Abstract
Frequent oncogenic alterations occur in the phosphoinositide 3-kinase (PI3K) pathway, urging identification of novel negative controls. We previously reported an original mechanism for restraining PI3K activity, controlled by the somatostatin G protein-coupled receptor (GPCR) sst2 and involving a ligand-regulated interaction between sst2 with the PI3K regulatory p85 subunit. We here identify the scaffolding protein filamin A (FLNA) as a critical player regulating the dynamic of this complex. A preexisting sst2-p85 complex, which was shown to account for a significant basal PI3K activity in the absence of ligand, is disrupted upon sst2 activation. FLNA was here identified as a competitor of p85 for direct binding to two juxtaposed sites on sst2. Switching of GPCR binding preference from p85 toward FLNA is determined by changes in the tyrosine phosphorylation of p85- and FLNA-binding sites on sst2 upon activation. It results in the disruption of the sst2-p85 complex and the subsequent inhibition of PI3K. Knocking down FLNA expression, or abrogating FLNA recruitment to sst2, reversed the inhibition of PI3K and of tumor growth induced by sst2. Importantly, we report that this FLNA inhibitory control on PI3K can be generalized to another GPCR, the mu opioid receptor, thereby providing an unprecedented mechanism underlying GPCR-negative control on PI3K.
INTRODUCTION
G protein-coupled receptors (GPCRs) are a large class of transmembrane receptors that represent major drug targets. GPCRs are crucial mediators of signaling pathways involved in numerous cellular responses including metabolism, secretion, growth, differentiation, and cell motility. Although GPCRs were originally thought to act solely through heterotrimeric G proteins which in turn regulate intracellular enzymes or ion channels, it is now well established that GPCRs directly interact, via their intracellular loops, with a variety of surface and cytoplasmic proteins that are often specific for a subset of receptors. These proteins have been implicated in regulating GPCR cell membrane stabilization, internalization, and desensitization as well as in scaffolding GPCR-mediated signaling (3, 25, 30, 40).
sst2 is an inhibitory receptor that belongs to the GPCR family of somatostatin receptors and transduces the majority of somatostatin actions, including inhibition of hormone and growth factor secretion, cell proliferation, survival, migration, and angiogenesis (38). Consistently, the critical role of sst2 in the negative regulation of endocrine processes and of tumor growth, and its overexpression in human endocrine tumors, has led to the wide use of sst2 agonists such as octreotide for therapeutic and diagnostic purposes (1, 53). Interestingly, sst2 behaves as a tumor suppressor gene for pancreatic cancer. Indeed, a selective loss of the sst2 receptor is observed in human pancreatic adenocarcinoma (7), and restoring sst2 expression in human pancreatic cancer cells results in inhibition of cell growth and tumorigenesis (2, 11, 18). In addition to G protein subunits, sst2 engages different protein partners to initiate inhibitory signaling pathways such as tyrosine phosphatases SHP-1 and SHP-2 and the tyrosine kinases src and JAK2 (16, 17, 20). More recently, we reported that sst2 inhibits the phosphoinositide 3-kinase (PI3K)/AKT pathway via an original mechanism that involves a direct interaction between the sst2 first intracellular loop and the PI3K regulatory p85 subunit (4). Indeed, upon ligand activation of sst2, p85 dissociates from a preexisting basal sst2-p85 complex which was shown to account for a significant PI3K activity in different cell models. p85 dissociation from sst2 results in the inhibition of PI3K activity. Importantly, somatostatin-mediated disruption of the sst2-p85 complex constitutes a critical step in transducing sst2 oncosuppressive effects (4). However, the molecular mechanisms involved in this disruption remain to be identified.
We hypothesized that ligand activation of the sst2 receptor induces the binding on the sst2 first intracellular loop of a yet-unidentified protein that, by competition, may force p85 dissociation from sst2. Interestingly, our precedent report indicated that the p85-binding YXXM motif is localized in the intracellular loop sequences of 40 GPCR retrieved from a nonexhaustive database of 780 GPCR entries (4). Among those, receptors of the opioid family, including the mu opioid receptor MOR, have been identified. Interestingly, when screening for proteins that might directly interact with the GPCRs sst2 and MOR, we came upon filamin A (FLNA), which has previously be shown to directly interact with MOR, regulating receptor trafficking (36).
Filamin A (FLNA) is an actin-binding and scaffolding protein for numerous cytosolic signaling proteins and transmembrane receptors including GPCRs (15, 36), regulating their cell organization, trafficking, activity, and downstream signals. We therefore hypothesized that FLNA orchestrates the disassembly of GPCR-p85 complexes. We here demonstrate that by directly interacting with a hydrophobic motif closely juxtaposed to the p85-binding Y71XXM motif present in the first intracellular loop of sst2 and MOR, FLNA competes with p85 for binding to GPCR and forces p85 dissociation from GPCR, thereby providing an original control mechanism for GPCR-mediated inhibition of the PI3K pathway. Interestingly, the switch of GPCR binding preference from p85 toward FLNA is determined by GPCR tyrosine phosphorylation status on p85- and FLNA-binding sites.
MATERIALS AND METHODS
Plasmids.
FLNA full-length cDNA was kindly provided by Y. Ohta (Tokyo Medical and Dental University). FLNA repeats 17 and 18 (amino acids 1863 to 2044), 19 and 20 (amino acids 2045 to 2235), and 21 to 24 (amino acids 2236 to 2647) were generated by PCR and subcloned into pGEX vectors (Amersham) for the expression of glutathione S-transferase (GST) fusion proteins. T7-tagged human sst2 was generated as described previously (49), and mutations of sst2-I65Y66V67 residues were performed using site-directed mutagenesis (Stratagene).
GST fusion proteins.
GST fusion proteins were affinity purified using gluthathione Sepharose 4B beads (Amersham). GST fusion proteins were eluted with elution buffer (10 mM reduced glutathione-50 mM Tris-HCL [pH 8.0] or 10 mM reduced glutathione-50 mM HEPES pH 8.0 for in vitro binding assays or surface plasmon resonance [SPR] analyses, respectively). GST was cleaved from p85 fusion protein using the Thrombin CleanCleave kit (Sigma).
SPR.
Real-time binding experiments were performed with a BIAcore 3000 biosensor instrument (BIAcore AB) and quantified in terms of resonance units (RU) (1,000 RU = 1 ng of protein bound/mm2 of flow cell surface) (4, 16). Synthetic biotinylated peptides covering sst2-il1 (residues T62 to T78), sst2-il2 (residues R140 to R157), sst2-il3 (residues F230 to T255), or the sst2 C-terminal domain (residues N304 to K321) were immobilized onto streptavidin-coated carboxymethylated dextran chips (BIAcore AB). Flow cells were coated with 50 RU of each peptide. GST fusion proteins were injected in the running buffer (HEPES 10 mM, NaCl 150 mM, EDTA 3 mM, polysorbate 0.005%). Kinetic constants (association constant [Ka], dissociation constant [Kd], and equilibrium dissociation constant [KD]) were evaluated using BIAevaluation 4.01 software (BIAcore AB).
For the competition assay, fusion GST-p85 protein was immobilized on a carboxymethylated dextran chip (chip CM5; BIAcore AB). Cell extracts (100 to 400 μg/ml) were injected together with increasing concentrations of GST-FLNA-19-20, and antibodies were injected when indicated (1 to 3% [vol/vol]).
Pulldown assays.
Pulldown assays were performed using biotinylated custom-synthesized peptides (NeoMPS) covering wild-type (WT) sst2-il1 or mutated I65YV67sst2-il1 (from residues T62 to T78, with residue Y71 phosphorylated), or covering wild-type MOR-il1 or mutated M92YV94-MOR-il1 (from residues F89 to A104 with residue Y98 phosphorylated). Streptavidin-coated beads were incubated at 4°C with the biotinylated peptide and then with the GST fusion proteins in binding buffer (10 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM dithiothreitol [DTT], 0.1% Triton X-100) in the presence of 1 mM EDTA and protease inhibitors for 2 to 3 h. Bound proteins were fractionated by SDS-PAGE and analyzed by Western blotting.
Molecular modeling.
sst2-il1, flanked with its transmembrane (TM) domains I and II (residues 42 to 105), was constructed using the X-ray crystal structure of bovine rhodopsin from entry 1HZX (47) as the template. A model structure of the whole sst2 receptor was also generated to reduce misinterpretations due to possible overlaps with other receptor parts (not shown). A structure model of FLNA repeat 19 was generated using the available X-ray structural data of FLNA repeat 17 in complex with glycoprotein IB (PDB entry 2PB3) (33). The more recently available structure information on FLNA repeat 19 from PDB entry 2J3S (23) supports this initial model. The docking of the sst2-il1-FLNA repeat 19 complex was achieved by manual docking using known FLNA interactions with other proteins. Each complex model from these docking was used as the target in detailed molecular refinements including optimization of backbone conformations, side chain conformations, and side chain interactions. Subsequent energy minimizations using insight II modules Homology, Discover, and Biopolymer (Accelrys, San Diego, CA) produced the final complex structures.
Cell culture and transfection.
M2 and A7 cells were kindly provided by T. P. Stossel (Harvard Medical School, Boston) and stably transfected with T7-tagged sst2. Pools were selected using 400 μg/ml hygromycin. Stable M2-sst2 and A7-sst2 transfectants were grown in minimal essential medium (MEM) supplemented with 8% newborn calf serum and 2% fetal calf serum (FCS). BON cells were maintained in (1:1) nutrient mixture of Dulbecco's modified Eagle medium (DMEM) and F12K with 10% FCS. CHO, HaCaT, SH-SY5Y, and BxPC-3 cells were grown in DMEM with 10% FCS as described previously (16). BxPC-3 cells were stably transfected with either the human T7-tagged wild-type sst2 or mutated I65Y66V67 sst2, and pools were selected using 400 μg/ml G-418. SH-SY5Y cells were stably transfected with the T7-tagged MOR receptor, as described previously (32), and provided by L. mouledous.
For RNA interference, cells were transfected with 100 nM FLNA small interfering RNA (siRNA) (55) or nontargeting siRNA (Qiagen) using DharmaFECT1 reagent (Dharmacon), according to the manufacturer's instructions. For transient transfection, we used FuGENE6 (Roche Diagnostics).
For cell treatment, cells were replaced with fresh media before the addition of somatostatin analog RC-160 (American Peptides) or DAMGO ([d-Ala2, (NMe)Phe4, Gly5-ol]-enkephalin; Bachem, Weil am Rhein, Germany) in the presence of FCS.
Immunoprecipitation and Western blotting.
Immunoprecipitation and Western blotting were performed as previously described (4).
Antibodies.
The following antibodies were purchased from the commercial suppliers indicated: monoclonal anti-GST (Sigma); polyclonal anti-p85 (Upstate Biotechnology); polyclonal phospho-(Tyr) p85 PI3K-binding motif, monoclonal anti-AKT, and polyclonal anti-S473-phospho-AKT (Cell Signaling); monoclonal antifilamin mAb1678 (Chemicon); monoclonal anti-T7-tag (Novagen); and polyclonal anti-sst2 (generated in our laboratory).
Cell proliferation.
Cells were counted with a Coulter counter model Z1 (Coulter Electronics) (24).
Cell viability assay.
Mitochondrial viability was measured using the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) (Sigma) colorimetric assay (4).
Caspase activity assay.
Executioner caspase activity was measured using the Quantipak kit (Biomol International) as described previously (18).
Immunocytofluorescence.
A7-sst2 and M2-sst2 cells were grown on poly-l-lysine-coated coverslips overnight. Following treatment with somatostatin 14 (SS-14), cells were fixed with 4% paraformaldehyde and 0.2% picric acid in phosphate buffer (pH 6.9) for 40 min at room temperature and washed several times. Specimens were permeabilized and then incubated with the anti-sst2 antibody followed by cyanine 3.18-conjugated secondary antibodies (Amersham, Braunschweig, Germany). Specimens were mounted and examined using a Leica TCS-NT laser scanning confocal microscope (Leica Microsystems, Nussloch, Germany).
BxPC3 cells expressing sst2-WT or mutated sst2EEE were grown on poly-l-lysine (Sigma-Aldrich)-coated glass coverslips in complete medium. Attached cells were washed in 1× phosphate-buffered saline (PBS) and then fixed in 4% paraformaldehyde for 10 min. After washing in PBS, the cells were blocked in PBS containing 0.1% bovine serum albumin for 1 h and incubated with a 1:100 dilution of he monoclonal anti-T7 antibody. After washing with PBS, cells were incubated with a 1:500 dilution of the Alexa Fluor 488 donkey anti-mouse IgG(H+L) (A21202; Invitrogen). Mounted cells were examined under Zeiss laser scanning confocal microscope LSM510 (26).
Quantification of receptor internalization by enzyme-linked immunosorbent assay (ELISA).
A7-sst2 and M2-sst2 cells were seeded onto poly-l-lysine-treated 24-well plates. The next day, cells were preincubated with the anti-T7 antibody for 2 h at 4°C. Following treatment with somatostatin 14 (SS-14), cells were fixed and incubated with peroxidase-conjugated anti-mouse antibody for 2 h at room temperature. After washing, the plates were developed with 2,2-azino-di[3-ethyl-benzthiazoline sulfate] (Roche Diagnostics, Manheim, Germany) solution and analyzed at 405 nm using a microplate reader (26).
Athymic mouse xenograft.
BxPC-3 cells expressing mock, wild-type, or mutated sst2EEE receptor were inoculated subcutaneously (s.c.) into athymic female mice (Swiss nude/nude). Tumor volumes were measured every week and calculated as previously described (4).
Statistical analysis.
Experiments were performed at least three times. Statistical analyses were performed with Student's t test. All values are mean ± standard error of the mean (SEM).
RESULTS
The actin-binding protein FLNA directly interacts with sst2 first intracellular loop.
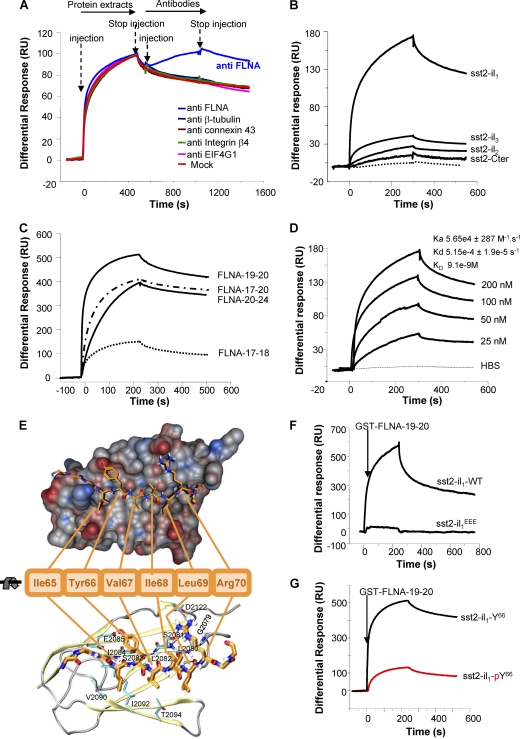
We investigated by surface plasmon resonance (SPR) whether FLNA directly interacts with sst2 first intracellular loop (sst2-il1). SPR was first designed by immobilizing a purified biotinylated peptide encompassing the sequence of the first intracellular loop of sst2 and injecting a protein extract that contains FLNA (CHO cells; see below) (Fig. 1A). A significant response was observed on the chip-immobilized sst2-il1 peptide, indicating that protein(s) present in the protein extract binds to sst2-il1. This response was further increased after injecting an anti-FLNA antibody, but not unrelated antibodies, indicating that FLNA is an interacting partner of sst2-il1. FLNA is a homodimeric protein, each monomer comprising an N-terminal actin-binding domain followed by a 24 highly homologous tandem repeats. Most of the FLNA-interacting proteins bind to FLNA between repeats 17 and 24 (37). Using the affinity-purified GST-FLNA repeats 17 to 24, we demonstrated by SPR that the direct interaction occurring between sst2-il1 and FLNA involves these repeats (Fig. 1B). This interaction was specific to sst2-il1 because it was not observed with purified biotinylated peptides encompassing the sequence of the other three cytoplasmic domains (2nd and 3rd intracellular loops, and C-terminal tail) of sst2 (Fig. 1B). To further refine the FLNA-sst2 interaction, we examined by SPR the ability of GST-FLNA repeats 17 and 18, 19 and 20, and 21 and 24 (FLNA-17-18, FLNA-19-20, and FLNA-21-24, respectively) to interact with sst2-il1. FLNA-19-20 and FLNA-21-24, but not FLNA-17-18, highly interacted with sst2-il1 (Fig. 1C). Moreover, FLNA-19-20 bound sst2-il1 in a dose-dependent manner and with a high affinity (KD = 9.1 nM) (Fig. 1D).
Fig 1.
Identification and characterization of a direct interaction between sst2 and FLNA in vitro. (A–D, F, G) SPR analyses: Results are expressed as a differential response (RU) after subtracting signals from running buffer. (A) SPR analysis using the biotinylated sst2-il1 peptide immobilized onto the Biacore chip and injection first of CHO protein extracts and then the anti-FLNA, anti-β-tubulin, anti-connexin 43, anti-integrin β4, or anti-G subunit of eukaryotic initiation factor 4 (anti-EIF4G) antibody (n = 3). (B) SPR analysis of the interaction of GST-FLNA repeats 17 to 24 with immobilized biotinylated peptides encompassing the sequence of each of the intracellular loops (sst2-il1 to sst2-il3) or the C-terminal tail of sst2 (sst2-Cter) (n = 2). (C) Mapping of sst2 receptor-binding site on FLNA: sst2-il1 peptide was immobilized onto the Biacore chip, and GST-FLNA repeat units 17 and 18, 19 and 20, or 21 to 24 (FLNA-17-18, FLNA-19-20, or FLNA-21-24, respectively) (100 nM) was injected (n = 3). (D) sst2-il1 peptide was immobilized onto the Biacore chip and increasing concentrations of GST-FLNA-19-20 were injected (25 to 200 nM). Calculated kinetic constants (Ka, Kd, KD) are indicated. (E) Molecular modeling of the sst2-il1-FLNA repeat 19 complex: van der Waals surface representation of FLNA repeat 19 shows contribution of the sst2-il1 hydrophobic site (electrostatic interactions) in the vicinity of the β-strand. Residues Y66 and R70 of sst2-il1 interact with E2085 and S2081/D2122 of FLNA repeat 19, respectively. This way, residues I65, V67, and I68 point toward L2080, L2082, I2084, V2090, I2092, and T2094 into the hydrophobic pocket of FLNA repeat 19, resulting in additional energy gain and stabilizing the interaction. (F) The sst2-I65Y66V67 motif is critical for FLNA-sst2-il1 interaction: SPR analysis of the interaction of GST-FLNA-19-20 to immobilized biotinylated peptides covering wild-type sst2-il1-WT or mutated sst2-il1EEE (n = 3). (G) phosphorylation of residue Y66 in sst2-IY66V motif abrogates FLNA-sst2-il1 interaction: SPR analysis of the interaction of GST-FLNA-19-20 to immobilized biotinylated peptides covering unphosphorylated (sst2-il1-Y66) or phosphorylated (sst2-il1-pY66) Y66 on sst2-il1 (n = 3).
Available data from the literature on FLNA-protein complexes (21, 27, 33, 46) suggest that the hydrophobic residues I65, Y66, and V67 present in a β-strand formed by sst2-il1 represents the motif for FLNA binding onto sst2-il1. The molecular model of the sst2-FLNA interaction obtained from the docking of sst2-il1 with FLNA repeat 19 is indeed consistent with this hypothesis (Fig. 1E). The main electrostatic interactions occur between the β-strand formed by sst2-il1 and the C strand of FLNA repeat 19. Moreover, Y66 and R70 residues in sst2-il1 interact with nearby hydrophilic side chains in FLNA. Hydrophobic residues V64, I65, and I68 in sst2-il1 point into the hydrophobic groove between strands C and D in FLNA repeat 19. V67 is located on the rather hydrophilic side of sst2-il1 β-strand and benefits from its short side chain, which is buried between neighboring side chains of Y66 and R70. To validate the proposed FLNA interaction motif in sst2-il1, we introduced negative charges by either exchanging residues I65, Y66, and V67 for three glutamate residues (sst2-il1EEE) or adding a phosphate group on Y66 (sst2-il1-pY66) to mimic a phosphorylated residue. SPR analyses indicated that FLNA interaction with sst2-il1EEE (Fig. 1F) or with sst2-il1-pY66 (Fig. 1G) is abolished.
FLNA competes with p85 for binding to sst2 first intracellular loop.
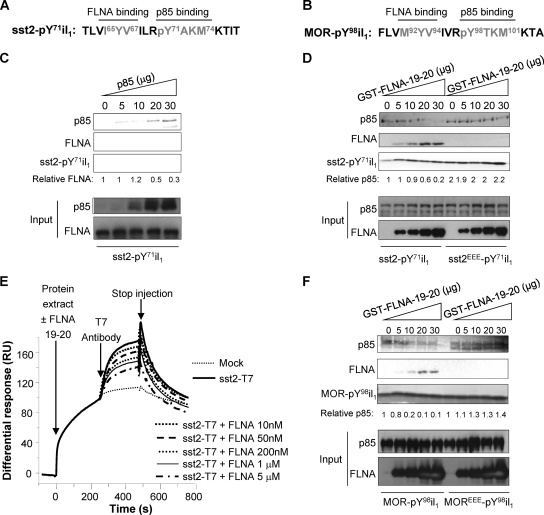
We previously reported that sst2-il1 physically interacts with p85, the regulatory subunit of PI3K. A prerequisite for this interaction is the phosphorylation of the Y71 residue in the p85-binding motif pY71AKM (4). The hypothesis that FLNA and p85 compete for binding to sst2-il1 was tested in vitro using a purified biotinylated peptide encompassing the sequence of the sst2-il1 where the Y66 residue is unphosphorylated (to allow FLNA interaction) and Y71 is phosphorylated (sst2-pY71il1) (Fig. 2A). The recombinant GST-FLNA-19-20 protein was able to bind to the biotinylated sst2-pY71il1 peptide, as assessed by pulldown (Fig. 2C). Challenging this interaction with increasing amounts of a purified GST-p85 recombinant protein caused a dose-dependent dissociation of FLNA-19-20 from sst2-pY71il1 (Fig. 2C). Likewise, GST-p85 binds to the sst2-pY71il1 peptide, but increasing amounts of FLNA-19-20 caused a dose-dependent inhibition of p85 binding to sst2-pY71il1 (Fig. 2D). Interestingly, this inhibition was abrogated when the FLNA binding-defective sst2EEE-pY71il1 peptide was used (Fig. 2D), confirming that p85 dissociation from the sst2-pY71il1 peptide relies on FLNA association specifically to the I65Y66V67 residues on sst2-il1. GST alone did not bind sst2-pY71il1 (not shown). These results argue for a competition between FLNA and p85 for binding to sst2-il1.
Fig 2.
FLNA and p85 compete for a direct binding to GPCR. (A and B) sst2 (A) and MOR (B) first intracellular loop sequences showing the identified p85-PI3K (4) and FLNA-binding sites. (C) Five micrograms of the GST-FLNA-19-20 was first mixed with the biotinylated sst2-pY71il1 peptide immobilized onto streptavidin beads, and increasing amounts (0 to 30 μg) of GST-p85 were added; bindings of GST-FLNA-19-20 and GST-p85 were assessed by Western blotting with the indicated antibodies. Loading of equal amounts of biotinylated sst2-pY71il1 peptide in each lane was checked by Coomassie blue gel staining. Quantification of the relative FLNA binding to the sst2-pY71il1 peptide was calculated at each concentration of added GST-p85, using ImageJ software (ratio FLNA blot/sst2-pY71il1 stain). An arbitrary value of 1 has been assigned to the quantity of FLNA associated to sst2-pY7il1 in the absence of added GST-p85. (D) Three micrograms of GST-p85 was first mixed with each of the biotinylated sst2-pY71il1 or mutated sst2EEE-pY71il1 peptide immobilized onto streptavidin beads, and increasing amounts (0 to 30 μg) of GST-FLNA-19-20 were added. Quantifications of the relative p85 binding to sst2-il1 peptides were calculated as described above. An arbitrary value of 1 has been assigned to the quantity of p85 associated to each sst2-il1 peptide in the absence of added GST-FLNA-19-20. (E) SPR analysis using GST-p85 protein immobilized onto the Biacore chip and injection of mock (dotted line) or T7-tagged sst2-transfected (solid line) CHO protein extracts. Competition was performed by coinjecting, together with the sst2-expressing CHO protein extracts, of increasing concentrations of GST-FLNA-19-20 (10 nM to 5 μM). When indicated, the anti-T7 antibody was injected. Results are expressed as a normalized differential response (RU), referring to the maximal response observed for each protein extract in the absence of the anti-T7 antibody and of GST-FLNA-19-20. (F) Three micrograms of GST-p85 was first mixed with each of the biotinylated MOR-pY98il1 or mutated MOREEE-pY98il1 peptide immobilized onto streptavidin beads, and increasing amounts of GST-FLNA-19-20 were added. Quantifications of the relative p85 binding to MOR-pY98il1 peptides were calculated as described above. An arbitrary value of 1 has been assigned to the quantity of p85 associated to each MOR-pY98il1 peptide in the absence of added GST-FLNA-19-20.
To further confirm these results, we analyzed the ability of FLNA to disrupt the sst2-p85 interaction by SPR analysis (Fig. 2E). Recombinant GST-p85 protein was immobilized onto the chip, and protein extracts either from mock- or sst2-expressing CHO cells were injected (sst2 is tagged with T7). Proteins present in both cell lines bind to GST-p85, as revealed by the response observed upon injection of both protein extracts. More specifically, binding to GST-p85 of the T7-tagged sst2 (present in sst2-expressing CHO protein extracts) is here demonstrated by the further increase in the response observed upon injection of an anti-T7 antibody, which is not observed when using mock CHO protein extracts. However, this T7 antibody-related response was dose-dependently abrogated when increasing concentrations of the GST-FLNA-19-20 (FLNA 10 nM to 5 μM) were coinjected as a competitor together with the CHO-T7-sst2 protein extract (Fig. 2E).
FLNA competes with p85 for binding to the mu opioid receptor.
Among the 40 GPCRs that we have previously identified to exhibit a p85-binding YXXM motif in one of their intracellular loop sequences, 26 of them, including MOR, also present a sequence similar to that of the FLNA-binding motif found in sst2-il1 and located just upstream of the YXXM motif (Table 1). This suggests that these receptors interact with both FLNA and p85 in a competitive manner. In MOR, the putative FLNA-binding (M92YV) and p85-binding (Y98TKM) motifs were identified in the first intracellular loop (MOR-il1) (Table 1). To investigate whether FLNA and p85 interact directly with MOR, and whether this interaction involves the predicted M92YV and Y98TKM motifs in MOR-il1, respectively, pulldown experiments using a MOR-il1 peptide containing a phosphorylated pY98 residue (MOR-pY98il1) (Fig. 2B) were performed. Interestingly, GST-p85 interacted with MOR-pY98il1 (Fig. 2F). GST alone did not bind MOR-pY98il1 (not shown). Remarkably, the addition of increasing amounts of GST-FLNA-19-20 dose-dependently inhibited GST-p85 binding to MOR-pY98il1. However, this inhibition was abrogated when the peptide MOREEE-pY98il1, encompassing the MOR first intracellular loop mutated on the putative M92Y93V94 FLNA-binding motif, was used (Fig. 2F). Altogether, these results demonstrate that the direct interaction of FLNA and p85 with MOR-il1 is mutually exclusive, as previously observed for sst2-il1.
Table 1.
Nonexhaustive list of GPCRs containing both the FLNA-binding motif and the YXXM sequence in close proximity in their il1, il2, il3, or C-ter domaina
| GPCR | FLNA and p85 binding sequences | Sequence localization |
|---|---|---|
| AGTR1_HUMAN type 1 angiotensin II receptor (AT1) | WIVIYFYMKLK | il1 |
| CCRL2_HUMAN C-C chemokine receptor-like 2 | LFIFTFLYVQMRKTLR | il3 |
| DRD5_HUMAN D(1B) dopamine receptor | IVFHKEIAAAYIHMMPN | C-ter |
| EDG1_HUMAN sphingosine 1-phosphate receptor Edg-1 | IAIERYITMLKMK | il2 |
| EDG7_HUMAN lysophosphatidic acid receptor Edg-7 | IIYSYKDEDMYGTMKKM | C-ter |
| GP153_HUMAN probable G-protein coupled receptor 153 | FSVTSLSYHRMWMV | il2 |
| GP162_HUMAN probable G-protein coupled receptor 162 | FTVASLSYHRMWM | il2 |
| KISSR_HUMAN KiSS-1 receptor | LLATCACYAAMLRHL | il3 |
| MPRB_HUMAN membrane progestin receptor beta | CYAKYRYRRPYPVMRKI | il2 |
| MTLR_HUMAN motilin receptor | MLIGRYRDMRTT | il1 |
| NMUR2_HUMAN neuromedin U receptor 2 | TVISVLYYLMALR | il3 |
| O13A1_HUMAN olfactory receptor 13A1 | AICHPLHYSSMMSK | il2 |
| OR4D6_HUMAN olfactory receptor 4D6 | LAIAKPLHYVTMMRK | il2 |
| OPRD_HUMAN opioid receptor delta type | VLVMFGIVRYTKMKTA | il1 |
| OPRK_HUMAN opioid receptor kappa type | FVIIRYTKMKTA | il1 |
| OPRM_HUMAN opioid receptor mu type | FLVMYVIVRYTKMKTA | il1 |
| OR5K1_HUMAN olfactory receptor f | VAICNPLQYHIMMSK | il2 |
| OR5K2_HUMAN olfactory receptor f | VAICNPLQYHIMMSK | il2 |
| OR8K5_HUMAN olfactory receptor 8K5 | VAICNPLLYYVIMSQR | il2 |
| PAR2_HUMAN proteinase-activated receptor 2 | VLIGFFYGNM | il2 |
| RDC1_HUMAN G-protein coupled receptor RDC1 homolog | SFINRNYRYELMKAF | C-ter |
| SSR1_HUMAN somatostatin receptor type 1 | MVIYVILRYAKMKTA | il1 |
| SSR2_HUMAN somatostatin receptor type 2 | LVIYVILRYAKMKTI | il1 |
| SSR4_HUMAN somatostatin receptor type 4 | LVIFVILRYAKMKTA | il1 |
| TRFR_HUMAN thyrotropin-releasing hormone receptor | SAINPVIYNLMSQK | C-ter |
| TS1R3_HUMAN taste receptor type 1 member 3 precursor | ILAAFHLLPRCYLLMRQP | C-ter |
il1, first intracellular loop domain; il2, second intracellular loop domain; il3, third intracellular loop domain; C-ter, COOH-terminal domain.
FLNA interaction with GPCR occurs in cellulo and is ligand regulated.
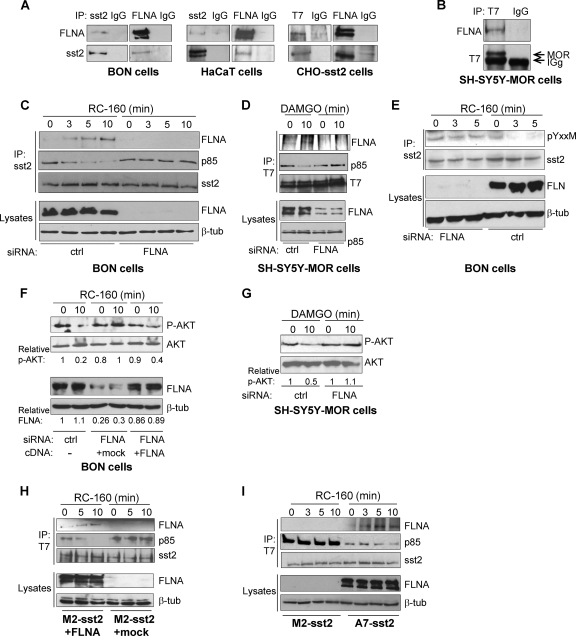
We then assessed whether FLNA interaction with sst2 and MOR occurs within a cellular context. Coimmunoprecipitation experiments show that FLNA specifically interacts with sst2 in cell lines that either express endogenous sst2 (neuroendocrine pancreatic BON and keratinocyte HaCaT cells) or that transiently express T7-tagged sst2 (CHO-sst2 cells) (Fig. 3A). Similarly, FLNA interacts with MOR in the neuroblastoma SH-SY5Y cell line that stably expresses the T7-tagged MOR (SH-SY5Y-MOR) (31, 32) (Fig. 3B). We next investigated whether the sst2 analog RC-160 and the MOR ligand DAMGO affect FLNA interaction with sst2 and MOR in BON and SH-SY5Y-MOR cells, respectively. Upon BON cell treatment with RC-160 for up to 10 min, FLNA association with sst2 was stimulated (Fig. 3C, top). Similarly, 10 min of SH-SY5Y-MOR cell treatment with DAMGO increased FNLA interaction with MOR (Fig. 3D, top).
Fig 3.
FLNA interaction with GPCR controls the dynamic of the GPCR-p85 complex and subsequent inhibition of PI3K. (A) Coimmunoprecipitation of FLNA and sst2 in BON, HaCaT, or sst2-expressing CHO cells. Immunoprecipitations (IP) were performed with anti-sst2, anti-T7, anti-FLNA, or IgG (control) antibody followed by immunoblotting (IB) with anti-FLNA or anti-sst2 antibody. (B) SHSY5Y cells stably transfected with the T7-MOR. Coimmunoprecipitation of FLNA and p85 with MOR using the indicated antibodies. (C to E) BON and SHSY5Y-MOR cells, transfected with either control (ctrl) or FLNA siRNA, were treated or not for the indicated times (0 to 10 min) with 10 nM RC-160 and DAMGO, respectively. Cell lysates were subjected to immunoprecipitation with the anti-sst2 or anti-T7 antibody, and immunoprecipitated proteins were analyzed by Western blotting with the indicated antibodies. Immunoblots of the starting cell lysates were done in parallel to assess total protein levels. (F and G) BON (F) and SHSY5Y-MOR (G) cells were transfected with control (ctrl) or FLNA siRNA. Twenty-four hours later, BON cells were transfected or not with the full-length FLNA cDNA or empty (mock) vector (F). Cells were treated for 10 min with 10 nM RC-160 (F) or DAMGO (G), and S473 AKT phosphorylation level was assessed by Western blotting. The relative ratio of S473 AKT phosphorylation/total AKT is indicated at each condition and is representative of three independent experiments. An arbitrary value of 1 has been assigned to the ratio of S473 AKT phosphorylation/total AKT in the unstimulated condition of control siRNA-transfected cells. Downregulation of FLNA expression has been controlled on total cell lysates immunoblotted with the anti-FLNA antibody. An arbitrary value of 1 has been assigned to the relative ratio of FLNA/β-tubulin, as calculated above. (H and I) A7 and M2 cells expressing sst2 (A7-sst2 and M2-sst2 cells, respectively) (I) and M2-sst2 transiently expressing FLNA (+FLNA) or a control vector (+mock) (H) were treated with 10 nM RC-160 for the indicated times. sst2-FLNA interaction was assessed as described above (panels C and D).
FLNA interaction with GPCR controls the dynamic of the GPCR-p85 complex and subsequent inhibition of PI3K.
We previously reported that sst2-il1 physically interacts with p85, the regulatory subunit of PI3K, which is dependent on the phosphorylation of the Y71 residue present in the sst2-il1 p85-binding Y71AKM motif. Ligand activation of sst2 disrupts a basal preexisting sst2-p85 complex, with the kinetic similar to that of the dephosphorylation of Y71. We therefore hypothesized that ligand-mediated sst2-p85 dissociation resulted from Y71 dephosphorylation. Importantly, p85 release from sst2 is a prerequisite for the subsequent sst2-mediated inhibition of the PI3K/AKT pathway and of cell survival (4).
We therefore explored whether FLNA binding to sst2-il1 impacts on the sst2-p85 interaction and on the phosphorylation of the Y71 residue present in the sst2-il1 p85-binding YAKM motif. Phosphorylation of the Y71 residue has been investigated using the phospho-(Tyr) p85 PI3K antibody, which recognizes peptides and proteins containing phosphorylated tyrosine at the consensus p85-binding YXXM motif and which has previously been shown to specifically recognize the sst2-il1-pY71 peptide only when the Y71 residue is phosphorylated (4). As previously reported in resting cells, p85 interacts with sst2 and the Y71 residue is phosphorylated, whereas treatment with RC-160 for up to 10 min resulted in a time-dependent dissociation of p85 from sst2 (Fig. 3C) and a concomitant dephosphorylation of Y71 (Fig. 3E). This resulted in the inhibition of S473 AKT phosphorylation, used as a readout for PI3K activity (Fig. 3F). Similarly, we demonstrated that p85 is present in a basal complex comprising MOR and that a 10-min SH-SY5Y-MOR cell treatment with DAMGO induced a dissociation of the MOR-p85 complex (Fig. 3D), resulting in S473 AKT dephosphorylation (Fig. 3G). Interestingly, in both cases, the kinetics of ligand-induced dissociation of p85 from GPCRs, and of FLNA association to GPCRs, are inversely correlated (Fig. 3C, sst2, and D, for MOR), suggesting that p85 and FLNA compete for binding to GPCR's first intracellular loop.
To test this hypothesis, BON and SH-SY5Y-MOR cells were transfected with a siRNA targeting FLNA. Abrogating FLNA expression resulted in a stabilized sst2-p85 complex, whereby cell treatment with RC-160 or with DAMGO is not able to dissociate p85 from sst2 (Fig. 3C) or from MOR (Fig. 3D), respectively, to inhibit S473 AKT phosphorylation (Fig. 3F and G), or to induce the dephosphorylation of the sst2 Y71 residue (Fig. 3E). Interestingly, in BON cells where FLNA expression had been silenced, transfection of a FLNA cDNA (which is not targeted by the FLNA siRNA) rescued inhibition of S473 AKT phosphorylation by RC-160 (Fig. 3F).
These results have been confirmed in a spontaneously FLNA-deficient cell line (melanoma M2 cells), where FLNA expression has been rescued by transient (Fig. 3H) or stable (melanoma A7) (Fig. 3I) transfection. M2 and A7 cells have also been stably transfected with the T7-tagged sst2 receptor (M2-sst2 and A7-sst2 cells) (Fig. 3H and I). Disruption of the sst2-p85 complex is induced by RC-160 only in FLNA-expressing cells, where FLNA association with sst2 is induced by the analog treatment. Altogether these results demonstrate that, upon cell treatment with the sst2 and MOR ligands, FLNA is recruited to the respective GPCR, whereas p85 dissociates from the receptors. Importantly, expression of FLNA is critical for the GPCR ligands to force the dissociation of p85 from sst2, to induce the dephosphorylation of the Y71 residue present in the sst2-il1 p85-binding Y71AKM motif, and to subsequently inhibit the PI3K pathway.
FLNA is required for sst2-mediated inhibition of cell survival.
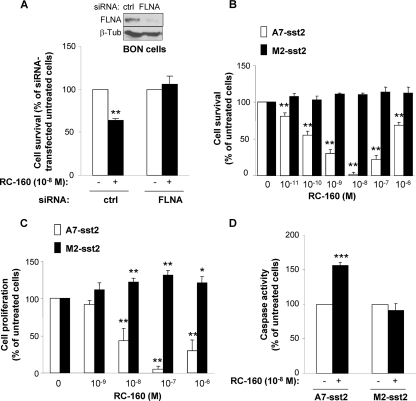
We have previously demonstrated that the somatostatin analog RC-160 inhibits cell survival of numerous cell types through inhibition of several pathways including PI3K (4, 6). Because FLNA plays a critical role for sst2-mediated inhibition of the PI3K pathway, we investigated whether it contributes to sst2 inhibitory action on cell survival in the endocrine pancreatic tumor cell line BON, which endogenously expresses sst2 and FLNA. BON cell treatment with RC-160 inhibited cell survival by 39% ± 3%, as compared to untreated cells (Fig. 4A). This inhibitory effect was however reversed when FLNA expression was silenced with a siRNA (Fig. 4A).
Fig 4.
FLNA is involved in sst2-mediated inhibition of cell growth and survival. (A) Viability of BON cells transfected with control (ctrl) or FLNA siRNA and treated or not for 48 h with 10 nM RC-160. The immunoblot shows FLNA knockdown obtained 72 h posttransfection. (B to D) A7-sst2 and M2-sst2 cells were treated or not with the indicated concentrations of RC-160. Cell viability (B) and cell proliferation (C) were measured after 48 h of treatment, and caspase activity after 24 h (D). Results represent mean ± SEM of three experiments (*, P < 0.05; **, P < 0.01 for comparisons with the siRNA-transfected untreated cells).
This result was then confirmed in the human malignant melanoma cell lines M2 (FLNA deficient) and A7 (FLNA transfected) (10), stably transfected with the T7-tagged sst2 (M2-sst2 and A7-sst2, respectively). M2-sst2 and A7-sst2 cells expressed comparable levels of cell surface sst2, and the absence of FLNA did not impact on basal sst2 membrane localization (Fig. 5A, time zero). Interestingly, in FLNA-deficient M2-sst2 cells, RC-160 was inefficient to inhibit cell survival. By contrast, the expression of FLNA in A7-sst2 cells enabled RC-160 inhibitory action (Fig. 4B). Consistently, expression of FLNA was also required for RC-160 to inhibit cell proliferation and to induce apoptosis (Fig. 4C and D), indicating a critical role for FLNA to mediate these sst2 inhibitory responses. RC-160 did not affect cell survival, cell proliferation, or apoptosis in parental A7 and M2 cells (not expressing sst2) (data not shown).
Fig 5.
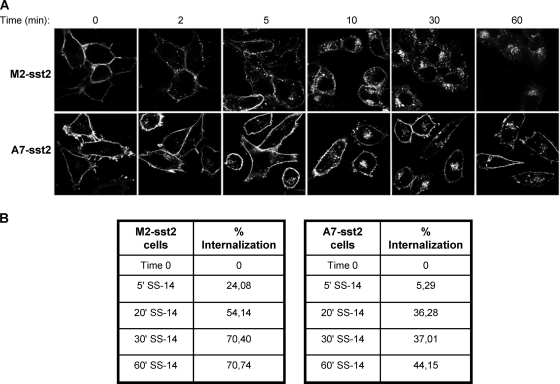
Role for FLNA in the control of sst2 endocytosis. (A and B) A7 and M2 cells stably expressing T7-sst2 (A7-sst2 and M2-sst2 cells) were treated or not with 10 nM SS-14 for 60 min. (A) The subcellular distribution of sst2 was analyzed by confocal microscopy using the anti-sst2 antibody and immunofluorescence. Representative results of one of three independent experiments are shown. Note that in untreated cells, sst2 was exclusively confined to the plasma membrane in both cell lines. SS-14 induced a robust internalization of sst2 in M2-sst2 cells, and to a lesser extent in A7-sst2 cells. (B) Cell surface sst2 receptors were labeled with the anti-T7 antibody followed by a peroxidase-conjugated secondary antibody. Receptor sequestration, quantified as the percentage of loss of cell surface receptors in agonist-treated cells, was measured by ELISA. Results are presented as the mean of four independent experiments performed in quadruplicate. SE values were smaller than 15% (not shown).
FLNA has been shown to regulate signaling and function of its protein partners by stabilizing their levels at the plasma membrane and/or modulating their endocytic trafficking (14, 28, 43, 48, 56). sst2 receptor internalization was therefore evaluated upon RC-160 treatment in A7-sst2 and M2-sst2 cells (Fig. 5). sst2 was predominantly confined to the plasma membrane in the absence of agonist in both cell lines. Sixty minutes of somatostatin treatment resulted in an enhanced sst2 internalization in M2-sst2 as compared to FLNA-expressing A7-sst2 cells (71% versus 44%, respectively) (Fig. 5A and B). These results demonstrated that expression of FLNA reduces the rate of sst2 internalization, which is consistent with its previously described function as a regulator of GPCR endocytosis (29, 43, 48). We therefore concluded that the presence of FLNA is critical for sst2 inhibitory action on cell survival. We suggest that once sst2 is activated by its ligand, FLNA binds to sst2 and forces the subsequent dissociation of p85 from sst2. It results in inhibition of the PI3K pathway and also in stabilization of sst2 at the cell membrane, where it transduces its inhibitory signal.
Direct FLNA-sst2 interaction is critical for sst2-mediated inhibition of cell survival and tumor growth.
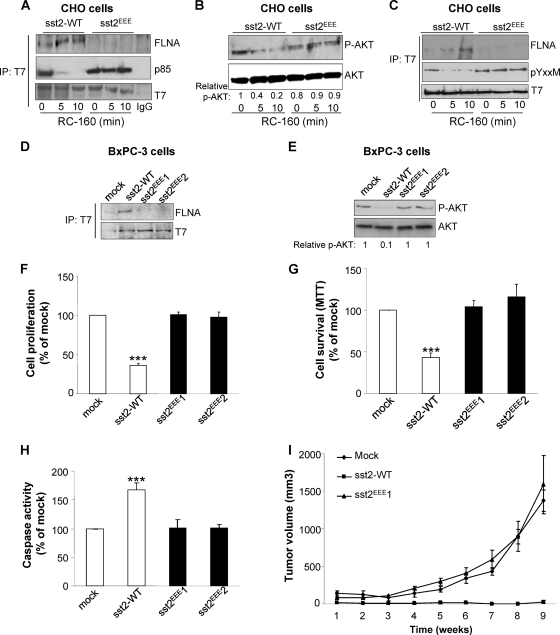
To demonstrate that the critical role of FLNA for transducing sst2 inhibitory signal on cell survival relies on its direct binding to sst2, we first transfected CHO cells with the T7-tagged wild type (sst2-WT) or mutated sst2 (sst2EEE) that was previously shown to be unable to bind FLNA in vitro (Fig. 1F and 2D). As expected, p85 was strongly present in sst2-WT and sst2EEE immunoprecipitates. FLNA was however only detected in sst2-WT and not in sst2EEE immunoprecipitates, confirming that mutation of the I65Y66V67 residues abrogates the interaction of FLNA with sst2 in cellulo (Fig. 6A). This result further demonstrated that the I65YV67 residues are critical for sst2 to directly interact with endogenous FLNA and that the specificity of this interaction is determined by these hydrophobic contacts. Cell treatment with RC-160 rapidly (5 to 10 min) stimulated FLNA binding to sst2-WT, but not to sst2EEE, which correlated with the dissociation of p85 from sst2-WT, but not from sst2EEE. In sst2EEE-expressing CHO cells, the preexisting sst2-p85 complex was indeed stable upon RC-160 cell treatment. Accordingly, S473 AKT phosphorylation was inhibited upon RC-160 cell treatment in WT-sst2- but not in sst2EEE-expressing CHO cells (Fig. 6B). Interestingly, CHO cell treatment with RC-160 induced the dephosphorylation of the p85-binding Y71AKM motif of sst2-WT but not of the mutated sst2EEE receptor (Fig. 6C). The kinetics of sst2-Y71 dephosphorylation parallel those of FLNA recruitment to, and p85 release from, sst2-WT (Fig. 6A and C).
Fig 6.
Direct FLNA-sst2 interaction through the sst2-I65Y66V67 motif is critical for sst2-mediated inhibition of the PI3K pathway, cell survival, and tumor growth. (A to C) CHO cells transfected with sst2-WT or mutated sst2EEE were treated as indicated with RC-160. (D and E) BxPC-3 cells were transfected with sst2-WT or mutated sst2EEE (pools 1 and 2). (A, C, and D) Cell lysates were subjected to anti-T7 immunoprecipitation and analyzed by Western blotting with the indicated antibodies. (B, E) Cell lysates were subjected to immunoblotting using the anti-phospho-AKT (P-AKT) antibody. Loading of equal amounts of cell lysates in each condition was checked by immunoblotting the same membrane with the anti-AKT antibody. The relative ratio of S473 AKT phosphorylation/total AKT is indicated at each condition and is representative of three independent experiments. An arbitrary value of 1 has been assigned to the ratio of S473 AKT phosphorylation/total AKT in the unstimulated condition of sst2-WT-transfected CHO (B) and BxPC-3 (E) cells. (F to I) BxPC-3 cells were stably transfected with WT or mutated sst2 (pool sst2EEE1) cDNA or with empty vector (mock). Cell proliferation (F), cell viability (G), and caspase activity (H) were measured. Results are expressed as the percentage of mock cells and represent the means ± SEM of three independent experiments. (I) BxPC-3 cells expressing either mock or sst2-WT or mutated sst2EEE1 were subcutaneously injected in athymic mice (eight animals per group), and tumor growth was monitored for up to 9 weeks. Results represent the means ± SEM of three independent experiments. (*, P < 0.05; **, P < 0.01 for sst2-WT and sst2EEE1 versus mock).
We have previously demonstrated that expressing sst2 in the human pancreatic BxPC-3 cells, which do not express endogenous sst2, results in an autocrine loop whereby sst2 induces somatostatin expression, which in turn continuously activates sst2. As a result, cell proliferation, cell survival, and tumor progression are inhibited (2, 4, 18, 50). Therefore, we used this cell model to investigate the role for direct FLNA binding to sst2 in transducing sst2 inhibitory effects on these parameters, in cellulo and in vivo. As previously observed in the other sst2-expressing cell lines, FLNA is present in the sst2 immunoprecipitate (sst2-WT) but is absent from this sst2 complex when the sst2EEE mutant receptor is expressed in BxPC-3 cells (Fig. 6D, pools sst2EEE1 and sst2EEE2). Accordingly, S473 AKT phosphorylation was shown to be inhibited in sst2-WT- but not in sst2EEE1- and sst2EEE2-expressing BxPC-3 cells (Fig. 6E). Importantly, sst2-mediated inhibition of cell proliferation (Fig. 6F) and of cell survival (Fig. 6G), induction of apoptosis (Fig. 6H), and inhibition of tumor growth in vivo (cell xenografts subcutaneously implanted in nude mice) (Fig. 6I) were reversed when the sst2EEE mutant receptor was expressed in BxPC-3 cells. These data demonstrated that the direct sst2-FLNA interaction, which triggers PI3K pathway inhibition, is critical for sst2 oncosuppressive activity. In contrast, levels of sst2-induced IP3 formation were comparable in BxPC-3-cells expressing sst2-WT or mutated sst2EEE (not shown), indicating that FLNA direct binding to sst2 specifically controls the PI3K survival pathway.
DISCUSSION
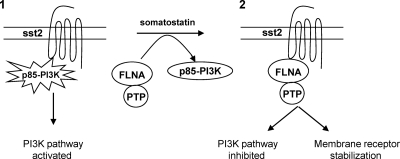
Our results describe an original molecular mechanism for inhibition of the PI3K pathway by GPCRs (sst2 and MOR), involving a ligand-mediated change in binding preference of receptors from the regulatory PI3K p85 toward the scaffolding protein FLNA. A critical question is how GPCR activation triggers this switch. In vitro data demonstrate that both proteins (p85 and FLNA) compete for direct binding onto two juxtaposed sites present in the GPCR's first intracellular loop (il1), their respective binding affinities being similarly high (nanomolar range). In cellulo results demonstrate that the phosphotyrosine state of the GPCR (sst2) dictates whether p85 or FLNA binds. We had previously demonstrated that direct p85 interaction with sst2 relies on the phosphorylation status of the tyrosine Y71 residue present in the p85-binding Y71AKM motif (sst2-il1). Using a phospho-(Tyr) p85 antibody (4), which recognizes peptides and proteins containing phosphorylated tyrosine at the consensus p85-binding YXXM motif, phosphorylation of sst2-Y71 was observed in cells cultured in normal conditions (10% serum). This was also consistent with our previous reports describing sst2 as a tyrosine-phosphorylated receptor in these same conditions (4, 16). Upon cell treatment with somatostatin, sst2 is dephosphorylated on tyrosines (16), including on the Y71 residue (4). This dephosphorylation occurs with a kinetic identical to that of p85 dissociation from and FLNA recruitment (as described here) to sst2. FLNA directly interacts with the I65Y66V67 motif (in sst2-il1), but only when the Y66 is not phosphorylated (Fig. 1G). Consistently, adding negative charges on the FLNA-binding I65Y66V67 motif (by introducing glutamic acid residues, sst2EEE) to mimic Y66 phosphorylation also abrogated FLNA recruitment to sst2-il1. This is consistent with the low binding of FLNA with sst2 at the basal unstimulated state, when sst2-il1 is shown to be tyrosine phosphorylated (16), including on Y71 (4), and to preferentially interact with p85. This is also consistent with the increased binding of FLNA to sst2-il1 upon cell stimulation with somatostatin. Therefore, an important question that arises is whether ligand-mediated dissociation of p85 from sst2 is induced by sst2 dephosphorylation on the Y71 residue, or by competitive recruitment of FLNA to sst2, or whether both events are related. Importantly, when FLNA expression is silenced or FLNA recruitment to sst2 inhibited, basal Y71 phosphorylation is here shown not to be diminished upon cell stimulation with somatostatin. These results therefore demonstrate that FLNA binding to sst2 is critical for the Y71 residue to be dephosphorylated upon ligand stimulation. One can therefore hypothesize that, by binding to sst2, FLNA allows the recruitment of a tyrosine phosphatase in close proximity with the phosphorylated p85-binding pY71AKM motif. Accordingly, FLNA has been shown to bind the Src homology 2-containing inositol 5′ phosphatase SHIP-2 and to allow its localization at the membrane in proximity with its substrates (12). We have previously demonstrated that upon stimulation with somatostatin, the sst2 inhibitory signal on cell survival relies on the sequential activation of two tyrosine phosphatases, SHP-2 and then SHP-1 (16). Whereas SHP-2 is rapidly recruited to the sst2 3rd intracellular loop and the C-terminal domain, the mechanisms for SHP-1 recruitment are not known. One can hypothesize that rapid activation of SHP-2 dephosphorylates the Y66 residue, which allows the recruitment to sst2 of FLNA, in competition with p85; a tyrosine phosphatase activity (SHP-1 is a candidate) is recruited with FLNA in close proximity with the phosphorylated Y71 residue, which upon dephosphorylation of this residue further facilitates p85 release from sst2 (Fig. 7).
Fig 7.
Model for GPCR-induced inhibition of the PI3K pathway: critical role for the switch in GPCR binding preference from p85 to FLNA. (1) In the presence of serum, p85 and a small fraction of FLNA are associated with sst2. By binding p85, sst2 is permissive for growth factor-induced PI3K activity. Therefore, cell survival, proliferation, and tumor growth are stimulated. (2) Upon ligand (somatostatin) treatment, GPCR (sst2) switches its binding preference from p85, which dissociates from GPCR, to FLNA, which associates to GPCR. This switch is facilitated by a FLNA-dependent recruitment of a protein tyrosine phosphatase (PTP) activity that dephosphorylates the p85-binding Y71AKM motif in the sst2-il1 loop. Competitive binding of p85 and FLNA to GPCR (sst2) is critical for subsequent inhibition of the PI3K pathway of cell survival and tumor growth. FLNA binding to GPCR stabilizes GPCR (sst2) expression at the cell membrane.
Interestingly, competition between FLNA and other proteins, including migfilin, talin, and 14-3-3, for binding to integrin cytoplasmic domains has been described, thereby regulating integrin activation (21, 23, 46). In several cases, phosphothreonine-mimicking mutations on integrin inhibited filamin but not talin or 14-3-3 binding, and authors hypothesized that kinases modulate these competitive bindings (46). We suggest that the concerted activation of kinases/phosphatases might rather be involved. More recently, a competitive binding between talin and FLNA has also been described on the conserved membrane-proximal NPXY integrin motif. The Y residue in this motif was shown to be critical for both talin and FLNA interactions. Interestingly, whereas talin binding induced integrin activity, FLNA abrogated it by recruiting FilGAP, a Rac inhibitor (13, 35), further supporting the role of FLNA as a scaffolding platform.
Importantly, the hydrophobic sequence critical for sst2-FLNA interaction and present in sst2 first intracellular loop just upstream of the p85-binding YAKM motif is retrieved in 26 (Table 1) of the 40 GPCRs, which we had previously identified to exhibit a YXXM motif (4), including sst2 and MOR. It suggests that the orchestrated competitive dissociation and association between p85 and FLNA may represent an original generalized mechanism for negative regulation of the PI3K pathway by these other 24 listed GPCRs. Strikingly, one of these hits corresponds to the protease-activated receptor 2 (PAR-2), which has been reported to inhibit PI3K by recruiting p85 into a scaffolding complex containing PAR-2 and β-arrestin (51). MOR had previously been reported to interact with FLNA, which regulates its trafficking. This interaction was shown to occur through the MOR C-terminal tail, whereas its third intracellular loop was not involved (36). We here provide the first demonstration for a direct interaction between the MOR-il1 and FLNA. Because of its high molecular weight and its organization in repeats, FLNA can interact simultaneously or sequentially with different intracellular regions of its partners (34).
Emerging evidences suggest that FLNA is a crucial regulator of GPCR signaling, acting as a scaffolding protein that links various intracellular signaling proteins to plasma membrane receptors and to cytoskeletal actin (19, 39, 42). Our results demonstrate that FLNA is critical to maintain the stability of the GPCR sst2 at the membrane in close proximity with actors of the lipid PI3K family. Future additional studies will however be needed to fully address how FLNA regulates sst2 trafficking. It has been suggested that FLNA presents an important role in vesicular transport and protein trafficking, including GPCR. By linking to the actin cytoskeleton, FLNA stabilizes its partners on the membrane (29, 43, 48, 56), whereas FLNA dissociation might facilitate the binding of adaptor proteins required for endocytosis (29).
The PI3K pathway is tightly controlled at multiple levels. Phosphatases including the tensin homolog PTEN and SHIP hydrolyze the PI3K product PI(3,4,5)P3, to PI(3,4)P2 and PI(4,5)P2, respectively, thereby terminating PI3K signaling. A further level of negative control exists involving the protein TORC1, which triggers a negative feedback regulation by promoting IRS-1 transcriptional repression and inhibitory phosphorylation in response to growth factors (8). The tyrosine phosphatases SHP2 and SHP1 have also been reported to negatively regulate specific growth factor-dependent PI3K activation by dephosphorylating Gab1 p85-binding sites and p85 on the Tyr688 residue, respectively (9, 57). Our data demonstrate that FLNA acts as a negative regulator of PI3K through a novel mechanism involving the competitive binding of FLNA and p85 with GPCR, which relies on FLNA-regulated phosphorylation events on crucial Y residues present in the two juxtaposed p85- and FLNA-binding motifs on sst2-il1.
FLNA repeats 19 to 24 are here shown to represent the interacting domains with sst2, as previously described for other GPCRs (34). FLNA strands C and D in repeat 19 are indeed binding sites for all FLNA-binding partners, for which atomic structures have been resolved (21, 33, 46). These FLNA-interacting proteins share a conserved hydrophobic amino acid motif (33), which we have also identified in sst2 and MOR first intracellular loops. Molecular modeling, consisting in a flexible docking of sst2-il1 to the homology-modeled FLNA repeat 19, has confirmed that this experimentally demonstrated complex is structurally and energetically relevant. Moreover, it has unraveled that this complex resembles those described between FLNA and migfilin and integrins, which also involve regions structured in β-strands and orientated in an antiparallel fashion in both proteins (21, 23). This finding implies that the sst2 regions, which comprise the first intracellular loop and the bottom of the first helix of this receptor, can adopt a β-strand structure following activation and interaction with intracellular signaling proteins. However, as yet, such GPCR dynamic behavior is particularly challenging for high-resolution structure analysis, and it is technically not feasible to obtain a high-resolution structure of a GPCR with current nuclear magnetic resonance (NMR) technology (22). Nevertheless, mutating the FLNA-binding IY66V motif into charged glutamic acid amino acids (sst2EEE) prevented sst2-FLNA interaction and abrogated somatostatin inhibitory action on cell survival and tumor growth, demonstrating that FLNA binding to sst2 is necessary and sufficient to trigger the sst2 oncosuppressive signal.
Numerous loss- or gain-of-function mutations and deletions in the FLN genes have been identified, and the striking phenotypic diversity generated by these mutations indicates that FLN present a broad range of biochemical functions critical during development (41). Interestingly, FLN mutations have also been found to be common in human breast cancers (44), but no functional relationships with cancer, nor with the PI3K pathway, have been reported yet. It has been described that FLNA presents anti-oncogenic functions through negative regulation of cell adhesion and migration (54), or of the PI3K pathway, as described here. It would therefore be interesting to evaluate whether the mutant FLN proteins found across the full range of human syndromes may cause altered binding to known interacting partners that would impact on tumorigenesis. Nevertheless, even if strongly expressed in cells, FLNA function can be altered by different mechanisms that affect the interactions with its partners (34). Moreover, recent studies have reported a correlation between low FLNA expression levels and breast or bladder cancer aggressiveness (45, 54). Why FLNA expression is decreased in these cancers is not known yet. Posttranslational mechanisms (phosphorylation and proteolysis) could be involved (34). Interestingly, the expression of a cleaved nuclear 90-kDa fragment of FLNA (encompassing repeats 16 to 23) has been shown to be necessary for androgen dependence in prostate cancer cells. In this model, cell sensitivity to an androgen receptor antagonist depends on negative control of the PI3K pathway by FLNA, thereby establishing a direct link between posttranslational modification of FLNA and PI3K-driven oncogenesis (52).
In summary, our findings demonstrate that by physically interacting with a subset of GPCRs, FLNA negatively impacts on the PI3K pathway through a scenario shown in Fig. 7. The PI3K/AKT signaling pathway plays a central role in cell survival and proliferation. Several components of this pathway are dysregulated in a wide spectrum of human cancers (i.e., PI3K, AKT, and PTEN), establishing its importance in oncogenesis (5). Future studies will undoubtedly be needed to delineate the role of FLNA and FLNA-GPCR interactions in the dysregulation of GPCR signaling and of the PI3K pathway, associated with tumor progression and/or with the large variety of GPCR functions.
ACKNOWLEDGMENTS
We thank T. P. Stossel and F. Nakamura (Harvard Medical School, Boston) for critical reading and suggestions. We are also grateful to T. P. Stossel for providing the A7 and M2 cells, Y. Ohta (Tokyo Medical and Dental University) for providing human FLNA cDNA, L. Mouledous (IPBS, Toulouse, France) for providing SH-SYSY-MOR cells, and J. Iacovoni (INSERM U1048, Toulouse, France) for his bioinformatic help screening the GPCR database.
S.N. was supported by INSERM, poste vert 000066129 and Fondation pour la Recherche Médicale, ACE 20061208677. This work was supported by the Association pour la Recherche contre le Cancer (grant no. 5000) and the Ligues Régionale et Nationale contre le Cancer.
Footnotes
Published ahead of print 27 December 2011
REFERENCES
- 1. Asnacios A, et al. 2008. Indium-111-pentetreotide scintigraphy and somatostatin receptor subtype 2 expression: new prognostic factors for malignant well-differentiated endocrine tumors. J. Clin. Oncol. 26:963–970 [DOI] [PubMed] [Google Scholar]
- 2. Benali N, et al. 2000. Inhibition of growth and metastatic progression of pancreatic carcinoma in hamster after somatostatin receptor subtype 2 (sst2) gene expression and administration of cytotoxic somatostatin analog AN-238. Proc. Natl. Acad. Sci. U. S. A. 97:9180–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bockaert J, Fagni L, Dumuis A, Marin P. 2004. GPCR interacting proteins (GIP). Pharmacol. Ther. 103:203–221 [DOI] [PubMed] [Google Scholar]
- 4. Bousquet C, et al. 2006. Direct binding of p85 to sst2 somatostatin receptor reveals a novel mechanism for inhibiting PI3K pathway. EMBO J. 25:3943–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bunney TD, Katan M. 2010. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat. Rev. Cancer. 10:342–352 [DOI] [PubMed] [Google Scholar]
- 6. Buscail L, et al. 1995. Inhibition of cell proliferation by the somatostatin analogue RC-160 is mediated by somatostatin receptor subtypes SSTR2 and SSTR5 through different mechanisms. Proc. Natl. Acad. Sci. U. S. A. 92:1580–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buscail L, et al. 1996. Loss of sst2 somatostatin receptor gene expression in human pancreatic and colorectal cancer. Cancer Res. 56:1823–1827 [PubMed] [Google Scholar]
- 8. Carracedo A, Pandolfi PP. 2008. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene 27:5527–5541 [DOI] [PubMed] [Google Scholar]
- 9. Cuevas BD, et al. 2001. Tyrosine phosphorylation of p85 relieves its inhibitory activity on phosphatidylinositol 3-kinase. J. Biol. Chem. 276:27455–27461 [DOI] [PubMed] [Google Scholar]
- 10. Cunningham CC, et al. 1992. Actin-binding protein requirement for cortical stability and efficient locomotion. Science 255:325–327 [DOI] [PubMed] [Google Scholar]
- 11. Delesque N, et al. 1997. sst2 somatostatin receptor expression reverses tumorigenicity of human pancreatic cancer cells. Cancer Res. 57:956–962 [PubMed] [Google Scholar]
- 12. Dyson JM, et al. 2001. The SH2-containing inositol polyphosphate 5-phosphatase, SHIP-2, binds filamin and regulates submembraneous actin. J. Cell Biol. 155:1065–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP. 2011. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 478:260–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng S, Lu X, Kroll MH. 2005. Filamin A binding stabilizes nascent glycoprotein Ibalpha trafficking and thereby enhances its surface expression. J. Biol. Chem. 280:6709–6715 [DOI] [PubMed] [Google Scholar]
- 15. Feng Y, Walsh CA. 2004. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat. Cell Biol. 6:1034–1038 [DOI] [PubMed] [Google Scholar]
- 16. Ferjoux G, et al. 2003. Critical role of Src and SHP-2 in sst2 somatostatin receptor-mediated activation of SHP-1 and inhibition of cell proliferation. Mol. Biol. Cell 14:3911–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Florio T. 2008. Somatostatin/somatostatin receptor signalling: phosphotyrosine phosphatases. Mol. Cell. Endocrinol. 286:40–48 [DOI] [PubMed] [Google Scholar]
- 18. Guillermet J, et al. 2003. Somatostatin receptor subtype 2 sensitizes human pancreatic cancer cells to death ligand-induced apoptosis. Proc. Natl. Acad. Sci. U. S. A. 100:155–160 Epub 2002 Dec 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hjalm G, MacLeod RJ, Kifor O, Chattopadhyay N, Brown EM. 2001. Filamin-A binds to the carboxyl-terminal tail of the calcium-sensing receptor, an interaction that participates in CaR-mediated activation of mitogen-activated protein kinase. J. Biol. Chem. 276:34880–34887 [DOI] [PubMed] [Google Scholar]
- 20. Hortala M, et al. 2003. Inhibitory role of the somatostatin receptor SST2 on the intracrine-regulated cell proliferation induced by the 210-amino acid fibroblast growth factor-2 isoform: implication of JAK2. J. Biol. Chem. 278:20574–20581 [DOI] [PubMed] [Google Scholar]
- 21. Kiema T, et al. 2006. The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell 21:337–347 [DOI] [PubMed] [Google Scholar]
- 22. Kobilka BK, Deupi X. 2007. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 28:397–406 [DOI] [PubMed] [Google Scholar]
- 23. Lad Y, et al. 2008. Structural basis of the migfilin-filamin interaction and competition with integrin beta tails. J. Biol. Chem. 283:35154–35163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lahlou H, et al. 2003. sst2 Somatostatin receptor inhibits cell proliferation through Ras-, Rap1-, and B-Raf-dependent ERK2 activation. J. Biol. Chem. 278:39356–39371 [DOI] [PubMed] [Google Scholar]
- 25. Lappano R, Maggiolini M. 2011. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat. Rev. Drug Discov. 10:47–60 [DOI] [PubMed] [Google Scholar]
- 26. Lesche S, Lehmann D, Nagel F, Schmid HA, Schulz S. 2009. Differential effects of octreotide and pasireotide on somatostatin receptor internalization and trafficking in vitro. J. Clin. Endocrinol. Metab. 94:654–661 [DOI] [PubMed] [Google Scholar]
- 27. Li C, et al. 2009. Binding of pro-prion to filamin A disrupts cytoskeleton and correlates with poor prognosis in pancreatic cancer. J. Clin. Invest. 119:2725–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin R, Karpa K, Kabbani N, Goldman-Rakic P, Levenson R. 2001. Dopamine D2 and D3 receptors are linked to the actin cytoskeleton via interaction with filamin A. Proc. Natl. Acad. Sci. U. S. A. 98:5258–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu G, et al. 1997. Cytoskeletal protein ABP-280 directs the intracellular trafficking of furin and modulates proprotein processing in the endocytic pathway. J. Cell Biol. 139:1719–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Magalhaes AC, Dunn H, Ferguson SS. 2011. Regulation of G protein-coupled receptor activity, trafficking and localization by GPCR-interacting proteins. Br. J. Pharmacol. doi:10.1111/j.1476-5381.2011.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mollereau C, Mazarguil H, Zajac JM, Roumy M. 2005. Neuropeptide FF (NPFF) analogs functionally antagonize opioid activities in NPFF2 receptor-transfected SH-SY5Y neuroblastoma cells. Mol. Pharmacol. 67:965–975 [DOI] [PubMed] [Google Scholar]
- 32. Mouledous L, et al. 2005. Long-term morphine treatment enhances proteasome-dependent degradation of G beta in human neuroblastoma SH-SY5Y cells: correlation with onset of adenylate cyclase sensitization. Mol. Pharmacol. 68:467–476 [DOI] [PubMed] [Google Scholar]
- 33. Nakamura F, et al. 2006. The structure of the GPIb-filamin A complex. Blood 107:1925–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakamura F, Stossel TP, Hartwig JH. 2011. The filamins: organizers of cell structure and function. Cell Adh. Migr. 5:160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nieves B, et al. 2010. The NPIY motif in the integrin beta1 tail dictates the requirement for talin-1 in outside-in signaling. J. Cell Sci. 123:1216–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Onoprishvili I, et al. 2003. Interaction between the mu opioid receptor and filamin A is involved in receptor regulation and trafficking. Mol. Pharmacol. 64:1092–1100 [DOI] [PubMed] [Google Scholar]
- 37. Popowicz GM, Schleicher M, Noegel AA, Holak TA. 2006. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem. Sci. 31:411–419 [DOI] [PubMed] [Google Scholar]
- 38. Pyronnet S, et al. 2008. Antitumor effects of somatostatin. Mol. Cell. Endocrinol. 286:230–237 [DOI] [PubMed] [Google Scholar]
- 39. Rey O, Young SH, Yuan J, Slice L, Rozengurt E. 2005. Amino acid-stimulated Ca2+ oscillations produced by the Ca2+-sensing receptor are mediated by a phospholipase C/inositol 1,4,5-trisphosphate-independent pathway that requires G12, Rho, filamin-A, and the actin cytoskeleton. J. Biol. Chem. 280:22875–22882 [DOI] [PubMed] [Google Scholar]
- 40. Ritter SL, Hall RA. 2009. Fine-tuning of GPCR activity by receptor-interacting proteins. Nat. Rev. Mol. Cell Biol. 10:819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Robertson SP. 2005. Filamin A: phenotypic diversity. Curr. Opin. Genet. Dev. 15:301–307 [DOI] [PubMed] [Google Scholar]
- 42. Scott MG, et al. 2006. Cooperative regulation of extracellular signal-regulated kinase activation and cell shape change by filamin A and beta-arrestins. Mol. Cell. Biol. 26:3432–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seck T, Baron R, Horne WC. 2003. Binding of filamin to the C-terminal tail of the calcitonin receptor controls recycling. J. Biol. Chem. 278:10408–10416 [DOI] [PubMed] [Google Scholar]
- 44. Sjoblom T, et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314:268–274 [DOI] [PubMed] [Google Scholar]
- 45. Smith SC, et al. 2007. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin. Cancer Res. 13:3803–3813 [DOI] [PubMed] [Google Scholar]
- 46. Takala H, et al. 2008. Beta2 integrin phosphorylation on Thr758 acts as a molecular switch to regulate 14-3-3 and filamin binding. Blood 112:1853–1862 [DOI] [PubMed] [Google Scholar]
- 47. Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. 2001. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs). 40:7761–7772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thelin WR, et al. 2007. Direct interaction with filamins modulates the stability and plasma membrane expression of CFTR. J. Clin. Invest. 117:364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tulipano G, et al. 2004. Differential beta-arrestin trafficking and endosomal sorting of somatostatin receptor subtypes. J. Biol. Chem. 279:21374–21382 [DOI] [PubMed] [Google Scholar]
- 50. Vernejoul F, et al. 2002. Antitumor effect of in vivo somatostatin receptor subtype 2 gene transfer in primary and metastatic pancreatic cancer models. Cancer Res. 62:6124–6131 [PubMed] [Google Scholar]
- 51. Wang P, DeFea KA. 2006. Protease-activated receptor-2 simultaneously directs beta-arrestin-1-dependent inhibition and Galphaq-dependent activation of phosphatidylinositol 3-kinase. Biochemistry 45:9374–9385 [DOI] [PubMed] [Google Scholar]
- 52. Wang Y, et al. 2007. A 90 kDa fragment of filamin A promotes Casodex-induced growth inhibition in Casodex-resistant androgen receptor positive C4-2 prostate cancer cells. Oncogene 26:6061–6070 [DOI] [PubMed] [Google Scholar]
- 53. Weckbecker G, et al. 2003. Opportunities in somatostatin research: biological, chemical and therapeutic aspects. Nat. Rev. Drug Discov. 2:999–1017 [DOI] [PubMed] [Google Scholar]
- 54. Xu Y, et al. 2010. Filamin A regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion. J. Exp. Med. 207:2421–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoshida N, et al. 2005. Filamin A-bound PEBP2beta/CBFbeta is retained in the cytoplasm and prevented from functioning as a partner of the Runx1 transcription factor. Mol. Cell. Biol. 25:1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang M, Breitwieser GE. 2005. High affinity interaction with filamin A protects against calcium-sensing receptor degradation. J. Biol. Chem. 280:11140–11146 [DOI] [PubMed] [Google Scholar]
- 57. Zhang SQ, et al. 2002. Receptor-specific regulation of phosphatidylinositol 3′-kinase activation by the protein tyrosine phosphatase Shp2. Mol. Cell. Biol. 22:4062–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]