Abstract
The imprinted Gnas cluster is involved in obesity, energy metabolism, feeding behavior, and viability. Relative contribution of paternally expressed proteins XLαs, XLN1, and ALEX or a double dose of maternally expressed Gsα to phenotype has not been established. In this study, we have generated two new mutants (Ex1A-T-CON and Ex1A-T) at the Gnas cluster. Paternal inheritance of Ex1A-T-CON leads to loss of imprinting of Gsα, resulting in preweaning growth retardation followed by catch-up growth. Paternal inheritance of Ex1A-T leads to loss of imprinting of Gsα and loss of expression of XLαs and XLN1. These mice have severe preweaning growth retardation and incomplete catch-up growth. They are fully viable probably because suckling is unimpaired, unlike mutants in which the expression of all the known paternally expressed Gnasxl proteins (XLαs, XLN1 and ALEX) is compromised. We suggest that loss of ALEX is most likely responsible for the suckling defects previously observed. In adults, paternal inheritance of Ex1A-T results in an increased metabolic rate and reductions in fat mass, leptin, and bone mineral density attributable to loss of XLαs. This is, to our knowledge, the first report describing a role for XLαs in bone metabolism. We propose that XLαs is involved in the regulation of bone and adipocyte metabolism.
INTRODUCTION
If the major modern day problems of obesity and diabetes are to be solved, a full understanding of genetic involvement in metabolism and food intake is needed. The imprinted Gnas/GNAS cluster located on chromosome 2 in mice and chromosome 20 in humans has an important role in metabolism (13, 60). Genomic imprinting results in the expression of a subset of genes according to parental origin. The Gnas cluster contains a number of different transcripts that are maternally, paternally, and/or biallelically expressed (47) (Fig. 1). The cluster contains three promoter regions giving rise to protein coding transcripts, Nesp, Gnasxl, and Gnas encoding the proteins Nesp55, XLαs, and Gsα, respectively (46). Nesp, Gnasxl, and Gnas all possess a unique first exon(s) which is spliced onto exon 2 of Gnas, and thus from this point on, all transcripts are identical in their sequence. Full-length transcripts extend through to exon 12 of Gnas, but there are also shortened neural transcripts of Gnas/Gnasxl that terminate prematurely before exon 4 called GsαN1/XLN1 (16, 44). Furthermore, a protein of unknown function, ALEX, is generated from an alternative reading frame of the Gnasxl transcript. Although the Gnasxl transcript encodes both proteins XLαs and ALEX, ALEX is generated only from the first Gnasxl exon, the remainder of the transcript is its 3′ untranslated region (UTR) (34).
Fig 1.
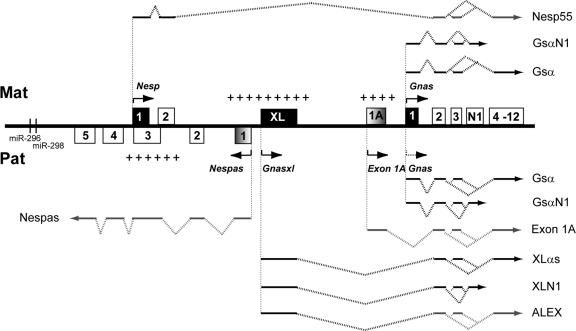
Schematic diagram of the mouse Gnas cluster. Maternal (Mat) and paternal (Pat) transcripts are shown above and below the line, respectively. Protein-coding first exons are shown as filled boxes, and noncoding first exons are shaded boxes. Noncoding sections of transcripts are shown as gray lines, whereas coding sections are black. Arrows show initiation and direction of transcription. The arrow corresponding to the paternal Gnas allele is shown as a dotted line to indicate that Gnas is only maternally expressed in some tissues. MicroRNAs are represented as vertical lines. Maternally and paternally methylated regions are indicated by plus signs above and below the line, respectively. Figure not to scale (adapted with permission from reference 45).
Of these transcripts, Nesp is exclusively maternally expressed (33, 47), Gnas is predominantly biallelically expressed in most tissues but is maternally expressed in tissues such as proximal renal tubules, paraventricular nucleus of the hypothalamus, and neonatal brown adipose tissue (BAT) (14, 61, 69). Gnas has also been shown to be maternally expressed in the pituitary in both humans (26) and neonatal mice (J. Skinner, unpublished data). Gnasxl is exclusively paternally expressed (1, 33, 39, 47). Both Gsα and XLαs function as the alpha subunit of the heterotrimeric Gs signaling protein. Gsα has been shown by both in vitro and in vivo studies, and XLαs by in vitro studies, to stimulate adenylyl cyclase and regulate receptor-stimulated cyclic AMP (cAMP) production (5, 25, 30, 35). There have, however, been reports that XLαs may act antagonistically to Gsα, as it represses adenylyl cyclase and cAMP signaling in vivo (48, 65). While Gsα is widely expressed, XLαs displays a more discrete expression pattern; it is expressed primarily in neuroendocrine tissues such as the pituitary and orexigenic neurons in the hypothalamuses of neonatal and adult rodents (28, 31, 43, 44, 48).
A number of mutations leading to loss of function within the Gnas cluster in mice have been described. A summary of some of these mutations is given in Table 1. From this table, it is clear that defects in maternal Gnas expression and paternal Gnasxl expression result in a series of opposite metabolic phenotypes that occur from the neonate to the adult. Mice that have a defect in maternal Gnas expression show increased adiposity 2 days after birth. As adults, they are obese and have a lower metabolic rate and lower sympathetic activity (12, 68). In contrast, neonates with a defect in paternal Gnasxl expression have decreased adiposity. These adults have less fat mass, a higher metabolic rate, and higher sympathetic activity (48, 65, 68). Furthermore, Gnasxl is also important in feeding behavior; loss of all Gnasxl transcripts is associated with poor suckling (48, 69). However, mice carrying the GnasOedsml-pat (Sml) allele suckle normally, although they have the raised metabolic rate characteristic of Gnasxl loss. These mice carry a point mutation in exon 6 which results in nonfunctional XLαs, but neural XLN1 and ALEX, which are not translated into protein at exon 6, are presumed to be unaffected, suggesting that the protein encoded by the full-length transcript controls the metabolic phenotype whereas suckling is controlled by the shortened proteins neural XLN1 and/or ALEX (32). The antagonistic biochemical and phenotypic effects of Gnas and Gnasxl and also the effect of Gnasxl on feeding behavior accord with the “kinship” or “parental conflict” theory of the evolution of imprinting, which predicts that paternal genes in offspring exert a high demand for maternal resources, whereas maternal genes in offspring are less demanding (41).
Table 1.
Summary of Gnas cluster mouse methods
| Mutation | Inheritance | Protein(s) affected | Phenotype |
References | |
|---|---|---|---|---|---|
| Neonate | Adult | ||||
| Deletion of Gnas exon 1 | Paternal | Gsα (paternal) | Normal body wt, normal metabolic rate, normal activity levels | 12, 25 | |
| Maternal | Gsα (maternal) | Perinatal lethality, edema | Severe adiposity, ↓ metabolic rate, ↓ length, normal food intake | 12, 25 | |
| Deletion of Gnas exon 2 | Paternal | Gsα (paternal), XLαs, XLN1, ALEX? | Perinatal lethality, poor suckling, ↓ adiposity, thin | ↓ Adiposity, ↑ metabolic rate, ↓ length, ↑ SNS activity | 68, 69 |
| Maternal | Gsα (maternal) | Perinatal lethality, edema, ↑ adiposity | ↑ Adiposity, ↓ metabolic rate, ↓ length, ↓ SNS activity | 68, 69 | |
| Deletion of Gnasxl exon | Paternal | XLαs, XLN1, ALEX | Perinatal lethality, poor suckling, adiposity | ↓ Adiposity, ↑ metabolic rate, ↓ length, ↑ SNS activity | 48, 65 |
| Duplication of distal chromosome 2 (paternal) | NAa | Gsα (maternal), Nesp55, ↑ XLαs, XLN1, ALEX? | Perinatal lethality, edema, wide body, hyperactivity, ↑ length (long bones) | 10, 62 | |
| Duplication of distal chromosome 2 (maternal) | NA | XLαs, XLN1, ALEX, ↑ Nesp55, Gsα | Perinatal lethality, no suckling, narrow body, hypoactivity | 10, 62 | |
| ENU point mutation of Gnas exon 6 (Oed/Sml) | Paternal | Gsα (paternal), XLαs | Prenatal lethality,b suckling unaffected, small body size | Adiposity unaffected by high-fat diet, ↑ metabolic rate, small body size, ↓ wt, ↓ BMI | 11, 32 |
NA, not applicable.
S. Ball, unpublished data.
Imprinted expression of the protein coding genes within the Gnas cluster is regulated by an imprinting control region (ICR), a differentially methylated region (DMR) that is maternally methylated and includes the promoter of a noncoding paternally expressed transcript, Nespas, that is transcribed antisense to Nesp (63, 64) (Fig. 1). A second maternally methylated DMR, the Exon 1A DMR, located just upstream of the Gnas promoter, specifically regulates the imprinted expression of Gnas (38, 61) (Fig. 1). Thus, the ICR must interact with the Exon 1A DMR, which in turn must act on Gnas to control its tissue-specific imprinting. The Exon 1A DMR encompasses a 2.5-kb region that contains a promoter for a noncoding RNA, Exon 1A, that is ubiquitous and exclusively paternally expressed, as well as the first exon of this transcript. This noncoding RNA, like other transcripts in the cluster, arises from a unique first exon that splices onto exon 2 of Gnas and extends through to exon 12 of Gnas. It is unknown how the unmethylated paternal Exon 1A DMR represses Gnas expression in some tissues. One possibility is transcriptional repression from the Exon 1A promoter; another is transcriptional interference from the Exon 1A transcript itself. In order to investigate further the regulation of Gnas by Exon 1A, or the Exon 1A promoter, we have generated a truncation of the Exon 1A transcript through insertion of a polyadenylation [poly(A)] cassette, designated Ex1A-T, as well as an inverted truncation control in which the poly(A) cassette was inserted in the opposite orientation, designated Ex1A-T-CON. Here we provide further insight into how Exon 1A regulates Gnas.
Studies of the phenotypes seen on paternal inheritance of Ex1A-T-CON and Ex1A-T have provided greater understanding of the role of the Gnas cluster in metabolism and suckling. Paternal inheritance of Ex1A-T-CON results in loss of imprinting of Gnas and postnatal growth retardation, indicating that overexpression of Gnas gives rise to a deleterious phenotype. Paternal inheritance of Ex1A-T gives rise to not only loss of imprinting of Gnas but also loss of expression of XLαs and XLN1. ALEX, however, is presumed to remain intact. Our results suggest that ALEX may play a role in suckling behavior. Gnasxl has previously been shown to play a role in fat metabolism, but here we show it also has a role in bone metabolism as well. In the past few years, it has become clear that adipocyte metabolism and bone metabolism are coregulated (18). For example, a number of key players in adipogenesis, such as leptin and peroxisome proliferator-activated receptor γ, have been shown to regulate bone remodeling while the osteoblast-specific hormone osteocalcin has been shown to regulate fat mass (29). Gnasxl may also be involved in the coregulation of bone and adipocytes. Although inactivating mutations of Gnas give rise to a number of bone phenotypes (6, 49, 51, 52, 59), this is the first report, as far as we are aware, of a bone phenotype that is specific to Gnasxl.
MATERIALS AND METHODS
Construction of the targeting vector.
The targeting constructs were designed to insert a poly(A) cassette derived from the rabbit β-globin gene (57) into the Exon 1A exon in both orientations at position 184080 (AL593857.10) (Fig. 2A). The constructs were generated by homologous recombination in yeast (58). Briefly, a 1.2-kb fragment (nucleotides 31392 to 32553; M18818) from the rabbit β-globin gene containing part of exon 2, complete intron 2, and exon 3 harboring the poly(A) signal was cloned in both orientations into an XhoI site, 5′ of the loxP site flanking the selection cassette, in pRAY-Cre (AJ627603). The 5′ and 3′ recombinogenic arms with homology to the Exon 1A exon, extending upstream and downstream of the site of insertion of the polyadenylation cassette, were amplified by PCR; the 5′ arm was cloned 5′ of the polyadenylation cassette, and the 3′ arm was cloned downstream of the 3′ loxP site. A linear fragment comprising the recombinogenic arms, poly(A) cassette, and selection cassette was cotransformed into yeast YPH501 with a 12-kb mouse genomic EcoRI fragment, encompassing the Exon 1A exon and Gnas exon 1, cloned in the yeast-Escherichia coli shuttle vector pRS414 (61) using a yeast transformation kit (Sigma). The recombined shuttle vector was recovered from yeast colonies using the Zymoprep II kit (Cambridge Bioscience) and electroporated into E. coli prior to targeting. All primer sequences are available on request. For both targeting constructs, the left arm was 9.2 kb and the right arm was 2.7 kb.
Fig 2.
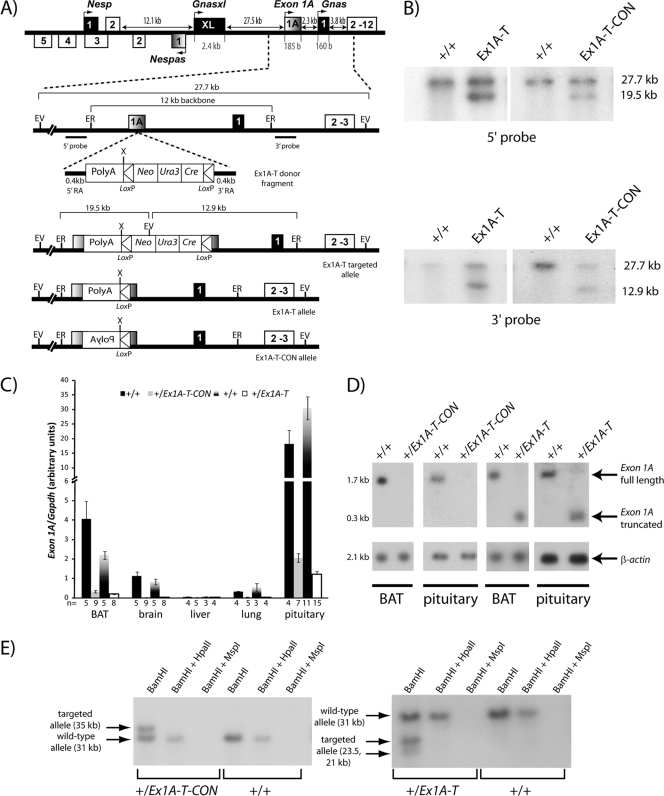
Insertion of a poly(A) cassette into the Exon 1A exon in both orientations. (A) Schematic overview of insertion in the targeted alleles. A 12-kb EcoRI fragment containing the Exon 1A exon and Gnas exon 1 was cloned into yeast shuttle vector pRS414. A donor fragment containing a rabbit β-globin poly(A) cassette (in either the forward or the reverse orientation); the Neo, Ura-3, and Cre genes between two loxP sites; and 0.4-kb recombinant arms on each side homologous with the 5′ and 3′ sequence of the insertion point in the Exon 1A exon, was also transformed into yeast. Both yeast constructs were then linearized and electroporated into ES cells, where they were screened for homologous recombination by Southern blot analysis. ER, EcoRI; EV, EcoRV; X, XhoI. (B) Screening for correct targeting by Southern blot assay. Genomic DNA from ES cells was digested with EcoRV and hybridized with 5′ and 3′ probes, respectively. The wild-type allele gives rise to a 27.7-kb fragment, while the targeted allele will give rise to a 19.5-kb or a 12.9-kb fragment for the 5′ and 3′ probes, respectively. (C) Real-time RT-PCR analysis of the Exon 1A transcript upon paternal transmission in newborn tissues. Primers were designed from the end of the Exon 1A exon to the start of Gnas exon 2. Expression was normalized to Gapdh. Error bars indicate the standard errors of the means. (D) Northern blot assay of Exon 1A and a β-actin loading control in newborn tissues. Pituitaries were pooled in groups of two, with a total of 4 animals being analyzed; BAT is representative of 10 to 14 samples analyzed per genotype. (E) Southern blot analysis of methylation of the Exon 1A promoter. Genomic DNA was first cut with BamHI to generate the 181728-to-184851 fragment (AL593857.10); this corresponds to the wild-type band of 3,124 bp. The inserted poly(A) cassette at 184080 of 1,313 bp [1,163 bp of poly(A) plus 150 bp of loxP] has a BamHI site at the start of the poly(A) cassette; thus, when the cassette is inserted in the forward orientation in the Ex1A-T mutation, two bands of 2,352 bp and 2,084 bp are observed, which correspond to the targeted allele, and when it is inserted in the reverse orientation in the Ex1A-T-CON mutation, a band of 3,515 bp is observed, which corresponds to the targeted allele, and the second band of 921 bp is too small to be seen on the blot.
Targeting of embryonic stem (ES) cells and mouse husbandry.
Both targeting constructs were linearized with NotI and electroporated into MAC3 ES cells derived from mouse strain 129/Sv/Ev (50, 61). Colonies surviving the G418 selection for both targeting constructs were screened for correct targeting by Southern blot analysis of EcoRV-digested genomic DNA probed with a 1.3-kb EcoRI fragment (nucleotides 18357 to 19683; AL593857.10) that was located 3′ of the right arm of the targeting constructs. Correct targeting at the 5′ end was confirmed by probing EcoRV-digested genomic DNA with a 1-kb PCR product (nucleotides 32979 to 33982; AL593857.10). Chimeras were generated by injecting targeted ES cells into C57BL/6J blastocysts, followed by transfer to pseudopregnant foster mothers (CD1M) (61). Excision of the floxed selection cassette, containing Ura3-Cre, occurred in the germ line of male chimeras by testis-specific expression of cre recombinase (8). Transmitting male chimeras were crossed with 129/SVEM mice, and heterozygotes were maintained on a 129/SVEM background. Proper excision of the cassette was confirmed by PCR amplification across the remaining loxP site.
Both the Ex1A-T and Ex1A-T-CON alleles were deposited with the Mouse Genome Informatics group of the Jackson Laboratory as Gnastm2Jop and Gnastm3Jop, respectively.
Mice were housed under specific-pathogen-free conditions in individually ventilated cages at 21 ± 2°C and a humidity of 55% ± 10% and subjected to a 12-h light/12-h dark cycle, in accordance with UK Home Office Welfare Guidelines. Mice had free access to water (25 ppm chloride) and food containing 11.5 kcal% fat, 23.93 kcal% protein, and 61.57 kcal% carbohydrate (SDS, RM3 diet).
Genotyping.
Genomic DNA was extracted from mouse biopsy specimens. Genotyping for both mutations was performed by duplex PCR using primers GGAAAGTGCAAAGGTGCAGAT, TTGCTTCAGGTGGCTGGTACCA, CTGTCTCATCATTTTGGCAAAG, and CTCAAGGGGCTTCATGATGT.
RNA analysis.
Total RNA was isolated using either the RNeasy or the RNeasy lipid kit (Qiagen). For Northern blot analysis of Gnas, Exon 1A, Gnasxl, and Actb, 1 to 3 μg of RNA was loaded onto an agarose gel, transferred to a nylon membrane, and hybridized to specific probes as described previously (61, 63). Gnas, Exon 1A, and Gnasxl riboprobes were each targeted to the unique first exon of each transcript (nucleotides 186498 to 186579, 183915 to 184090, and 155917 to 156264; AL593857.10, respectively). Primers pairs for real-time reverse transcription (RT)-PCR were designed as described previously (24). cDNA was synthesized from 4 to 5 μg of RNA primed by random hexamers with a SuperScript III First Strand cDNA Synthesis kit (Invitrogen). Real time RT-PCR was performed on an ABI Prism 7500 Fast system (Applied Biosystems) using Fast SYBR green PCR Master Mix (Applied Biosystems). Each reaction was done in triplicate with approximately 10 ng of cDNA per reaction. Samples were normalized to glyceraldehyde 3-phosphate dehydrogenase gene (Gapdh) levels. Analysis of data was carried out with ABI Prism 7500 system software (version 1.4). Primers are shown in Table 2; Gapdh primers were described previously (17).
Table 2.
Primers used in real-time RT-PCR
| Description | Sequence |
|---|---|
| Gnas exon 1A forward | AGCTAACCCAAGGAGCACCTAA |
| Gnas exon 2 reverse | GTTAAACCCATTAACATGCAGGA |
| Gnas exon 1 forward | AGAAGGACAAGCAGGTCTACCG |
| Gnas XL exon forward | ATAAGAAACGCAGCAAGCTCATC |
| Gnas XL exon reverse | CATGTAGTCCATCTTCTCCTCCT |
| Gnas N1 exon reverse | GGACTGTAGCCATCATCTAGTGG |
| Leptin exon 2 forward | TGCCTATCCAGAAAGTCCAGGAT |
| Leptin exon 3 reverse | TCATTGGCTATCTGCAGCACAT |
| Ucp1 exon 3 forward | ACCACAGAAAGCTTGTCAACAC |
| Ucp1 exon 4 reverse | CCCCTTCATGAGGTCATATGTTA |
Methylation analysis.
Genomic DNA was isolated from neonatal brain and BAT using the AllPrep DNA/RNA kit (Qiagen). Methylation of the Exon 1A DMR was then assessed as described previously (61).
Western blotting.
Total cell lysates were extracted using radioimmunoprecipitation assay buffer (phosphate-buffered saline with 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS). A 50-μg sample of cell lysate was loaded into each lane of a 4 to 12% gradient Bis-Tris NuPAGE gel (Invitrogen). Proteins were transferred to a polyvinylidene difluoride membrane (GE Healthcare) and probed with specific primary antibodies, followed by horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Sigma) and then detected using the ECL plus Western blotting detection kit (GE Healthcare). The anti-rabbit Gsα antibody (1:1,000; Calbiochem) (7, 23) is directed against the C-terminal Gsα epitope RMHLRQYELL. The rat anti-rabbit XLαs antibody (1:1,000; gift from W. B. Huttner) is directed against a glutathione S-transferase-tagged XL exon fragment. Anti-rabbit glyceraldehyde 3-phosphate dehydrogenase (Sigma G9295) was used as a loading control.
Mouse weights.
Individual mice of both sexes on a standard diet were weighed at birth and then weekly for 12 weeks. For each litter, which contained both wild-type and mutant mice, at each time point the average wild-type weight of each sex was calculated and then each individual mouse's weight within the litter was taken as a percentage of the average wild-type weight of the corresponding sex.
Dual-energy X-ray absorptiometry (DEXA) analysis.
At 12 weeks of age, male mice were weighed and given a nonrecoverable general anesthetic before scanning with a Lunar PIXImus Mouse Densitometer (Wipro; GE Healthcare). Fat mass, lean mass, bone mineral density, and body length were all measured.
Metabolic caging.
At 13 weeks of age, male mice were weighed and then individually housed in metabolic cages for 24 h, during which time they had free access to preweighed food and water. After the 24-h time period, the amounts of food and water consumed were measured. After metabolic housing, the mice were then returned to their home cage.
Metabolic rate measurements.
At 12 weeks of age, male mice were weighed and then individually housed in indirect calorimetry cages (Oxymax; Columbus Instruments) for 22 h. Cages allowed free access to food and water. Oxygen consumption, carbon dioxide consumption, respiratory exchange ratio, and heat production were all analyzed.
Plasma leptin analysis.
Plasma samples were collected from 12-week-old male mice. Leptin was then quantified using the mouse and rat leptin enzyme-linked immunosorbent assay kit (BioVendor) and analyzed using the FLUOstar OPTIMA ABS absorbance microplate reader (BMG LABTECH).
Suckling activity.
On postnatal day 5, suckling activity was assessed as described previously (32). Briefly, the mother was removed from the litter for 2 h and then returned. Each mouse in the litter was weighed before separation, just before the mother was returned (starved weight), and 2 h after the mother had been returned (fed weight). The difference in the starved and fed weights was calculated in terms of 75% of the starved weight (53) and was taken as a measure of suckling ability.
Statistical methods.
All comparisons were between cohorts of mutant and wild-type age-matched siblings and were made using an unpaired two-tailed Student t test.
RESULTS
Truncation of Exon 1A.
A poly(A) cassette from the rabbit β-globin gene (15, 54, 57) was inserted within the Exon 1A exon at position 184080 (AL593857.10), 21 bp 5′ from the end of the Exon 1A exon (as described in reference 39) in both a 5′-to-3′ orientation to truncate the Exon 1A transcript (Ex1A-T) and in a 3′-to-5′ orientation as a control (Ex1A-T-CON) by homologous recombination in ES cells (Fig. 2A and B).
After germ line transmission of the targeted alleles, one single line of each of the targeted alleles was analyzed and expression of Exon 1A was assessed following paternal transmission in both transgenic lines. To test that the Exon 1A transcript had been successfully truncated in the +/Ex1A-T (the maternal allele precedes the paternal allele in all of the genotypes described here) mice, we designed primers for real-time RT-PCR at the 3′ end of the Exon 1A exon, after the poly(A) cassette insertion site and at the 5′ end of Gnas exon 2, so that only transcript downstream of the poly(A) cassette would be analyzed. We then assayed transcript levels in a number of different neonatal tissues in both the +/Ex1A-T and +/Ex1A-T-CON mutants, as well as their wild-type siblings (Fig. 2C). Exon 1A levels were also assayed using Northern blot analysis with a probe specific for the Exon 1A exon (Fig. 2D). Real-time RT-PCR showed that Exon 1A transcript levels were much reduced 3′ of the poly(A) cassette in the +/Ex1A-T mouse compared with those in the wild-type mouse. Upon Northern blot analysis, a single transcript shorter than full-length Exon 1A was detected, consistent with a form of the Exon 1A transcript truncated within the Exon 1A exon. Thus, we conclude that truncation of the Exon 1A transcript in +/Ex1A-T mice has occurred as designed.
When the poly(A) cassette was inserted in the reverse orientation in the +/Ex1A-T-CON line, the Exon 1A levels were also much reduced (Fig. 2C) and undetected by Northern blot assay (Fig. 2D). One explanation for this low level of expression is that incorporation of a 1.2-kb poly(A) cassette destabilized the full-length Exon 1A transcript, which is only 17.8 kb. Methylation of the Exon 1A DMR was assessed by methylation-sensitive restriction enzyme Southern blot assays in +/Ex1A-T and +/Ex1A-T-CON mice (Fig. 2E). This revealed that the Exon 1A DMR containing the Exon 1A promoter remained unmethylated on the paternal allele of both +/Ex1A-T and +/Ex1A-T-CON mice. Thus, loss of the Exon 1A transcript in +/Ex1A-T-CON mice was not due to gain of methylation of the Exon 1A promoter. In addition, insertion of the poly(A) cassette did not alter the methylation pattern at the Exon 1A DMR. In conclusion, both mutant lines showed loss of the full-length Exon 1A transcript.
Imprinted Gnas is upregulated.
We next examined the expression of Gnas transcripts by real-time RT-PCR and Northern blotting. The protein product Gsα was assayed by Western blotting. Gnas/Gsα was assayed in both +/Ex1A-T and +/Ex1A-T-CON individuals and their wild-type littermates in neonatal BAT and pituitary tissue, in which Gnas is predominantly maternally expressed, as well as in neonatal tissues such as those of the brain, lung, and liver, in which Gnas is biallelically expressed (Fig. 3A to C).
Fig 3.
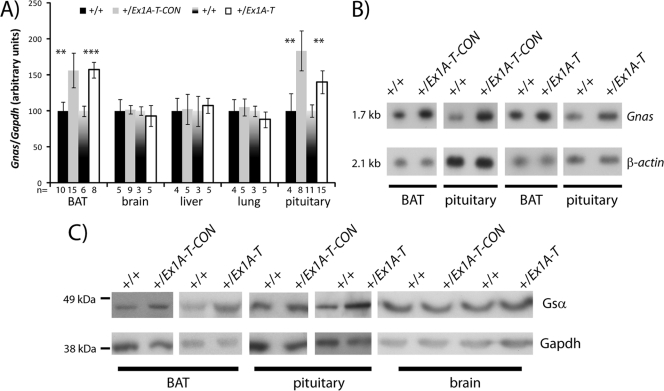
Upregulation of Gnas. (A) Real-time RT-PCR analysis of Gnas transcript in newborn tissues. Primers were designed from the end of Gnas exon 1 to the start of Gnas exon 2. Expression was normalized to Gapdh. Error bars indicate the standard errors of the means; **, P < 0.05; ***, P < 0.01. (B) Northern blot assay of Gnas and a β-actin loading control in newborn tissues. Pituitaries were pooled in groups of two, with a total of five animals analyzed per genotype. BAT is representative of 10 to 14 samples analyzed. (C) Western blot assay of Gsα and a GAPDH loading control in newborn tissues. Nine to thirteen pituitaries were pooled for each genotype, and other tissues are representative of at least two samples analyzed per genotype.
Similar to previous reports on mice with a paternal deletion of Exon 1A (38, 61), upon loss or truncation of the Exon 1A transcript, we observed raised expression levels of Gnas in tissues such as neonatal BAT and pituitary tissue, where Gnas is normally maternally expressed and paternally repressed, but detected no change in transcript levels in brain, liver, and lung tissues, which are tissues in which Gnas normally shows biallelic expression. We attribute the raised levels of Gnas in BAT and pituitary tissue to loss of imprinted expression and derepression of Gnas on the paternal allele.
Expression of Gnasxl transcripts.
We also assayed the effect on production of Gnasxl transcripts, as the poly(A) cassette lies in the first intron of Gnasxl. Real-time RT-PCR primers were designed at the 3′ end of the Gnas XL exon and at the 5′ end of Gnas/Gnasxl exon 2 to assay both the full-length Gnasxl transcript and the neural transcript XLN1 but not forms truncated by the poly(A) cassette. Upon paternal transmission of the Ex1A-T mutation, there was a significant reduction of Gnasxl transcripts, consistent with truncation of both the full-length and neural XLN1 Gnasxl transcripts due to insertion of the poly(A) cassette (Fig. 4A). We also designed real-time RT-PCR primers at the 5′ and 3′ ends of the Gnas XL exon which showed no significant change in expression of the Gnas XL exon amplicon (Fig. 4B). This confirmed that the Gnasxl transcript was indeed truncated between the Gnas XL (1st) exon and Gnas/Gnasxl exon 2 rather than loss of expression resulting from ablation of the Gnasxl transcript. Lastly, we designed primers at the 3′ end of the Gnas XL exon and the 5′ end of the neural N1 exon and carried out real-time RT-PCR analysis specifically for XlN1 in the neonatal brain, which further confirmed the loss of the full-length neural transcript (data not shown). Northern blot assays of embryonic day 15.5 (E15.5) embryos probed for the Gnas XL exon also confirmed a loss of full-length Gnasxl (Fig. 4C).
Fig 4.
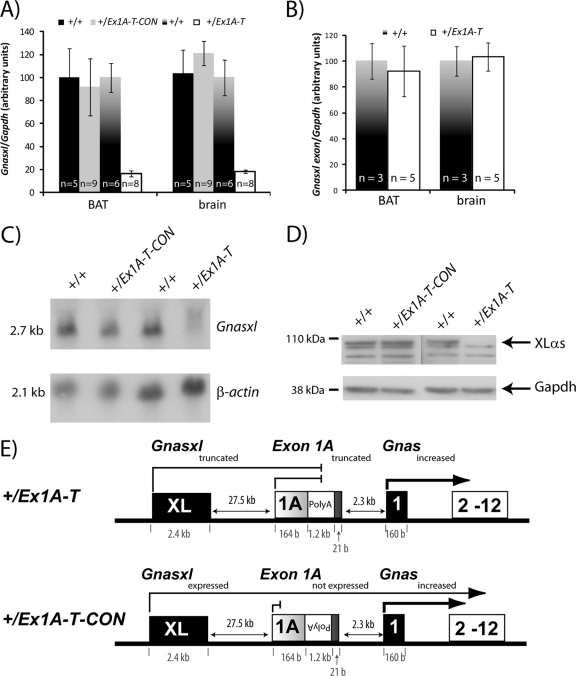
Truncation of Gnasxl. (A) Real-time RT-PCR analysis of Gnasxl transcript in newborn tissues. Primers were designed at the end of the Gnas XL exon and at the start of Gnas exon 2. Expression was normalized to Gapdh. Error bars indicate the standard errors of the means; P < 0.005. (B) Real-time RT-PCR analysis of Gnasxl transcript in newborn tissues. Primers were designed at the start and end of the Gnas XL exon. Expression was normalized to Gapdh. Error bars indicate the standard errors of the means. (C) Northern blot assay of Gnasxl and a β-actin loading control in E15.5 whole embryos. Results shown are representative of 3 to 5 samples per genotype. (D) Western blot assay of XLαs and a GAPDH loading control in E15.5 whole embryos. Samples shown are representative of at least two samples analyzed per genotype. (E) Schematic diagram summarizing the outcomes of the poly(A) cassette insertion in the +/Ex1A-T and +/Ex1A-T-CON mutations on Gnas cluster transcripts on the paternal allele.
In contrast, in the +/Ex1A-T-CON mutation, there was no obvious difference in the level of Gnasxl, indicating that insertion of the polyadenylation cassette had not affected Gnasxl expression (Fig. 4A and C). We next investigated whether the two mutations affected XLαs protein levels by Western blotting. Truncation of the Gnasxl transcript in +/Ex1A-T but not +/Ex1A-T-CON mice resulted in loss of full-length XLαs protein (Fig. 4D).
The poly(A) truncation site is located downstream of the Gnas XL exon, and although both the Gnasxl and XlN1 transcripts were truncated and nonfunctional in the +/Ex1A-T mutation, we surmise that the alternative reading frame protein, ALEX, which contains its protein coding region solely within the Gnas XL exon, would be intact and functional.
In light of these results, we are presented with two different mouse models (Fig. 4E). Paternal transmission of the Ex1A-T mutation results in tissue-specific upregulation of Gsα and loss of XLαs (and XLN1) protein but ALEX is presumed to be at wild-type levels. Paternal transmission of the Ex1A-T-CON mutation also results in tissue-specific upregulation of Gsα, but XLαs and presumably ALEX are equivalent to the wild type. Next we investigated the phenotype of these mice.
Both mutations show reductions in growth.
Within 24 h of birth, neonatal pups were recorded and genotyped. The +/Ex1A-T-CON mice were found at expected Mendelian frequencies at birth (50.6% of 316 neonates). The +/Ex1A-T mice occurred with a frequency of 45.5% (of 444 neonates), which is not significantly different from the expected 50% when analyzed by the chi-squared test (P > 0.05).
Mice that paternally inherited Ex1A-T and Ex1A-T-CON and their wild-type siblings of both sexes were weighed at birth and then weekly for 12 weeks to assess growth. The weights of the mutant mice are shown as a percentage of that of their same-sex wild-type siblings, and only data from mice that survived to 12 weeks were considered (Fig. 5A). Both the +/Ex1A-T-CON and +/Ex1A-T mice were smaller at birth, the +/Ex1A-T-CON mice had 95% (P = 0.009) and the +/Ex1A-T mice had 92% (P = 1 × 10−4) of the weight of their wild-type siblings. Eighty-five percent of +/Ex1A-T and 93% of +/Ex1A-T-CON mice survived past weaning. Most losses (75%) were the smallest members of their litters and died between 1 and 2 weeks of age, when their weight gain was at its lowest. Both the +/Ex1A-T and +/Ex1A-T-CON mice were growth retarded. This was most evident at 2 weeks of age.
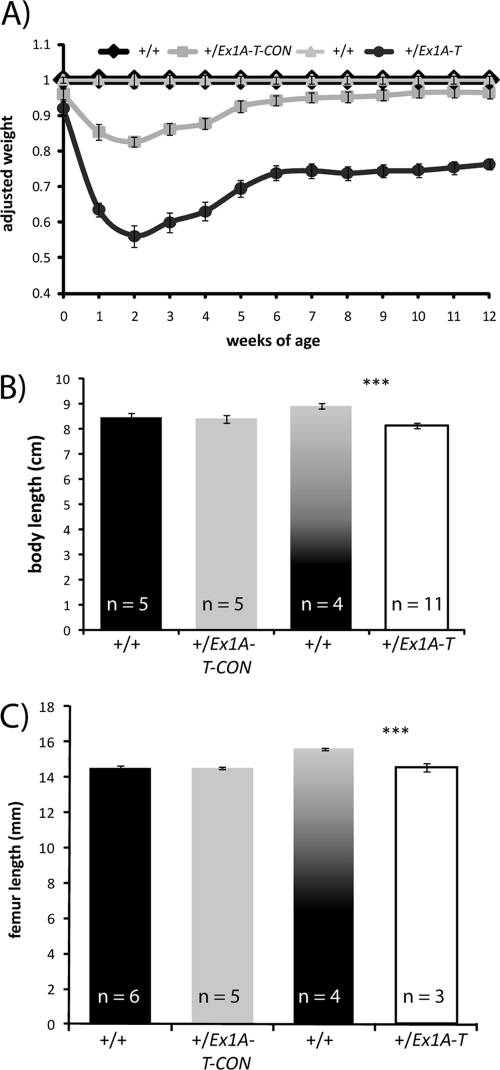
Fig 5.
Growth retardation. (A) Growth curve of +/Ex1A-T and +/Ex1A-T-CON mice and their wild-type littermates of both sexes over 12 weeks. Wild-type littermate weights have been normalized to 1 at each time point, and weights of the transgenic mice have been taken as a percentage of wild-type weights at each time point. Error bars indicate the standard errors of the means; n = 14 to 34. (B) Body lengths of 12-week-old-mice, measured from the nose to the start of the tail. Error bars indicate the standard errors of the means; ***, P < 0.01. (C) Femur lengths of 12-week-old-mice. Error bars indicate the standard errors of the means; ***, P < 0.01.
The +/Ex1A-T-CON mice had 83% (P = 1 × 10−13) of the weight of their wild-type siblings at 2 weeks, after which point they started to recover to near wild-type weights by 7 weeks (96%; P = 0.01 to 0.1; 7 to 12 weeks). Weights of +/Ex1A-T mice were considerably more reduced at 56% (P = 1 × 10−11) of those of their wild-type siblings at 2 weeks of age; they too then started to make a recovery but only to 75% (P = 1 × 10−11 to 1 × 10−13; 7 to 12 weeks) of the wild-type weight. This indicates that both Gsα and XLαs affect growth both postnatally and prenatally. Thus, upregulation of Gnas in +/Ex1A-T-CON mice gives rise to a growth retardation, which is almost fully reversible, but upregulation of Gnas together with loss of Gnasxl/XlN1 in +/Ex1A-T mice results in severe growth retardation, which is only partially reversible.
+/Ex1A-T mice have shorter body lengths.
A shorter body length has previously been reported in the Gnasxl knockout mouse (65). Body and femur lengths were measured at 12 weeks of age in both +/Ex1A-T and +/Ex1A-T-CON mice (Fig. 5B and C). Mice that paternally inherited Ex1A-T had significantly shorter body lengths (P = 1 × 10−4) and also had shorter femur lengths than their wild-type littermates (P = 0.006), but there was no significant difference in either body or femur length in the mice that paternally inherited Ex1A-T-CON compared to their wild-type siblings. This confirms the role of Gnasxl in the regulation of body length and reveals for the first time that femur length is also regulated by Gnasxl.
+/Ex1A-T mice have less fat mass.
At 12 weeks of age both +/Ex1A-T and +/Ex1A-T-CON mice were analyzed by DEXA. The +/Ex1A-T mice had less total fat and less total lean mass than their wild-type siblings (2.7 versus 4.9 g of fat mass [P = 1 × 10−6], 15.4 versus 20.8 g of lean mass [P = 1 × 10−7]). Furthermore, compared to their body weight, the +/Ex1A-T mice had a significantly lower percentage of fat mass (P = 0.009) and a correspondingly higher percentage of lean mass (P = 0.0005) (Fig. 6A). Thus, the +/Ex1A-T mice are smaller because they have an absolute reduction of both lean and fat masses with a disproportionately greater reduction of fat mass. Similar effects on total fat and lean mass, as well as percentages of fat and lean mass, were seen in +/Ex1A-T-CON mice compared with their wild-type siblings, but these differences were not statistically significant. The body mass index (BMI) of the +/Ex1A-T mice, but not that of the +/Ex1A-T-CON mice, was also found to be significantly reduced at 2 and 12 weeks (2.3 versus 2.8 [P = 0.03, n = 3 to 6] at 2 weeks; 2.2 versus 2.9 [P = 1 × 10−6, n = 6 to 11] at 12 weeks).
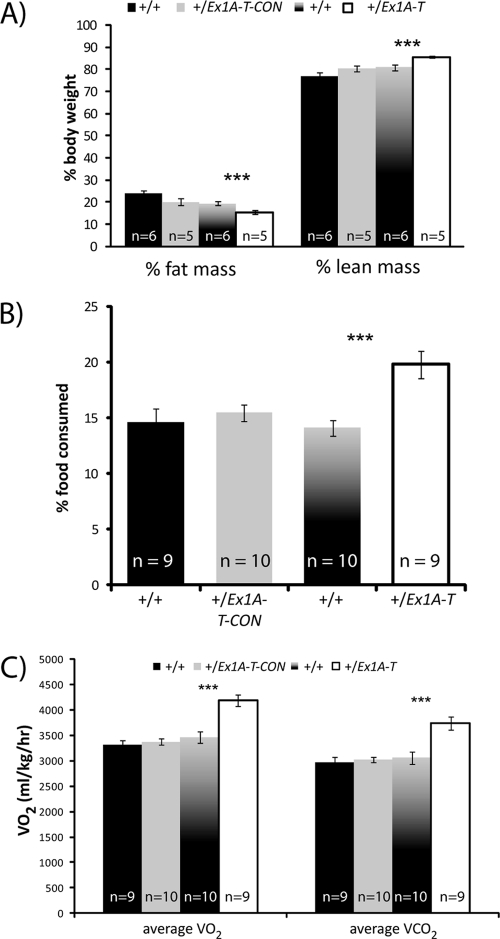
Fig 6.
Analysis of adiposity. (A) Percentages of fat and lean masses of 12-week-old male mice from DEXA analysis. Error bars indicate the standard errors of the means; ***, P < 0.01. (B) Percentages of food intake by 12-week-old male mice. Error bars indicate the standard errors of the means; **, P < 0.05; ***, P < 0.01. (C) Rates of oxygen consumption and carbon dioxide output of 12-week-old male mice. Error bars indicate the standard errors of the means; ***, P < 0.01.
Reduction of fat mass is not due to diet.
To ascertain the cause of the observed lower fat mass, 12-week-old-mice were housed in metabolic cages, where their food and water intake was measured over a 24-h period. There was no difference in food intake in the +/Ex1A-T-CON mice. However, intriguingly the +/Ex1A-T mice in fact ate significantly more food than their wild-type siblings (4.1 versus 3.6 g; P = 0.04), an increase that was even more pronounced when measured as a proportion of body weight (P = 0.0007) (Fig. 6B). We conclude that the reduction of fat mass of the adult +/Ex1A-T mice was not in any way due to reduced food intake.
No reduction in suckling in +/Ex1A-T pups.
Previous studies have shown that neonatal mice that lack all Gnasxl transcripts have reduced suckling ability (10, 48, 69). Thus, we measured suckling ability in 5-day-old +/Ex1A-T and +/Ex1A-T-CON mice and their wild-type littermates after a 2-h separation from the mother. Suckling ability was assessed in 5-day-old mice by the difference between starved and fed body weights measured in terms of body weight to account for the smaller weight of the mutant mice. We observed no difference in the suckling abilities of 20 +/Ex1A-T-CON mice and 16 wild-type littermates (3.66% versus 4.31%, P = 0.41), indicating that overexpression of Gsα has no effect on suckling ability. There was also no significant difference in the suckling ability of the 26 +/Ex1A-T mice and that of 21 wild-type controls (3.34% versus 4.23%, P = 0.28). This is comparable to results found in the Sml mice, which have nonfunctional XLαs but in which XLN1 and ALEX are presumed to be functional (32).
+/Ex1A-T mice have a higher metabolic rate.
Twelve-week-old +/Ex1A-T but not +/Ex1A-T-CON mice showed significant increases in both oxygen consumption and carbon dioxide output (Fig. 6C). This is indicative of an increased metabolic rate in the +/Ex1A-T mice, which is consistent with a reduction of fat mass.
+/Ex1A-T mice have a lower bone mineral density.
Bone mineral density was also assessed during analysis of 12-week-old +/Ex1A-T and +/Ex1A-T-CON mice by DEXA (Fig. 7A). +/Ex1A-T mice were found to have a significant reduction in bone mineral density. In contrast, +/Ex1A-T-CON mice showed no difference in bone mineral density.
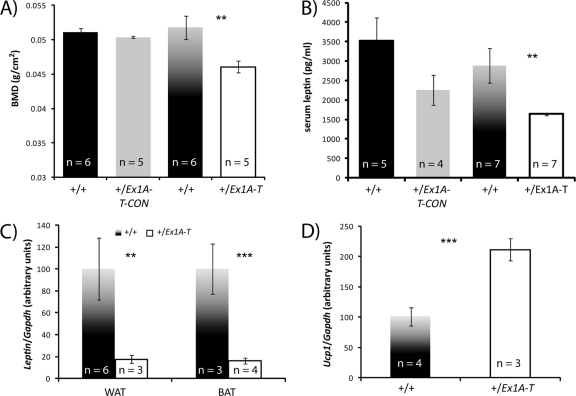
Fig 7.
(A) Bone mineral densities of 12-week-old male mice from DEXA analysis. Error bars indicate the standard errors of the means; **, P < 0.05. (B) Serum leptin levels of 12-week-old male mice. Error bars indicate the standard errors of the means; **, P < 0.05. (C) Leptin levels in 12-week-old male BAT and WAT by real-time RT-PCR. Error bars indicate the standard errors of the means; **, P < 0.05, ***, P < 0.01. (D) Ucp1 levels in 12-week-old male BAT by real-time RT-PCR. Error bars indicate the standard errors of the means; ***, P < 0.01.
Leptin is downregulated.
Serum leptin levels were measured in 12-week-old +/Ex1A-T and +/Ex1A-T-CON mice and their wild-type siblings (Fig. 7B). Leptin mRNA levels were also measured in white adipose tissue (WAT) and BAT by real-time RT-PCR in +/Ex1A-T mice and their wild-type littermates (Fig. 7C). Leptin levels in serum and mRNA levels in WAT and BAT were found to be significantly reduced in +/Ex1A-T mice. This is consistent with their lean phenotype, as leptin circulates at levels proportional to adiposity (40), and indeed, when leptin levels are calculated relative to fat mass, there is no difference between the +/Ex1A-T mice and their wild-type littermates. Low leptin levels could also explain why the +/Ex1A-T mice are hyperphagic as low leptin results in increased hyperphagia (21). There was no significant difference between the serum leptin levels of +/Ex1A-T-CON mice and those of their wild-type littermates.
Ucp1 is upregulated.
Ucp1 is positively regulated by sympathetic, β-adrenergic, activity. Ucp1 levels were found to be upregulated in adult BAT of +/Ex1A-T mice (Fig. 7D), and this is indicative of an increase in sympathetic nervous system (SNS) activity. There was no significant difference between Ucp1 levels in +/Ex1A-T adult WAT and the wild type.
DISCUSSION
We initially set out to further elucidate the mechanisms whereby Exon 1A brings about silencing of Gnas on the paternal allele. Proposed mechanisms include competition between the Gnas and Exon 1A promoters for shared transcription elements, binding of tissue-specific methylation-sensitive silencing or activating factors to the Exon 1A DMR, or transcriptional interference of the Exon 1A transcript across the Gnas promoter or the Exon 1A transcript itself (45).
In both +/Ex1A-T and +/Ex1A-T-CON mice, loss or truncation of the Exon 1A transcript resulted in loss of imprinted expression of Gnas. Promoter competition is unlikely to account for these findings, as the Exon 1A promoter sequence was not disrupted in either the Ex1A-T or the Ex1A-T-CON allele. Moreover, as shown in the Northern blot assays of Exon 1A (Fig. 2D), truncated Exon 1A transcript was transcribed in +/Ex1A-T mice, indicating a functional Exon 1A promoter.
Our findings are consistent with a transcriptional interference model whereby transcription of Exon 1A across the Gnas promoter can bring about silencing of Gnas. Evidence supporting the transcriptional interference model includes the finding that Exon 1A levels are highest in tissues in which Gnas is paternally repressed, i.e., neonatal BAT and pituitary tissue (Fig. 2C) and also that Exon 1A-mediated repression of Gnas occurs in cis (63).
However, neither the silencer nor the enhancer model can be ruled out, as both mutations involve insertion of exogenous DNA into the Gnas locus. Although the methylation status of the Exon 1A DMR remains unchanged in both mutants (Fig. 2E) and no binding sites have been removed, the Exon 1A DMR covers the region from position −3400 to position −939 upstream of the Gnas exon 1 transcriptional start site and includes the Exon 1A exon itself, as well as approximately 1 kb upstream and 1.5 kb downstream of the Exon 1A exon (39). Thus, insertion of a 1.2-kb cassette within this DMR may well have disrupted binding to silencer or enhancer blocker elements.
Here we have shown that the +/Ex1A-T-CON mice present with postnatal growth retardation, but no change in bone mineral density or metabolic rate at 12 weeks of age when they have recovered to 96% of the wild-type weight. This can be attributed solely to loss of imprinting of Gnas, as Gnasxl expression, as well as all other Gnas cluster protein-coding transcripts, is normal. In contrast, the +/Ex1A-T mice show loss of imprinting of Gnas combined with loss of Gnasxl. This gives rise to severe growth retardation, which is not fully reversible. The +/Ex1A-T adult mice are also shorter, have a lower bone mineral density, a reduced fat mass, and an increased metabolic rate compared to their wild-type siblings. Leptin was found to be downregulated in +/Ex1A-T mice, while Ucp1 was upregulated.
The parental conflict hypothesis for the evolution of imprinting predicts that maternally expressed imprinted genes will be growth inhibiting whereas paternally expressed imprinted genes will be growth promoting (41). Thus, the finding that upregulation of a maternally expressed gene, Gnas, gives rise to mice that are smaller than their wild-type siblings, as seen in the +/Ex1A-T-CON mice, is consistent with the hypothesis, and so is the finding that when both maternally expressed Gnas is upregulated and paternally expressed Gnasxl is ablated, as is the case in +/Ex1A-T mice, there is an additive effect and these mice are even smaller than those which only have an upregulation of Gnas.
Much of the phenotype observed in +/Ex1A-T mice corresponds to that seen in other mouse models that have a loss of functional XLαs such as Gnasxl knockout mice, MatDp(dist2) mice, and Sml mice (10, 11, 32, 48, 62, 65, 68, 69). However, whereas there are considerable prenatal losses of Sml (S. Ball, unpublished data) and MatDp(dist2) mice and Gnasxl knockout mice show profound perinatal lethality, +/Ex1A-T mice have relatively few preweaning losses and are essentially fully viable and thus provide a good model for investigation of the effects of loss of Gnasxl. In addition to the above, we have reported here two other novel findings upon loss of Gnasxl transcripts.
Unlike all other models with deficient Gnasxl expression, except Sml mice, +/Ex1A-T mice do not show suckling defects. Suckling defects are characteristic of mice with paternally derived deletions of either the Gnas XL exon (48) or Gnas exon 2 (69) or mice with maternal duplication/paternal deficiency of distal chromosome 2, MatDp(dist2) (10). There are three well-characterized paternal transcripts that encompass and originate at the Gnas XL exon (Fig. 1). The first of these is Gnasxl, a transcript which splices from the Gnas XL exon into exon 2 of Gnas and is transcribed through to exon 12 to generate the XLαs protein. The second is XLN1, which is very similar to Gnasxl, except that it contains an alternate exon, N1, located after Gnas exon 3 and results in a shortened, neurally specific transcript and encodes the XLN1 protein. The third transcript is ALEX, which is identical in nucleotide sequence to Gnasxl, but the protein ALEX is generated from an alternate reading frame within the Gnas XL exon and is only protein coding within this exon.
The protein products XLαs, XLN1, and ALEX are all predicted to be absent in MatDp(dist2) mice and mice with paternal deletions of either the Gnas XL exon or Gnas exon 2. The Gnasxl promoter is silent in MatDp(dist2), and an exon essential for all three proteins is missing in Gnas XL exon deletion mice. Loss of paternal Gnas exon 2 would certainly result in loss of XLαs and XLN1, as transcripts for both of these proteins are in part encoded by this exon. Gnas exon 2 is also part of the 3′ UTR of the ALEX transcript, and its deletion is likely to destabilize the Gnasxl/ALEX transcript and result in loss of ALEX protein. Sml mice have a point mutation in Gnas exon 6 (56) and thus have disrupted XLαs, but XLN1 and ALEX are thought to be unaffected. Here we have shown that the +/Ex1A-T mice, which have a poly(A) truncation site inserted between the Gnas XL exon and Gnas exon 2, lack XLαs and XLN1 but the ALEX protein is probably intact. Thus, in Gnasxl mutants with suckling failure, ALEX is absent but in Gnasxl mutants that suckle normally, ALEX is theoretically present. Thus, we have shown that the suckling defects observed in previously described Gnasxl mutants cannot be due to loss of XLαs or XLN1 and propose that this is due to loss of ALEX. Interestingly, Gnasxl is expressed in brain nuclei that can affect suckling (48) and as ALEX's RNA is indistinguishable from Gnasxl RNA, ALEX may be expressed in these regions as well. It also remains a possibility that the suckling defects are due to poorly characterized XXLb1, XXLb2, or some as-yet-undescribed transcript (1, 27).
The other phenotypes observed in +/Ex1A-T mice (and not +/Ex1A-T-CON mice), namely; reduced body weight, reduced adiposity, reduced body length, increased food consumption, increased metabolic rate, increased Ucp1, and reduced leptin, have also been found in other adult mouse models lacking Gnasxl (32, 65, 68). Thus, these phenotypes can be attributed to loss of XLαs/XLN1. While we did not analyze locomotor activity, previous work has shown that loss of XLαs has no effect on locomotor activity in adult mice (65).
The second novel finding is the presence of a bone phenotype, namely, that loss of Gnasxl results in a lowered bone mineral density. To our knowledge no previous study has identified a bone density phenotype specifically associated with Gnasxl, although bone phenotypes such as shortened bones and growth plate defects have been observed in mice when Gnas expression is reduced or ablated (6, 9, 51, 52). We propose here that XLαs may provide a new link between bone and adipocyte metabolism.
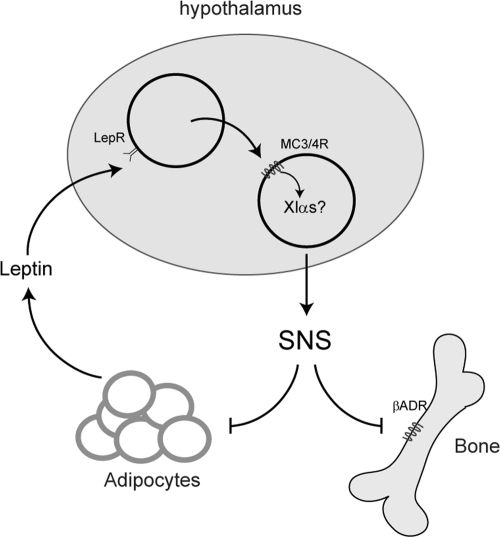
Understanding of the relationship between bone and adipocyte metabolism has increased over recent years. Although the exact mechanism of how bone and adipocytes are coregulated remains controversial (37, 66), a substantial amount of this mechanism is known. Leptin has been shown to inhibit bone mass through hypothalamic regulation of the SNS (19, 20). Leptin binds to leptin receptors in the arcuate nucleus to release melanocortin peptides, which then bind to melanocortin receptors, which signal downstream to regulate SNS activity (42). Both melanocortin 3 and 4 receptors (MC3R/MC4R) are Gs-coupled receptors which are expressed in the hypothalamus and are involved in regulating energy homeostasis (42). The SNS regulates bone metabolism through β-adrenergic pathways (20, 22) and also innervates WAT to positively regulate lipolysis in this organ (3, 4, 55, 67). In a wild-type mouse, leptin is expressed from adipocytes at a rate proportional to fat mass. High levels of circulating leptin act at the hypothalamus through the melanocortin pathway to increase β-adrenergic SNS activity to reduce bone and adipocyte mass, which in turn reduces the amount of circulating leptin (Fig. 8). How the melanocortin pathway regulates SNS activity downstream of the melanocortin receptors is as yet unknown.
Fig 8.
Model of how leptin regulates the SNS and thus bone and adipocyte formation through the hypothalamus. We hypothesize that XLαs may be involved in this regulatory loop by coupling to melanocortin receptors 3 and 4 in the hypothalamus.
In adults, Gnasxl is expressed primarily in neuroendocrine tissues such as the orexigenic neurons of the hypothalamus (28, 31, 43, 44). It is not expressed in adult WAT and BAT (65) and was not detectable in adult bone by real-time RT-PCR in the present study (data not shown).
We hypothesize that XLαs acts in the hypothalamus downstream of the MC3R/MC4R to repress SNS activity and thus repress bone formation and adipogenesis. In the absence of XLαs, leptin is unable to mediate its effects through MC3R/MC4R at the hypothalamus to repress SNS activity due to loss of XLαs. Consequently, SNS activity is upregulated, resulting in a decrease in bone and WAT mass. Thus, we propose that the bone and adipose phenotype observed due to loss of XLαs can be explained by XLαs acting downstream of leptin in the hypothalamus to mediate SNS activity. There is evidence that regulation of the SNS by XLαs occurs from birth, as newborn pups that lack XLαs have been shown to have increased cAMP levels in BAT (48) which is consistent with an upregulation of SNS activity. Thus, XLαs may regulate bone formation through the SNS throughout life.
The decrease in WAT is expected to lead to a decrease in leptin, resulting in an increased appetite. Regulation of appetite by leptin through the melanocortin pathway does not occur through the SNS; instead, this appears to be regulated through Sim1 downstream of MC4R in the paraventricular nucleus of the hypothalamus (2, 36). In animals without XLαs, low levels of leptin would act to increase appetite and thus food intake in an XLαs-independent (possibly Sim1) pathway.
ACKNOWLEDGMENTS
This work was supported by the UK Medical Research Council and the Wellcome Trust.
We thank Denise Barlow and Frank Sleutels for providing a cloned fragment of the rabbit β-globin gene. We also thank Bruce Cattanach for comments on the manuscript; Chris Esapa and Rosario Romero for technical advice; staff of the Mary Lyon Centre (MLC) for animal husbandry, in particular, Lynn Jones, Diane Napper, Jackie Harrison, Kelly Hunt, Lucie Vizor, and Sara Wells; the MLC transgenic service; and the MLC necropsy service.
Footnotes
Published ahead of print 3 January 2012
REFERENCES
- 1. Abramowitz J, Grenet D, Birnbaumer M, Torres HN, Birnbaumer L. 2004. XLalphas, the extra-long form of the alpha-subunit of the Gs G protein, is significantly longer than suspected, and so is its companion Alex. Proc. Natl. Acad. Sci. U. S. A. 101:8366–8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balthasar N, et al. 2005. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell 123:493–505 [DOI] [PubMed] [Google Scholar]
- 3. Bamshad M, Aoki VT, Adkison MG, Warren WS, Bartness TJ. 1998. Central nervous system origins of the sympathetic nervous system outflow to white adipose tissue. Am. J. Physiol. 275:R291–R299 [DOI] [PubMed] [Google Scholar]
- 4. Bartness TJ, Song CK. 2007. Thematic review series: adipocyte biology. Sympathetic and sensory innervation of white adipose tissue. J. Lipid Res. 48:1655–1672 [DOI] [PubMed] [Google Scholar]
- 5. Bastepe M, et al. 2002. Receptor-mediated adenylyl cyclase activation through XLalpha(s), the extra-large variant of the stimulatory G protein alpha-subunit. Mol. Endocrinol. 16:1912–1919 [DOI] [PubMed] [Google Scholar]
- 6. Bastepe M, et al. 2004. Stimulatory G protein directly regulates hypertrophic differentiation of growth plate cartilage in vivo. Proc. Natl. Acad. Sci. U. S. A. 101:14794–14799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baxendale RW, Fraser LR. 2003. Immunolocalization of multiple Galpha subunits in mammalian spermatozoa and additional evidence for Galphas. Mol. Reprod. Dev. 65:104–113 [DOI] [PubMed] [Google Scholar]
- 8. Bunting M, Bernstein KE, Greer JM, Capecchi MR, Thomas KR. 1999. Targeting genes for self-excision in the germ line. Genes Dev. 13:1524–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castrop H, et al. 2007. Skeletal abnormalities and extra-skeletal ossification in mice with restricted Gsalpha deletion caused by a renin promoter-Cre transgene. Cell Tissue Res. 330:487–501 [DOI] [PubMed] [Google Scholar]
- 10. Cattanach BM, Kirk M. 1985. Differential activity of maternally and paternally derived chromosome regions in mice. Nature 315:496–498 [DOI] [PubMed] [Google Scholar]
- 11. Cattanach BM, Peters J, Ball S, Rasberry C. 2000. Two imprinted gene mutations: three phenotypes. Hum. Mol. Genet. 9:2263–2273 [DOI] [PubMed] [Google Scholar]
- 12. Chen M, et al. 2005. Alternative Gnas gene products have opposite effects on glucose and lipid metabolism. Proc. Natl. Acad. Sci. U. S. A. 102:7386–7391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen M, Nemechek NM, Mema E, Wang J, Weinstein LS. 2011. Effects of deficiency of the G protein Gsα on energy and glucose homeostasis. Eur. J. Pharmacol. 660:119–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen M, et al. 2009. Central nervous system imprinting of the G protein G(s)alpha and its role in metabolic regulation. Cell Metab. 9:548–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chotalia M, et al. 2009. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 23:105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crawford JA, et al. 1993. Neural expression of a novel alternatively spliced and polyadenylated Gs alpha transcript. J. Biol. Chem. 268:9879–9885 [PubMed] [Google Scholar]
- 17. de Bovis B, et al. 2005. clc is co-expressed with clf or cntfr in developing mouse muscles. Cell Commun. Signal 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Paula FJ, Horowitz MC, Rosen CJ. 2010. Novel insights into the relationship between diabetes and osteoporosis. Diabetes Metab. Res. Rev. 26:622–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ducy P, et al. 2000. Leptin inhibits bone formation through a hypothalamic relay: a central control of bone mass. Cell 100:197–207 [DOI] [PubMed] [Google Scholar]
- 20. Elefteriou F, et al. 2005. Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature 434:514–520 [DOI] [PubMed] [Google Scholar]
- 21. Friedman JM, Halaas JL. 1998. Leptin and the regulation of body weight in mammals. Nature 395:763–770 [DOI] [PubMed] [Google Scholar]
- 22. Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. 2005. The molecular clock mediates leptin-regulated bone formation. Cell 122:803–815 [DOI] [PubMed] [Google Scholar]
- 23. Fukao M, Mason HS, Kenyon JL, Horowitz B, Keef KD. 2001. Regulation of BK(Ca) channels expressed in human embryonic kidney 293 cells by epoxyeicosatrienoic acid. Mol. Pharmacol. 59:16–23 [DOI] [PubMed] [Google Scholar]
- 24. Funnell AP, et al. 2007. Erythroid Kruppel-like factor directly activates the basic Kruppel-like factor gene in erythroid cells. Mol. Cell. Biol. 27:2777–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Germain-Lee EL, et al. 2005. A mouse model of Albright hereditary osteodystrophy generated by targeted disruption of exon 1 of the Gnas gene. Endocrinology 146:4697–4709 [DOI] [PubMed] [Google Scholar]
- 26. Hayward BE, et al. 2001. Imprinting of the G(s)alpha gene GNAS1 in the pathogenesis of acromegaly. J. Clin. Invest. 107:R31–R36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holmes R, Williamson C, Peters J, Denny P, Wells C. 2003. A comprehensive transcript map of the mouse Gnas imprinted complex. Genome Res. 13:1410–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ivanova E, Kelsey G. 2011. Imprinted genes and hypothalamic function. J. Mol. Endocrinol. 47:R67–R74 [DOI] [PubMed] [Google Scholar]
- 29. Kawai M, Devlin MJ, Rosen CJ. 2009. Fat targets for skeletal health. Nat. Rev. Rheumatol. 5:365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaya AI, Ugur O, Oner SS, Bastepe M, Onaran HO. 2009. Coupling of beta2-adrenoceptors to XLalphas and Galphas: a new insight into ligand-induced G protein activation. J. Pharmacol. Exp. Ther. 329:350–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kehlenbach RH, Matthey J, Huttner WB. 1994. XL alpha s is a new type of G protein. Nature 372:804–809 [DOI] [PubMed] [Google Scholar]
- 32. Kelly ML, et al. 2009. A missense mutation in the non-neural G-protein alpha-subunit isoforms modulates susceptibility to obesity. Int. J. Obes. (Lond.) 33:507–518 [DOI] [PubMed] [Google Scholar]
- 33. Kelsey G, et al. 1999. Identification of imprinted loci by methylation-sensitive representational difference analysis: application to mouse distal chromosome 2. Genomics 62:129–138 [DOI] [PubMed] [Google Scholar]
- 34. Klemke M, Kehlenbach RH, Huttner WB. 2001. Two overlapping reading frames in a single exon encode interacting proteins—a novel way of gene usage. EMBO J. 20:3849–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klemke M, et al. 2000. Characterization of the extra-large G protein alpha-subunit XLalphas. II. Signal transduction properties. J. Biol. Chem. 275:33633–33640 [DOI] [PubMed] [Google Scholar]
- 36. Kublaoui BM, Holder JL, Jr, Gemelli T, Zinn AR. 2006. Sim1 haploinsufficiency impairs melanocortin-mediated anorexia and activation of paraventricular nucleus neurons. Mol. Endocrinol. 20:2483–2492 [DOI] [PubMed] [Google Scholar]
- 37. Lam DD, et al. 2011. Leptin does not directly affect CNS serotonin neurons to influence appetite. Cell Metab. 13:584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu J, et al. 2005. Identification of the control region for tissue-specific imprinting of the stimulatory G protein alpha-subunit. Proc. Natl. Acad. Sci. U. S. A. 102:5513–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu J, Yu S, Litman D, Chen W, Weinstein LS. 2000. Identification of a methylation imprint mark within the mouse Gnas locus. Mol. Cell. Biol. 20:5808–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maffei M, et al. 1995. Leptin levels in human and rodent: measurement of plasma leptin and ob RNA in obese and weight-reduced subjects. Nat. Med. 1:1155–1161 [DOI] [PubMed] [Google Scholar]
- 41. Moore T, Haig D. 1991. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 7:45–49 [DOI] [PubMed] [Google Scholar]
- 42. Oswal A, Yeo GS. 2007. The leptin melanocortin pathway and the control of body weight: lessons from human and murine genetics. Obes. Rev. 8:293–306 [DOI] [PubMed] [Google Scholar]
- 43. Pasolli HA, Huttner WB. 2001. Expression of the extra-large G protein alpha-subunit XLalphas in neuroepithelial cells and young neurons during development of the rat nervous system. Neurosci. Lett. 301:119–122 [DOI] [PubMed] [Google Scholar]
- 44. Pasolli HA, Klemke M, Kehlenbach RH, Wang Y, Huttner WB. 2000. Characterization of the extra-large G protein alpha-subunit XLalphas. I. Tissue distribution and subcellular localization. J. Biol. Chem. 275:33622–33632 [DOI] [PubMed] [Google Scholar]
- 45. Peters J, et al. 2006. Imprinting control within the compact Gnas locus. Cytogenet. Genome Res. 113:194–201 [DOI] [PubMed] [Google Scholar]
- 46. Peters J, Williamson CM. 2007. Control of imprinting at the Gnas cluster. Epigenetics 2:207–213 [DOI] [PubMed] [Google Scholar]
- 47. Peters J, et al. 1999. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc. Natl. Acad. Sci. U. S. A. 96:3830–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Plagge A, et al. 2004. The imprinted signaling protein XL alpha s is required for postnatal adaptation to feeding. Nat. Genet. 36:818–826 [DOI] [PubMed] [Google Scholar]
- 49. Plagge A, Kelsey G, Germain-Lee EL. 2008. Physiological functions of the imprinted Gnas locus and its protein variants Galpha(s) and XLalpha(s) in human and mouse. J. Endocrinol. 196:193–214 [DOI] [PubMed] [Google Scholar]
- 50. Ramírez-Solis R, Davis AC, Bradley A. 1993. Gene targeting in embryonic stem cells. Methods Enzymol. 225:855–878 [DOI] [PubMed] [Google Scholar]
- 51. Sakamoto A, Chen M, Kobayashi T, Kronenberg HM, Weinstein LS. 2005. Chondrocyte-specific knockout of the G protein G(s)alpha leads to epiphyseal and growth plate abnormalities and ectopic chondrocyte formation. J. Bone Miner Res. 20:663–671 [DOI] [PubMed] [Google Scholar]
- 52. Sakamoto A, et al. 2005. Deficiency of the G-protein alpha-subunit G(s)alpha in osteoblasts leads to differential effects on trabecular and cortical bone. J. Biol. Chem. 280:21369–21375 [DOI] [PubMed] [Google Scholar]
- 53. Schmidt-Nielsen K. 1990. Energy metabolism, metabolic rate and body size, p 192–201. In Animal physiology, 4th ed. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 54. Shin JY, Fitzpatrick GV, Higgins MJ. 2008. Two distinct mechanisms of silencing by the KvDMR1 imprinting control region. EMBO J. 27:168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shrestha YB, et al. 2010. Central melanocortin stimulation increases phosphorylated perilipin A and hormone-sensitive lipase in adipose tissues. Am. J. Physiol. Regul. Integr. Comp. Physiol. 299:R140–R149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Skinner JA, Cattanach BM, Peters J. 2002. The imprinted oedematous-small mutation on mouse chromosome 2 identifies new roles for Gnas and Gnasxl in development. Genomics 80:373–375 [DOI] [PubMed] [Google Scholar]
- 57. Sleutels F, Zwart R, Barlow DP. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415:810–813 [DOI] [PubMed] [Google Scholar]
- 58. Storck T, Kruth U, Kolhekar R, Sprengel R, Seeburg PH. 1996. Rapid construction in yeast of complex targeting vectors for gene manipulation in the mouse. Nucleic Acids Res. 24:4594–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weinstein LS. 2006. G(s) alpha mutations in fibrous dysplasia and McCune-Albright syndrome. J. Bone Miner Res. 21(Suppl 2):P120–P124 [DOI] [PubMed] [Google Scholar]
- 60. Weinstein LS, Xie T, Qasem A, Wang J, Chen M. 2010. The role of GNAS and other imprinted genes in the development of obesity. Int. J. Obes. (Lond.) 34:6–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Williamson CM, et al. 2004. A cis-acting control region is required exclusively for the tissue-specific imprinting of Gnas. Nat. Genet. 36:894–899 [DOI] [PubMed] [Google Scholar]
- 62. Williamson CM, et al. 1998. Imprinting of distal mouse chromosome 2 is associated with phenotypic anomalies in utero. Genet. Res. 72:255–265 [DOI] [PubMed] [Google Scholar]
- 63. Williamson CM, et al. 2006. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat. Genet. 38:350–355 [DOI] [PubMed] [Google Scholar]
- 64. Wroe SF, et al. 2000. An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc. Natl. Acad. Sci. U. S. A. 97:3342–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xie T, et al. 2006. The alternative stimulatory G protein alpha-subunit XLalphas is a critical regulator of energy and glucose metabolism and sympathetic nerve activity in adult mice. J. Biol. Chem. 281:18989–18999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yadav VK, et al. 2009. A serotonin-dependent mechanism explains the leptin regulation of bone mass, appetite, and energy expenditure. Cell 138:976–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Youngstrom TG, Bartness TJ. 1995. Catecholaminergic innervation of white adipose tissue in Siberian hamsters. Am. J. Physiol. 268:R744–R751 [DOI] [PubMed] [Google Scholar]
- 68. Yu S, et al. 2000. Paternal versus maternal transmission of a stimulatory G-protein alpha subunit knockout produces opposite effects on energy metabolism. J. Clin. Invest. 105:615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu S, et al. 1998. Variable and tissue-specific hormone resistance in heterotrimeric Gs protein alpha-subunit (Gsalpha) knockout mice is due to tissue-specific imprinting of the gsalpha gene. Proc. Natl. Acad. Sci. U. S. A. 95:8715–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]