candidate kinase appears as a doublet by SDS-9% PAGE (Fig. 2), suggesting the occurrence of either two proteins or two isoforms resulting from posttranslational modification.
FIG. 2.
A 118-kDa candidate axonemal kinase, demonstrated by an in-gel kinase assay, comigrates with RSP2. (A) Prominent bands, revealed by in-gel kinase assay of wild-type axonemes, include a doublet that migrates at 118 kDa (double arrowheads) and a 39-kDa band (single arrowhead) previously determined to be CK1 (62). (B) In contrast to the 39-kDa axonemal CK1, the 118-kDa kinase is not soluble in 0.6 M NaCl but is soluble in 0.6 M KI (compare the KI extract and ppt lanes). (The prominent bands that migrate at about 60 kDa [second and third lanes from left] are likely degradation products of RSP2; in data not shown, the bands are not present in extracts from pf14 axonemes.) Numbers at right are molecular masses in kilodaltons. (C) Axonemal proteins analyzed by in-gel kinase assays (upper panel) and Western blot assays, with an antibody to RSP2 (lower panel), demonstrated comigration of the 118-kDa candidate kinase with RSP2 (wild-type axonemes, lane 1). Both the 118-kDa kinase and RSP2 are missing in axonemes from pf14 (mutant lacking the entire radial spoke) (lane 2), present in axonemes from pf17 (mutant lacking only the radial spoke head) (lane 3), greatly reduced in axonemes from pf24 (mutant with a mutation in the RSP2 gene, expressing only a small amount of RSP2 [see the text]) (lane 4), and reduced in axonemes from pf27 (radial spoke mutant with reduced levels of spoke phosphoproteins [22]) (lane 5). Both the 118-kDa candidate kinase and RSP2 were soluble in 0.6 M KI (compare lanes 6, axonemal ppt, with lane 7, KI extract), similar to the solubility of radial spokes. WT, wild type.
Based on the molecular mass of 118 kDa, resistance to NaCl extraction (40), and presence in the KI extract (63), we predicted that the 118-kDa candidate kinase was RSP2. Based on analysis of radial spoke mutants and biochemical fractionation of radial spokes, RSP2 is a 118-kDa phosphorylated radial spoke subunit located in the radial spoke stalk (40, 63). To test this, axonemes from several well-defined radial spoke mutants (Fig. 2C) (see reference 22) and axonemal fractions including the 20S radial spoke and 15S radial spoke stalk (Fig. 3) were assessed by both in-gel kinase assay and Western blotting with the antibodies to RSP2. The prediction was that the presence and position of the 118-kDa candidate kinase would exactly correspond to the presence and position of RSP2.
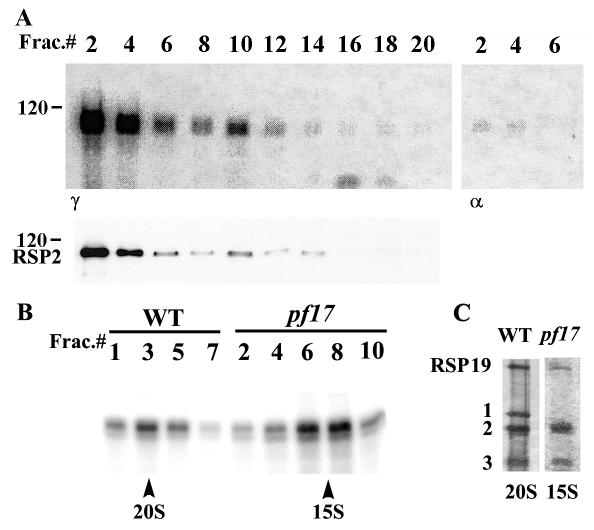
FIG. 3.
(A) The 118-kDa candidate kinase in the 20S radial spoke fraction (in-gel kinase assay, upper panel) comigrated with RSP2 revealed by polyclonal antibodies (Western blotting, lower panel). In this experiment, KI extracts derived from pf28pf30 axonemes were fractionated by velocity sedimentation on 5 to 20% sucrose gradients, and even-numbered fractions, 2 to 20, were analyzed from bottom to top of the gradient. In controls (right panel), incubation of the gel with [α-32P]ATP resulted in distinct, but relatively minor, labeling. (B) The 118-kDa candidate kinase also cosedimented with the 15S radial spoke stalk prepared from pf17 axonemes (compare 20S peak, wild-type fraction 3, with 15S peak, pf17 fractions 6 to 8). Sucrose gradient fractions were separated in 9% (A) and 6% (B) acrylamide gels. (C) Representative silver-stained protein gel of the peak spoke fractions from wild type and pf17 revealed the clear separation of RSP1, RSP2, and RSP3. Notably, as expected, RSP1 is missing in pf17, facilitating band purification of RSP2. These data, along with the data in Fig. 2, strongly indicate that the 118-kDa candidate kinase is RSP2. WT, wild type.
As illustrated in Fig. 2C, in every instance the in-gel candidate kinase pattern of the 118-kDa band (upper panel) directly matched the Western blot revealed by a monoclonal antibody to RSP2 (lower panel). For example, the 118-kDa candidate kinase revealed by in-gel analysis matches the RSP2 position revealed by Western blotting (lane 1, Fig. 2C). In contrast, both the 118-kDa candidate kinase and RSP2 are missing in axonemes from pf14, a mutant lacking the radial spokes (lane 2, Fig. 2C). Moreover, the candidate kinase and RSP2 are present in axonemes from pf17, a mutant lacking only the radial spoke heads (lane 3, Fig. 2C). This result excludes RSP1, a 120-kDa radial spoke protein located in the spoke head, as the candidate kinase since, in axonemes from pf17, RSP2 is assembled but RSP1 is missing (Fig. 3) (see references 40 and 63). The candidate kinase and RSP2 are greatly reduced in axonemes from pf24, an RSP2 mutant containing only trace amounts of RSP2 (lane 4, Fig. 2C) (see reference 22), and in axonemes from pf27, a mutant containing a reduced amount of RSP2 (lane 5, Fig. 2C) (see reference 40). As further support, both the 118-kDa candidate kinase and RSP2 were extracted with 0.6 M KI (compare WTKIppt and WTKIext, Fig. 2C). These results strongly indicate that the 118-kDa candidate protein kinase is RSP2.
As a further test, the isolated 20S radial spoke and 15S spoke stalk fractions were analyzed by both in-gel kinase assays and Western blotting with antibodies to RSP2. As anticipated, the 118-kDa candidate kinase and RSP2 precisely cosedimented and comigrated in the 20S radial spoke fraction (Fig. 3A). As a control, addition of [α-32P]ATP in place of [γ-32P]ATP resulted in less labeling (right panel, Fig. 3A), indicating that much, but not all, of the labeling of the 118-kDa protein doublet is the result of genuine phosphotransferring in the in-gel assay. Furthermore, the 118-kDa candidate spoke kinase cosedimented with the 15S radial spoke stalk fraction, derived from pf17 axonemes (right panel, Fig. 3B). Notably, the 118-kDa protein, revealed in 6% polyacrylamide gels (Fig. 3C), resolved as a major single band in the in-gel kinase assay (Fig. 3B) and is well separated from other proteins (Fig. 3C). Thus, it seems unlikely that RSP2 serves as a substrate of another protein kinase that happened to (i) cofractionate, (ii) cosediment, and (iii) comigrate with RSP2. Together, the results strongly indicate that the 118-kDa candidate kinase is RSP2.
Cloning and characterization of RSP2.
For further characterization, RSP2 cDNA was cloned by reverse genetics starting with the peptide sequence from RSP2 followed by RT-PCR, PCR, and library screening (summarized in Fig. 4A). To avoid contamination from RSP1, RSP2 was band purified from the 15S spoke stalk fraction from pf17 axonemes (Fig. 3C). Six peptide sequences, listed in Materials and Methods, were obtained by microsequencing. RT-PCR with the degenerate primers derived from peptides 1 and 5 resulted in a specific 850-bp product. The predicted amino acid sequence contained four of six peptides (peptides 1 to 3 and 5), indicating the recovery of an RSP2 cDNA. Peptides 4 and 6 were found in the subsequently recovered cDNA and genomic library clones (Fig. 4). The cDNA probe was used for screening cDNA and genomic libraries and Northern, Southern, and RFLP mapping analysis. The 5′ end of the cDNA was confirmed by RT-PCR and a sequence from an expressed sequence tag database (accession no. BE238327). Both strands of all clones were sequenced.
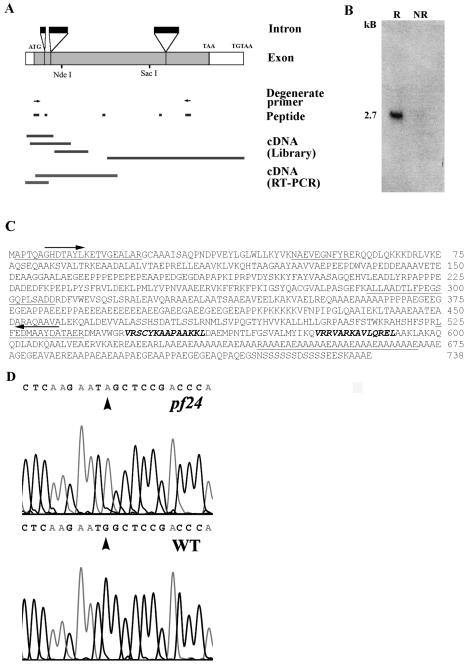
FIG. 4.
(A) Strategy for cloning RSP2 starting with peptide sequences from band-purified RSP2 (see Materials and Methods). PCR, using the degenerate oligonucleotide primers (arrows) derived from two peptides, resulted in a probe for screening cDNA and genomic libraries. Additional RT-PCR was carried out to recover entire cDNA. The coding sequence (gray boxes), untranslated regions (open boxes), and introns (black boxes) were deduced by comparing complete genomic, cDNA, and peptide sequences. (B) Northern blot analysis, with the RSP2 RT-PCR probe, revealed a 2.7-kb message that became greatly amplified in cells 45 min following deflagellation (R, RNA from flagellar regenerating cells; NR, RNA from control, nonregenerating cells). (C) Predicted amino acid sequences of RSP2 (accession no. AY373262). Sequences obtained by peptide microsequencing are underlined. Arrows indicate the peptide sequences used for designing degenerate oligonucleotide primers for successful RT-PCR, and boldface italic indicates amphipathic helical domains predicted to bind calmodulin (Fig. 5 and 6). (D) Comparison of wild-type and pf24 RSP2 genomic sequences, revealing that the pf24 mutation results from a single base pair replacement and conversion of the initial ATG to ATA. WT, wild type.
As predicted, Northern analyses revealed a single 2.7-kb message for RSP2, which was amplified dramatically following deflagellation (compare R with NR lanes, Fig. 4B; deflagellation commonly results in amplification of transcripts for flagellar proteins). Southern analysis revealed a single RSP2 gene (data not shown). Moreover, as described below, recombinant RSP2 comigrates with axonemal RSP2, binds to RSP2 antibodies, and contains all six of the peptides derived from the band-purified RSP2 (underlined, Fig. 4C), together confirming the cloning of the RSP2 gene. The RSP2 gene consists of four exons and three introns (Fig. 4A), and both a potential TATA box and TGTAA, the potential polyadenylation signal, were found in the 5′ and 3′ untranslated regions, respectively (GenBank accession no. AY373262).
Based on dikaryon rescue, intragenic reversion, and genetic mapping analysis, it was established that RSP2 is the protein product of the PF24 gene located on linkage group X (22). Only a trace amount of RSP2 is produced, and flagella are paralyzed in pf24 (Fig. 3C). Using RFLP mapping (see Materials and Methods), we confirmed that RSP2 maps near the PF24 locus as reported previously (28). Moreover, to further confirm that the cloned gene is RSP2 and to determine the genetic defect in pf24, we compared the genomic sequence for the wild type and pf24. The sequences were identical with only one difference in pf24: the ATG initiation codon was changed to ATA (Fig. 4D). PCR products derived from different primer pairs and the reverse-strand sequence confirmed the finding. It appears that translation initiation still takes place at ATA despite the very low efficiency, since trace levels of wild-type-sized RSP2 protein can be detected in pf24 axonemes by very sensitive methods including 35S labeling and Western blotting (22) (Fig. 2C, lane 4) and there are no nearby alternative initiation codons. (For discussion of alternative translation initiation codons in other eukaryotic organisms and Chlamydomonas chloroplasts, see references 10 and 29.)
Thus, multiple independent lines of evidence reveal that RSP2 is the protein product of the PF24 gene, including identification of a mutation in the RSP2 gene in pf24 and dikaryon and reversion analysis of pf24 (22). Additional evidence, not shown here, includes that (i) RFLP analysis mapping of both the RSP2 gene and the nearby pf6 gene (accession no. AF327876) shows that they are located at the genetic loci PF24 and PF6, respectively, in linkage group X (28; http://www.biology.duke.edu/chlamygenome/nuclear_maps.html) and (ii) sequences for RSP2 and adjacent molecular mapping markers, including katanin p60 (accession no. AF205377), in linkage group X are found in Chlamydomonas genome scaffold 25 (http://genome.jgi-psf.org/chlre1/chlre1.home.html).
GAF and Ca2+-dependent calmodulin binding domains in RSP2.
The pI of the RSP2 protein is predicted to be 4.48, consistent with its position in two-dimensional gels (40, 63). The predicted mass of 77 kDa is considerably less than the 118 kDa observed by SDS-PAGE, a phenomenon reported for numerous axonemal proteins including RSP3 (13, 15). Nonetheless, the recombinant RSP2, expressed in bacteria, comigrated with axonemal RSP2 at ∼118 kDa in SDS-polyacrylamide gels and bound to the RSP2 antibodies (see below). Interestingly, the sequence neither contains the signature sequence domains found in conventional protein kinases (20) nor shares homology to atypical kinases (11, 14, 30, 46, 60).
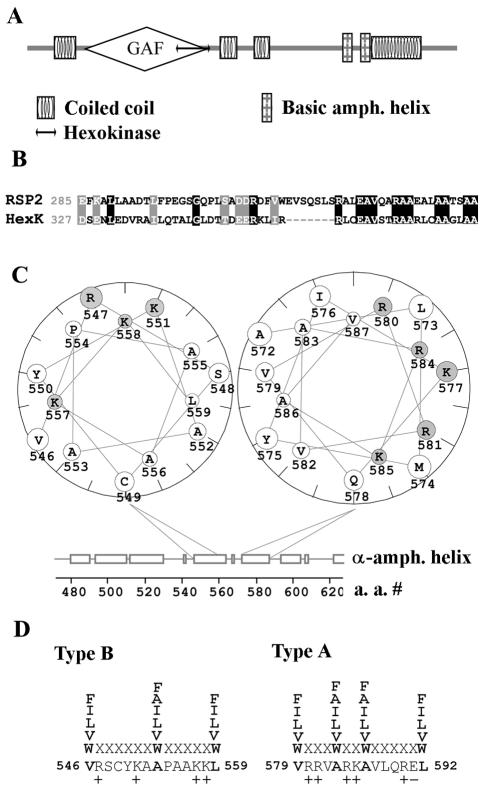
Sequence analysis of RSP2, using SMART, predicted four coiled-coil domains (located at residues 48 to 85, 333 to 365, 388 to 412, and 588 to 676) and a GAF domain (after cGMP; adenylyl cyclase; FhlA [1, 24]), located between residues 102 and 333 (Fig. 5A). Analysis using Pfam also revealed the GAF domain at residues 102 to 328. The GAF domain is a ubiquitous small molecule binding domain present in numerous signal transduction proteins found in organisms from all phyla and thought to bind small molecules like cGMP to allosterically influence catalytic activity. Blast-PSI analysis revealed that the C-terminal half of the GAF domain is homologous to part of the hexokinase catalytic domain (residues 285 to 340, Fig. 5A and B).
FIG. 5.
Predicted tertiary structure of RSP2. (A) Sequence analysis with the SMART program predicted four coiled-coil domains, a GAF domain, a hexokinase-like domain, and two basic amphipathic helical structures predicted to bind calmodulin. (B) Sequence comparison of the homologous regions in hexokinase and the C-terminal half of the RSP2 GAF domain predicted by the Blast-PSI program; similar residues are shown by gray boxes, and identical residues are shown by black boxes. (C) The two basic amphipathic helices were identified based on the Eisenberg moment and plotted as a helical wheel with the Protean program. Basic amino acid residues are designated by gray circles. (D) The basic amphipathic helices contain the conserved hydrophobic residues (boldface) and multiple basic residues characteristic of type A (residues 579 to 592) and type B (residues 546 to 559) 1-8-14 calcium-dependent calmodulin binding motifs (45).
In addition, Eisenberg moment and helical wheel analysis revealed a region containing basic amphipathic helices located approximately 50 amino acids N terminal to the last coiled coil (Fig. 4C, boldface amino acid letters; Fig. 5A and C), a common structural feature of calmodulin binding proteins (36). Further evaluation revealed that the sequences are structural homologs of the “1-8-14” Ca2+-dependent calmodulin binding motifs (45), including type B and type A (residues 546 to 559 and 579 to 592, respectively) (Fig. 5D), compiled from sequences in calmodulin binding proteins.
RSP2 binds calmodulin in a calcium-dependent manner.
For functional studies, native as well as N-terminally six-His-tagged, recombinant RSP2 was expressed in E. coli. The recombinant proteins were soluble and were recognized as 118 kDa by both monoclonal (left panel, Fig. 6A) and polyclonal (right panel, Fig. 6A) anti-RSP2 antibodies, further confirming that RSP2 was cloned. Importantly, the recombinant RSP2 comigrated with axonemal RSP2. In contrast to axonemal RSP2, the polyclonal antibody bound to the recombinant RSP2 much better than the monoclonal antibody did, indicating an unusual posttranslational modification in the bacterial expression system. To test whether the recombinant RSP2 contains kinase activity, [γ-32P]ATP was added to Ni column-purified six-His-tagged RSP2; however, the purified, recombinant RSP2 revealed only weak autophosphorylation (data not shown). Moreover, none of the recombinant RSP2 proteins displayed activity in the in-gel kinase assay. These negative results may be due to the posttranslational modification of the bacterially expressed RSP2 discussed above. We postulated that the bacterially expressed recombinant RSP2 was abnormally phosphorylated; however, treatment of the protein with exogenously added eukaryotic or prokaryotic phosphatases failed to alter protein migration in gels or restore in vitro kinase activity (data not shown).
FIG. 6.
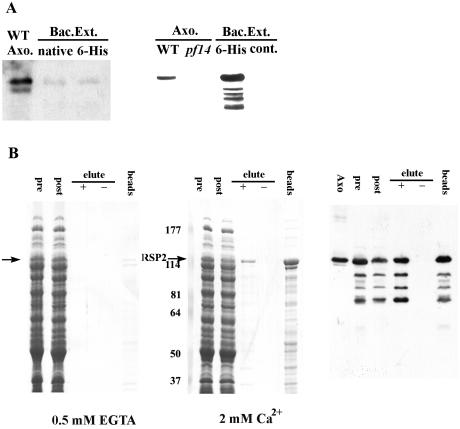
Recombinant RSP2 binds calmodulin in a Ca2+-dependent manner. (A) Extracts from bacteria transformed with the expression constructs for either native or six-His-tagged RSP2 were analyzed by Western blotting with monoclonal (left panel) and polyclonal (right panel) anti-RSP2 antibodies. Axonemes from wild-type and pf14 cells (lane 2, right panel) and the bacterial extracts without the fusion proteins (lane 4, right panel) served as controls. Compared to the polyclonal antibody (right panel), the monoclonal antibody (left panel) recognized the recombinant protein with significantly lower affinity. WT, wild type. (B) Calmodulin affinity chromatography was performed on extracts from the bacteria expressing native RSP2 in the presence (middle panel, Coomassie blue protein stain; right panel, parallel Western blot with RSP2 antibody) and absence (left panel) of calcium, and then the proteins were eluted from the calmodulin matrix with 2 mM EGTA (“+” symbol, all panels). In the presence of EGTA, no binding of RSP2 was detected (arrow, left panel). In contrast, when Ca2+ was present, RSP2 bound to the matrix (compare “pre” and “post” columns, middle and right panels), and in comparison, no bacterial proteins were visibly reduced following extraction with the calmodulin-Sepharose matrix. A fraction of the bound RSP2 can be eluted with EGTA (compare “−” and “+” lanes, middle and right panels), including degradation products of the expressed RSP2 revealed by Western blotting (right panel). However, a large fraction of the bound RSP2 remained associated with the calmodulin matrix following EGTA elution (“beads,” middle and right panels), indicating strong affinity between a fraction of RSP2 and calmodulin. Numbers at left of the middle panel are molecular masses in kilodaltons.
To test whether RSP2 could bind calmodulin, bacterial extracts containing recombinant, untagged RSP2 were incubated with calmodulin-Sepharose in the presence of EGTA (left panel, Fig. 6B) or Ca2+ (middle and right panels, Fig. 6B). The binding was evaluated by SDS-PAGE, Coomassie blue staining, and Western blotting of extracts before binding to calmodulin-Sepharose (“pre,” Fig. 6B), following extraction with the calmodulin-Sepharose beads (“post,” Fig. 6B). Binding was also evaluated following elution from calmodulin bead fractions (“elute”) by using elution buffer either containing (“+”) or lacking (“−”) EGTA.
In the presence of calcium, recombinant RSP2 is significantly reduced from the extracts following incubation with calmodulin-Sepharose (middle and right panels, arrow; compare “pre” and “post” lanes in Fig. 6B). In contrast, very little RSP2 binds to the calmodulin-Sepharose beads in the presence of EGTA (arrow, left panel, Fig. 6B). Furthermore, elution of RSP2 from the calmodulin-Sepharose matrix occurs only with the EGTA-containing elution buffer (“elute +,” lanes 3, middle and right panels, Fig. 6B). These results indicate that RSP2 binds calmodulin, and as predicted from the sequence, calmodulin binding is mostly calcium dependent. However, notably, a significant fraction of RSP2 remains bound to the calmodulin matrix following EGTA-dependent elution, suggesting a strong affinity of RSP2 once bound to calmodulin (“beads,” middle and right panels, Fig. 6B).
DISCUSSION
Our goal is to determine how the radial spokes control ciliary and flagellar bending and to test the hypothesis that the spokes act as a signal transducer involved in local control of dynein activity. The focus on RSP2 as a key regulatory protein was developed initially through evidence that RSP2 is a prominent phosphoprotein (40) and that RSP2 displays kinase-like activity in vitro by in-gel kinase assays. However, RSP2 does not contain signature sequences of the known protein kinases; thus, further experimentation will be required to determine the basis for in vitro in-gel kinase activity of RSP2. Of more immediate interest, the in-gel kinase analysis led to our focused study of RSP2, and these studies indicate that RSP2 binds calmodulin and contains a GAF domain known to mediate signaling of small ligands including cyclic nucleotides (see below). The results support a model in which RSP2 mediates calcium control of flagellar motility through an RSP2-calmodulin complex and possibly controls axonemal protein phosphorylation (Fig. 1).
Structural-functional domains in RSP2.
The RSP2 sequence predicts several coiled-coil domains, a GAF domain, and tandem calcium-dependent calmodulin binding motifs (Fig. 5). The multiple domain structure suggests that RSP2 is a scaffold designed for multiple molecular interactions crucial for both spoke assembly and function in the control of motility. For example, coiled-coil domains in RSP2 may help mediate protein-protein interactions, including dimerization, responsible for radial spoke assembly. The GAF domain and the discovery that RSP2 binds calmodulin further support a model in which RSP2 interacts with several protein partners.
The function of the GAF domain in RSP2 is intriguing but far from clear. This domain, recently identified by using sensitive alignment programs, is among the largest of all classes of domains found, particularly in proteins involved in cell signaling (1, 24). It has been clearly demonstrated that some GAF domains allosterically regulate enzymatic activity by binding small ligands (1, 24). Crystallography also revealed that some of these domains mediate homodimerization (21, 31). Based on analysis of relative stoichiometry, it is highly likely that each radial spoke contains two molecules of each major spoke protein (22; P. Yang, unpublished data). The major proteins include RSP2, and predictably, the GAF domain in RSP2 may mediate homodimerization. Such a model would be consistent with the symmetry of radial spoke and spoke head morphology (18), the mass anticipated for the 20S spoke particles, and the mass derived from gel filtration (38).
Though the ligands for most of the GAF proteins are unknown, the best characterized are cyclic nucleotide monophosphates (cNMPs). Both cAMP and cGMP modulate ciliary beating, presumably through cNMP-dependent kinases (4), but the mechanism remains unknown. Alternatively, cNMP binding to the GAF domain in RSP2 could account for cyclic nucleotide control of motility. In addition, the GAF domain in RSP2 may bind ATP and account for a fraction of the signal observed in the in-gel kinase assay. This speculation is based on similar tertiary structures in GAF and PAS domains (21), and in phytochromes the PAS domain mediates phosphorylation (64).
RSP2-calmodulin interaction and control of flagellar motility.
The sequence of RSP2 also revealed two 1-8-14 Ca2+-dependent calmodulin binding motifs. The motifs are defined as described in Fig. 5D and in the work of Rhoads and Friedberg (45) and are based on the predicted amphipathic helical structure with conserved residues at positions 1, 8, and 14 in the helix. We also demonstrated that recombinant RSP2 binds to calmodulin-Sepharose when Ca2+ is present (Fig. 6). Further, only a fraction of the RSP2 bound to calmodulin could be eluted by incubation with 0.5 mM EGTA (compare RSP2 eluted, Fig. 6B, middle and right panels, with RSP2 retained on the calmodulin-Sepharose matrix, Fig. 6B, far right lane). This observation is consistent with the high affinity of calmodulin for 1-8-14-type motifs found in other proteins (39, 45, 58). The high affinity may partly account for the stable association of calmodulin with axonemes following extraction with EDTA or 0.6 M NaCl (37) and with isolated radial spokes and the spoke stalk following KI extraction (63).
Calmodulin may mediate specific calcium-induced changes in axonemal motility, including altered waveform (5, 7, 8, 37). For example, as discussed in the introduction, in vitro reactivation of axonemes from Chlamydomonas has revealed that calcium alters waveform by direct interaction with structural components of the axoneme (3, 25, 27, 34). Identification of RSP2 as an axonemal calmodulin receptor provides a new focus and opportunity to investigate the mechanism.
One model, implied in Fig. 1, is that calcium binding to the calmodulin-RSP2 complex modifies the physical structure of the radial spoke stalk, altering the physical interactions between the asymmetric central pair apparatus and subsets of outer doublet microtubules, ultimately inducing local control of dynein-driven microtubule sliding. This model is consistent with recent functional studies indicating that a change in calcium can locally alter microtubule sliding and that the central pair-radial spoke system plays a central role in the control process (33, 48, 49, 53). The model is also generally consistent with the postulated role of the radial spokes in control of the size and shape of the axonemal bend (6, 47). Taking advantage of the isolated radial spokes (63) and single particle analysis (e.g., reference 9), it is possible to test the idea that calcium alters radial spoke structure. It is also possible that calcium binding to the radial spoke locally alters the activity of axonemal kinases and phosphatases in position to locally control dynein-driven microtubule sliding (47-50, 53, 62). This model is not exclusive of a structural model. Most importantly, the model can be tested in vivo by mutating RSP2, rather than calmodulin; transforming the RSP2 mutant pf24; and analyzing the molecular and motility phenotypes.
Acknowledgments
We thank Zaggum Bhatti (Marquette University Summer Research Program) for technical support; Carolyn Silflow (University of Minnesota) for RFLP mapping of RSP2; Pete Lefebvre (University of Minnesota), Elizabeth Smith (Dartmouth College), and Greg Pazour (University of Massachusetts) for DNA libraries; Dennis Diener and Joel Rosenbaum (Yale University) for antibodies to RSP2; and Triscia Hendrickson (Emory University) for helpful comments.
This work was supported by grants from the March of Dimes (FY02-416) and the NIH (GM51173 to W.S.S. and GM68101 to P.Y.).
REFERENCES
- 1.Aravind, L., and C. P. Ponting. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22:458-459. [DOI] [PubMed] [Google Scholar]
- 2.Bannai, H., M. Yoshimura, K. Takahashi, and C. Shingyoji. 2000. Calcium regulation of microtubule sliding in reactivated sea urchin sperm flagella. J. Cell Sci. 113:831-839. [DOI] [PubMed] [Google Scholar]
- 3.Bessen, M., R. B. Fay, and G. B. Witman. 1980. Calcium control of waveform in isolated flagellar axonemes of Chlamydomonas. J. Cell Biol. 86:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonini, N. M., and D. L. Nelson. 1988. Differential regulation of Paramecium ciliary motility by cAMP and cGMP. J. Cell Biol. 106:1615-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brokaw, C. J., and S. M. Nagayama. 1985. Modulation of the asymmetry of sea urchin sperm flagellar bending by calmodulin. J. Cell Biol. 100:1875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brokaw, C. J., D. J. L. Luck, and B. Huang. 1982. Analysis of the movement of Chlamydomonas flagella: the function of the radial-spoke system is revealed by comparison of wild-type and mutant flagella. J. Cell Biol. 92:722-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brokaw, C. J. 1987. Regulation of sperm flagellar motility by calcium and cAMP-dependent phosphorylation. J. Cell. Biochem. 35:175-184. [DOI] [PubMed] [Google Scholar]
- 8.Brokaw, C. J. 1991. Calcium sensors in sea urchin sperm flagella. Cell Motil. Cytoskelet. 18:123-130. [DOI] [PubMed] [Google Scholar]
- 9.Burgess, S. A., M. Walker, H. Sakakibara, P. Knight, and K. Oiwa. 2003. Dynein structure and power stroke. Nature 421:715-718. [DOI] [PubMed] [Google Scholar]
- 10.Chen, X., K. L. Kindle, and D. B. Stern. 1995. The initiation codon determines the efficiency but not the site of translation initiation in Chlamydomonas chloroplast. Plant Cell 7:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cote, G. P., X. Luo, M. B. Murphy, and T. T. Egelhoff. 1997. Mapping of the novel protein kinase catalytic domain of Dictyostelium myosin II heavy chain kinase. J. Biol. Chem. 272:6846-6849. [DOI] [PubMed] [Google Scholar]
- 12.Curry, A. M., and J. L. Rosenbaum. 1993. Flagellar radial spoke: a model molecular genetic system for studying organelle assembly. Cell Motil. Cytoskelet. 24:224-232. [DOI] [PubMed] [Google Scholar]
- 13.Diener, D. R., L. H. Ang, and J. L. Rosenbaum. 1993. Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of the axoneme binding domain of radial spoke protein 3. J. Cell Biol. 123:183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Futey, L. M., Q. Medley, G. P. Cote, and T. T. Egelhoff. 1995. Structural analysis of MHCK A from Dictyostelium. J. Biol. Chem. 270:523-529. [DOI] [PubMed] [Google Scholar]
- 15.Gaillard, A. R., D. R. Diener, J. L. Rosenbaum, and W. S. Sale. 2001. Flagellar radial spoke protein 3 is an A-kinase anchoring protein (AKAP). J. Cell Biol. 153:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner, L. C., E. O'Toole, C. A. Perrone, T. Giddings, and M. E. Porter. 1994. Components of a “dynein regulatory complex” are located at the junction between the radial spokes and the dynein arms in Chlamydomonas flagella. J. Cell Biol. 127:1311-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gingras, D., D. White, J. Garin, J. Cosson, P. Huitorel, H. Zingg, C. Cibert, and C. Gagnon. 1998. Molecular cloning and characterization of a radial spoke head protein of sea urchin sperm axonemes: involvement of the protein in the regulation of sperm motility. Mol. Biol. Cell 9:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodenough, U. W., and J. E. Heuser. 1985. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J. Cell Biol. 100:2008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamasaki, T., T. J. Murtaugh, B. H. Satir, and P. Satir. 1989. In vitro phosphorylation of Paramecium axonemes and permeabilized cells. Cell Motil. Cytoskelet. 12:1-11. [DOI] [PubMed] [Google Scholar]
- 20.Hanks, S. K., and T. Hunter. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9:576-596. [PubMed] [Google Scholar]
- 21.Ho, Y. S., L. M. Burden, and J. H. Hurley. 2000. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 19:5288-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, B., G. Piperno, Z. Ramanis, and D. J. Luck. 1981. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell Biol. 88:80-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, B., Z. Ramanis, and D. J. Luck. 1982. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell 28:115-124. [DOI] [PubMed] [Google Scholar]
- 24.Hurley, J. H. 7 January 2003, posting date. GAF domains: cyclic nucleotides come full circle. Sci. STKE 164:PE1. [Online.] http://www.stke.org/cgi/content/full/sigtrans;2003/164/pe1. [DOI] [PubMed] [Google Scholar]
- 25.Hyams, J. S., and G. G. Borisy. 1978. Isolated flagellar apparatus of Chlamydomonas: characterization of forward swimming and alteration of waveform and reversal of motion by calcium ions in vitro. J. Cell Sci. 33:235-253. [DOI] [PubMed] [Google Scholar]
- 26.Kameshita, I., and H. Fujisawa. 1989. A sensitive method for detection of calmodulin-dependent protein kinase II activity in SDS PAGE. Anal. Biochem. 183:139-143. [DOI] [PubMed] [Google Scholar]
- 27.Kamiya, R., and G. B. Witman. 1984. Submicromolar levels of calcium control the balance of beating between the two flagella in demembranated models of Chlamydomonas. J. Cell Biol. 98:97-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kathir, P., M. LaVoie, W. Brazelton, N. A. Hass, P. A. Lefebvre, and C. D. Silflow. 2003. Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell 2:362-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko, F. C. F., and K. L. Chow. 2003. A mutation at the start codon defines the differential requirement of dpy-11 in C. elegans body hypodermis and male tail. Biochem. Biophys. Res. Commun. 309:201-208. [DOI] [PubMed] [Google Scholar]
- 30.Kurvari, V., Y. Zhang, Y. Luo, and W. J. Snell. 1996. Molecular cloning of a protein kinase whose phosphorylation is regulated by genetic adhesion during Chlamydomonas fertilization. Proc. Natl. Acad. Sci. USA 93:39-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez, S. E., A. Y. Wu, N. A. Glavas, X.-B. Tan, S. Turley, W. G. J. Hol, and J. A. Beavo. 2002. The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc. Natl. Acad. Sci. USA 20:13260-13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mastronarde, D. N., E. T. O'Toole, K. L. McDonald, J. R. McIntosh, and M. E. Porter. 1992. Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas. J. Cell Biol. 118:1145-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakano, I., T. Kobayashi, M. Yoshimura, and C. Shingyoji. 2003. Central-pair-linked regulation of microtubule sliding by calcium in flagellar axonemes. J. Cell Sci. 116:1627-1636. [DOI] [PubMed] [Google Scholar]
- 34.Omoto, C. K., and C. J. Brokaw. 1985. Bending patterns in Chlamydomonas flagella: II. Calcium effects on reactivated Chlamydomonas flagella. Cell Motil. 5:53-60. [DOI] [PubMed] [Google Scholar]
- 35.Omoto, C. K., I. R. Gibbons, R. Kamiya, C. Shingyoji, K. Takahashi, and G. B. Witman. 1999. Rotation of the central pair microtubules in eukaryotic flagella. Mol. Biol. Cell 10:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Neil, K. T., and W. F. DeGrado. 1990. How calmodulin binds its targets: sequence independent recognition of amphiphilic α helices. Trends Biol. Sci. 15:59-64. [DOI] [PubMed] [Google Scholar]
- 37.Otter, T. 1989. Calmodulin and the control of flagellar movement, p. 281-298. In F. D. Warner, P. Satir, and I. R. Gibbons (ed.), Cell movement, vol. 1. Alan R. Liss, Inc., New York, N.Y.
- 38.Padma, P., Y. Satouh, K. Wakabayashi, A. Hozumi, Y. Ushimara, R. Kamiya, and K. Inaba. 2003. Identification of a novel leucine-rich repeat protein as a component of the flagellar radial spoke in ascidian Ciona intestinalis. Mol. Biol. Cell 14:774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Picton, C., C. B. Klee, and P. Cohen. 1980. Phosphorylase kinase from rabbit skeletal muscle: identification of the calmodulin-binding subunits. Eur. J. Biochem. 111:553-558. [DOI] [PubMed] [Google Scholar]
- 40.Piperno, G., B. Huang, Z. Ramanis, and D. J. Luck. 1981. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 88:73-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piperno, G., Z. Ramanis, E. F. Smith, and W. S. Sale. 1990. Three distinct inner dynein arms in Chlamydomonas flagella: molecular composition and location in the axoneme. J. Cell Biol. 110:379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piperno, P., K. Mead, and W. Shestak. 1992. The inner dynein arms I2 interact with a “dynein regulatory complex” in Chlamydomonas flagella. J. Cell Biol. 118:1455-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porter, M. E., and W. S. Sale. 2000. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151:F37-F42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ranum, L. P., M. D. Thompson, J. A. Schloss, P. A. Lefebvre, and C. Silflow. 1988. Mapping flagellar genes in Chlamydomonas using restriction fragment length polymorphisms. Genetics 120:109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhoads, A. R., and F. Friedberg. 1997. Sequence motifs for calmodulin recognition. FASEB J. 11:331-340. [DOI] [PubMed] [Google Scholar]
- 46.Ryazanov, A. G. 2002. Elongation factor-2 kinase and its newly discovered relatives. FEBS Lett. 514:26-29. [DOI] [PubMed] [Google Scholar]
- 47.Satir, P. 1985. Switching mechanisms in the control of ciliary motility, p. 1-46. In P. Satir (ed.), Modern cell biology. Alan R. Liss, Inc., New York, N.Y.
- 48.Smith, E. F. 2002. Regulation of flagellar dynein by the axonemal central apparatus. Cell Motil. Cytoskelet. 52:33-42. [DOI] [PubMed] [Google Scholar]
- 49.Smith, E. F. 2002. Regulation of flagellar dynein by calcium and a role for an axonemal calmodulin and calmodulin-dependent kinase. Mol. Biol. Cell 13:3303-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, E. F., and W. S. Sale. 1992. Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science 257:1557-1559. [DOI] [PubMed] [Google Scholar]
- 51.Sturgess, J. M., J. Chao, J. Wong, N. Aspin, and J. A. Turner. 1979. Cilia with defective radial spokes: a cause of human respiratory disease. N. Engl. J. Med. 300:53-56. [DOI] [PubMed] [Google Scholar]
- 52.Wakabayashi, K., T. Yagi, and R. Kamiya. 1997. Ca2+-dependent waveform conversion in the flagellar axoneme of Chlamydomonas mutants lacking the central-pair/radial spoke system. Cell Motil. Cytoskelet. 38:22-28. [DOI] [PubMed] [Google Scholar]
- 53.Wargo, M. J., and E. F. Smith. 2003. Asymmetry of the central apparatus defines the location of active microtubule sliding in Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA 100:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warner, F. D., and P. Satir. 1974. The structural basis of ciliary bend formation: radial spoke positional changes accompanying microtubule sliding. J. Cell Biol. 63:35-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams, B. D., M. A. Velleca, A. M. Curry, and J. L. Rosenbaum. 1989. Molecular cloning and sequence analysis of the Chlamydomonas gene coding for radial spoke protein 3: flagellar mutation pf-14 is an ochre allele. J. Cell Biol. 109:235-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Witman, G. B., J. Plummer, and G. Sander. 1978. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76:729-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Witman, G. B. 1986. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134:280-290. [DOI] [PubMed] [Google Scholar]
- 58.Xia, X.-M., B. Fakler, G. Wayman, T. Johnson-Pais, J. E. Keen, T. Ishii, B. Hirschberg, C. T. Bond, S. Lutsenko, J. Maylie, and J. P. Adelman. 1998. Mechanism of calcium gating in small-conductance calcium-activated potassium channels. Nature 395:503-507. [DOI] [PubMed] [Google Scholar]
- 59.Yagi, T., and R. Kamiya. 2000. Vigorous beating of Chlamydomonas axonemes lacking central pair/radial spoke structures in the presence of salts and organic compounds. Cell Motil. Cytoskelet. 46:190-199. [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi, H., M. Matsushita, A. C. Nairn, and J. Kuriyan. 2001. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol. Cell 7:1047-1057. [DOI] [PubMed] [Google Scholar]
- 61.Yang, P., and W. S. Sale. 1998. The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol. Biol. Cell 9:3335-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, P., and W. S. Sale. 2000. Casein kinase I is anchored on axonemal doublet microtubules and regulates flagellar dynein phosphorylation and activity. J. Biol. Chem. 275:18905-18912. [DOI] [PubMed] [Google Scholar]
- 63.Yang, P., D. R. Diener, J. L. Rosenbaum, and W. S. Sale. 2001. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J. Cell Biol. 153:1315-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeh, K. C., and J. C. Lagarias. 1998. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95:13976-13981. [DOI] [PMC free article] [PubMed] [Google Scholar]