Abstract
AU-rich elements (AREs), residing in the 3′ untranslated region (UTR) of many labile mRNAs, are important cis-acting elements that modulate the stability of these mRNAs by collaborating with trans-acting factors such as tristetraprolin (TTP). AREs also regulate translation, but the underlying mechanism is not fully understood. Here we examined the function and mechanism of TTP in ARE-mRNA translation. Through a luciferase-based reporter system, we used knockdown, overexpression, and tethering assays in 293T cells to demonstrate that TTP represses ARE reporter mRNA translation. Polyribosome fractionation experiments showed that TTP shifts target mRNAs to lighter fractions. In murine RAW264.7 macrophages, knocking down TTP produces significantly more tumor necrosis factor alpha (TNF-α) than the control, while the corresponding mRNA level has a marginal change. Furthermore, knockdown of TTP increases the rate of biosynthesis of TNF-α, suggesting that TTP can exert effects at translational levels. Finally, we demonstrate that the general translational repressor RCK may cooperate with TTP to regulate ARE-mRNA translation. Collectively, our studies reveal a novel function of TTP in repressing ARE-mRNA translation and that RCK is a functional partner of TTP in promoting TTP-mediated translational repression.
INTRODUCTION
Posttranscriptional regulation is an important mechanism for controlling gene expression (20, 44), which occurs not only through RNA stability but also through pre-mRNA splicing, polyadenylation, and translation. AU-rich elements (AREs) are key posttranscriptional regulatory elements residing in the 3′ untranslated region (3′ UTR) of many short-lived mRNAs (referred to as ARE-mRNAs). These mRNAs encode many inflammatory factors, cytokines, and oncoproteins (7, 36, 42, 54). AREs have been well characterized as regulators of ARE-mRNA instability (13, 38). Targeted deletion of the ARE in the tumor necrosis factor alpha (TNF-α) mRNA in mice (ΔARE mice) results in overproduction of TNF-α and development of severe inflammatory responses (28). Tristetraprolin (TTP, also known as TIS11 or Zfp36) is an intensively studied ARE-binding protein, which contains two CCCH zinc finger domains required for ARE binding (12, 30, 31). TTP deficiency in mice causes systemic inflammatory syndrome (43). This syndrome results from increased production of TNF-α and granulocyte-macrophage colony-stimulating factor (GM-CSF) (2, 3). TTP destabilizes many ARE-mRNAs in macrophages and HeLa cells (6, 23, 30, 35). In addition to TTP, the ARE-binding proteins AUF1 and KH-type splicing regulatory protein (KSRP) also participate in ARE-mRNA decay (33, 50). In contrast, HuR serves to increase ARE-mRNA stability (52).
In addition to dictating mRNA turnover, AREs are also involved in translation repression. Repression was first observed in Xenopus oocytes (29) by microinjection of polyadenylated reporter ARE-mRNAs containing the 3′ UTR of the genes encoding GM-CSF, interferon, or c-Fos. Subsequently, Han et al. demonstrated that the core TNF-α ARE impairs the burst in production of TNF-α induced by endotoxin at the translation level in macrophages (22). Further studies revealed that the ARE can switch from repression to activation of translation for TNF-α mRNA under serum starvation conditions (46, 47). The ARE-binding protein TIA-1 contributes to translational regulation of ARE-mRNA. For example, macrophages from TIA-1−/− mice produce more TNF-α than those from wild-type mice upon lipopolysaccharide (LPS) stimulation (40). TIA-1 deletion increases association of TNF-α mRNA with polyribosomes, suggesting that TIA-1 functions as a repressor of TNF-α mRNA translation (40). In addition, AUF1 and TIAR control translation of MYC mRNA (32). Recently, the ELAV protein HuD and KSRP were implicated in ARE-mRNA translational regulation (14, 19).
In contrast to our understanding of mRNA stability regulation, much less is understood of the mechanism underlying translational control of ARE-mRNAs. New insights into ARE-mRNA translation control have recently been provided by demonstration of localization of ARE-mRNAs into cellular processing bodies (P bodies) (18). P bodies are cytoplasmic aggregates of messenger ribonucleoproteins (mRNPs) containing proteins involved in translation repression and mRNA degradation, such as the miRISC core components Ago2 and GW182, the general translation inhibitor RCK/P54, decapping-related enzymes, and deadenylases (8, 15, 16). RCK/P54 is a member of the evolutionarily conserved DEAD box helicase family (49). With their ATP-dependent RNA-unwinding activity (49), RCK and its homologs are presumed to remodel mRNPs and are involved in multiple processes of RNA metabolism, including transcription, splicing, transport, storage, and translation (34). RCK is a general translation repressor along with its yeast homolog Dhh1p (11). A recent report showed that RCK interacts with Ago2 in microRNA (miRNA)-mediated pathways and that the interaction is required for translation repression promoted by miRNA but not small interfering RNA (siRNA) (9). Given that ARE-mRNAs localize in P bodies (18), it is possible that P-body components may be involved in ARE-mRNA translational repression. In addition, evidence shows that TTP is crucial for ARE-mRNA accumulation in P bodies (18), suggesting a potential role for TTP in governing ARE-mRNA translation. Previous studies indicated that TTP closely associates with multiple P-body proteins, prompting us to speculate that TTP regulates ARE-mRNA translation through P-body components. Here, we demonstrate that TTP represses ARE-RNA translation. Furthermore, we screened several P-body components and found that RCK promotes ARE-mRNA translational repression mediated by TTP and that RCK's function depends on its helicase activity.
MATERIALS AND METHODS
Plasmid constructs.
Hemagglutinin-tagged TTP (HA-TTP), Myc-Ago2, and HA-septin 1 were developed in our lab previously (23, 41). To construct plasmids encoding Myc-tagged TTP or septin 1, the full-length TTP or septin 1 gene was amplified and inserted between the EcoRI and XhoI sites of pCMV-Myc. The TTP-EGFP (enhanced green fluorescent protein) gene was constructed by inserting a DNA fragment encoding TTP into the XhoI and EcoRI sites of pEGFP-N3. Myc-RCK was a gift from Jens Lykke-Andersen. siRNA-resistant versions were generated by site-directed mutagenesis with plasmid HA-TTP or Myc-RCK as the template, respectively, and the QuikChange mutagenesis kit (Stratagene). Oligonucleotide sequences for mutagenesis will be provided upon request.
To construct luciferase reporter plasmids, the firefly luciferase-coding region was amplified by PCR from pGL3 basic vector (Promega) and inserted between the NotI and XhoI sites of plasmid pcDNA3 to form pcDNA3-FL (FL) and pcDNA3-RL (RL). The human GM-CSF 3′ UTR was amplified by PCR from 293T cell genomic DNA. Its core ARE sequence (46 bp) was then deleted by overlapping PCR. The GM-CSF 3′ UTR or its deletion mutant was cloned downstream of the firefly luciferase coding region between the XhoI and XbaI sites of plasmid pcDNA3-FL to form FL-GM-CSF or FL-GM-CSFΔARE (Fig. 1A). The GM-CSF 3′ UTR was inserted in the antisense direction between the XhoI and XbaI sites of plasmid pcDNA3-FL to form FL-Control.
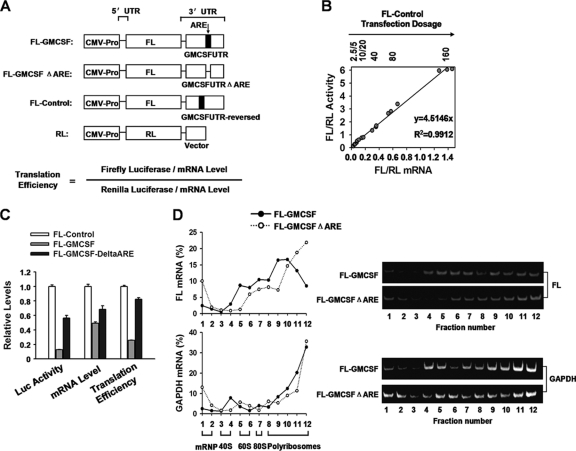
Fig 1.
ARE-mRNA is translationally repressed by the core ARE. (A) Diagram of luciferase reporter plasmids. FL, firefly luciferase; RL, Renilla luciferase; ΔARE, deletion of the AU-rich element. (B) Different dosages (2.5 to 160 ng/well) of FL-Control were transfected into 293T cells, together with 2 ng of RL plasmid. FL mRNA level and enzyme activity were normalized to those of the RL transfection control. Three dosage experiments were performed. The equation of FL/RL activity (y) versus corresponding mRNA level (x) is y = 4.5146x (R2 = 0.9912). Hence, translation efficiency is defined in panel A. (C) The indicated reporter plasmids were cotransfected with the RL plasmid into 293T cells. FL mRNA level, activity, and translation efficiency were normalized to those of the RL transfection control. The relative levels of FL mRNA, activity, and translation efficiency were normalized to those of FL-Control. Mean values and SD from three independent experiments are shown. (D) The core ARE sequence reduced the FL-GM-CSF reporter mRNA loading with polyribosomes. Four micrograms of FL-GM-CSF or FL-GM-CSFΔARE was transfected into 293T cells in 100-mm dishes. Forty-eight hours later, cytoplasmic lysates were prepared and loaded onto a 10 to 50% sucrose gradient as described in Materials and Methods. The RNA in individual fractions was extracted. Percentages of FL and GAPDH mRNA in the sucrose gradient fractions were analyzed by qRT-PCR (left). The relative distribution of FL and GAPDH mRNA was also determined by semiquantitative RT-PCR with the same primer pairs as in qRT-PCR and were visualized by native 10% PAGE (right). Representative data from three independent experiments are shown, with ribosome subunit distributions indicated below the bottom graph (Fig. 2A).
The tethering assay plasmids pCIneo-RL-5BoxB and pCIneo-λN were gifts from Elisa Izaurralde (Max Planck Institute for Developmental Biology). To construct pCIneo-λN-TTP and pCIneo-λN-septin 1, the cDNAs of TTP and septin 1 were amplified from the vectors described above and inserted downstream of λN between the EcoRI and NotI sites of plasmid pCIneo-λN. To construct pCIneo-FL, which served as a transfection control, the firefly luciferase coding region was amplified as described above and ligated into pCIneo-λN, which had been digested with NheI and NotI to remove the fragment encoding the λN tag.
Cell culture and luciferase reporter assay.
For analysis of translation efficiency, transfections of 293T cells were performed in 24-well plates with Lipofectamine 2000 (Invitrogen). Transfection mixtures contained 50 ng of FL reporter plasmid. Plasmids expressing various proteins were cotransfected in specific experiments, as follows: 10 ng of HA/Myc-TTP or HA/Myc-septin 1, 50 ng of Myc-RCK or its mutants, or Myc-septin 1. pcDNA3.0 was added to make a total of 200 ng of plasmid in each experiment; 2 ng of RL was included in all transfection mixes. In all experiments, FL and RL activities were measured 48 h after transfection with the dual-luciferase reporter assay system (Promega). FL activity was normalized to that of RL.
Quantitative real-time RT-PCR (qRT-PCR) for FL, RL, and GAPDH mRNA.
Total RNA was isolated by TRIzol (Invitrogen), and DNA was removed from RNA samples with RNase-free DNase I (TaKaRa). Total RNA was reverse transcribed into cDNA with Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega) and oligo(dT18) according to the manufacturer's instructions. Fluorescence real-time PCR was performed with the double-stranded DNA dye SYBR green real-time PCR Master Mix (ToYoBo) with the ABI Prism 7900 system (Perkin-Elmer, Torrance, CA). The detailed experimental procedure and analyses were as described previously (55). The following primers were used: for FL, 5′-TGAGTACTTCGAAATGTCCGTTC-3′ (forward) and 5′-GTATTCAGCCCATATCGTTTCAT-3′ (reverse); for RL, 5′-GCAGCATATCTTGAACCATTC-3′ (forward) and 5′-TTGTACAACGTCAGGTTTACC-3′ (reverse); for human glyceraldehyde 3-phosphate dehydrogenase (GAPDH), 5′-AGCCACATCGCTCAGACAC-3′ (forward) and 5′-GCCCAATACGACCAAATCC-3′ (reverse); for mouse TNF-α, 5′-TCTTCTCATTCCTGCTTGTGG-3′ (forward) and 5′-GGTCTGGGCCATAGAACTGA-3′ (reverse); and for mouse GAPDH, 5′-CCTTGAGATCAACACGTACCAG-3′ (forward) and 5′-CGCCTGTACACTCCACCAC-3′ (reverse). The qRT-PCR products were visualized by fractionation in 2% agarose gels to ensure correct product size. The FL mRNA level was normalized to that of RL. The equation for translation efficiency is defined in Fig. 1A.
Semiquantitative reverse transcription-PCR.
Total RNA isolation, DNase I treatment, and reverse transcription (RT) were performed as described above. PCRs to amplify FL, RL, and GAPDH cDNA were performed with their specific primers as described above. PCR consisted of 28 to 33 cycles of denaturing at 95°C for 20 s, annealing at 60°C for 20 s, and extension at 72°C for 20 s. Amplification cycles were preceded by a denaturation step (95°C for 5 min) followed by an elongation step (72°C for 10 min). After amplification, PCR products were analyzed on a 10% native polyacrylamide gel.
Transfection of siRNAs.
293T cells were split to a density of 3 × 105/well in 6-well plates 24 h before transfection. The following siRNAs were used: control siRNA, 5′-CUCGUAAUGCAAUGGGUCCTT-3′; Ago2 siRNA, 5′-GCA CGG AAG UCC AUC UGA ATT-3′; GW182 siRNA, 5′-GAA AUG CUC UGG UCC GCU AUU-3′; Lsm1 siRNA, 5′-GUG ACA UCC UGG CCA CCU CAC UU-3′; siRCK, 5′-GCAGAAACCCUAUGAGAUUUU-3′; siTTP-1, 5′-GCGCUACAAGACUGAGCUATT-3′; and siTTP-2, 5′-CGCUGCCACUUCAUCCACAUU-3′. On the following day, each cell sample was incubated for 6 h in a 1-ml transfection mixture containing 2 μl of Lipofectamine 2000 reagent (Invitrogen) and siRNAs at a concentration of 50 nM according to the manufacturer's protocols. Twenty-four hours later, cells were split into 24-well plates (0.9 × 105/well) and further incubated for 24 h. Seventy-five nanomolar siRNA, 200 ng of total plasmids, and 1 μl of Lipofectamine 2000 were mixed in 500 μl Opti-MEM medium in the second transfection. Plasmids included 50 ng of FL reporter plasmid, 10 ng of plasmid encoding HA-tagged TTP or septin 1, and 50 ng of plasmid encoding Myc-RCK or its mutations or septin 1. pcDNA3.0 was added to a total of 200 ng of plasmid in each experiment. Forty-eight hours later, cells were harvested for mRNA and protein analysis.
Macrophage culture, transfection, and ELISA.
RAW264.7 cells (murine macrophage cell line) were obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium (DMEM) containing penicillin (100 U/ml), 100 μg/ml streptomycin, and 10% heat-inactivated fetal calf serum (FCS) at 37°C and 5% CO2. The cells were split to a density of 3 × 105/well in 12-well plates 24 h before transfection. The following siRNAs were used: simRCK, 5′-GCUUUAUCUGGUAGGGAUATT-3′, and simTTP, 5′-UCGGAGGACUUUGGAACAUTT-3′. About 50 μM siRNA and control siRNA were transfected into cells with Oligofectamine (Invitrogen) according to the manufacturer's recommendations. Twenty-four hours after transfection, the medium was replaced with fresh complete medium for an additional 24 h of incubation. The supernatants were collected for cytokine detection by enzyme-linked immunosorbent assay (ELISA), and the cells were subject to TRIzol for mRNA purification and quantification by qRT-PCR. For plasmid transfection, 500 ng of HA-tagged septin 1 or TTP plasmid was transfected into cells in 12-well plates with Fugene HD (Roche) according to the manufacturer's recommendations.
Polyribosome profile analysis.
Polyribosome profile analyses were performed as described previously (32). Briefly, 5 ×107 cells that had been transfected with plasmids as indicated in the figure legends were cultured in medium with 100 μg/ml cycloheximide (CHX) at 37°C for 5 min. Cells were washed twice with cold phosphate-buffered saline (PBS) containing 100 μg/ml cycloheximide and harvested in lysis buffer [20 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.3% (vol/vol) Triton X-100, 100 μg/ml cycloheximide, 1 mM dithiothreitol (DTT), 200 U/ml RNase Out (Invitrogen), 1 mM phenylmethylsulfonyl fluoride (PMSF), complete protease inhibitor mixture (Roche)] on ice for 10 min. Lysed cells were centrifuged at 13,000 rpm for 10 min at 4°C. The protein concentration of the cytoplasmic lysate was measured by a Bradford assay. Approximately 2.5 mg cytoplasmic lysate proteins were layered on top of linear 10%-to-50% (wt/vol) sucrose gradients (11 ml). Tubes were centrifuged in a Beckman SW41 rotor at 40,000 rpm for 3 h at 4°C. Fractions (900 μl) were collected, starting from the top of the gradient. RNA in each fraction was extracted with TRIzol and treated with DNase I (TaKaRa). RNA samples were assayed by qRT-PCR and semiquantitative RT-PCR as described above. For the experiment whose results are shown in Fig. 2, EDTA (20 mM) was added to the cell lysis buffer instead of CHX where indicated. The following primers were used for 18S and 28S rRNA detection by qRT-PCR: 18S, 5′-GCAATTATTCCCCATGAACG-3′ (forward) and 5′-GGGACTTAATCAACGCAAGC-3′ (reverse), and 28S, 5′-TTACCCTACTGATGATGTGTTGTTG-3′ (forward) and 5′-CCTGCGGTTCCTCTCGTA-3′ (reverse).
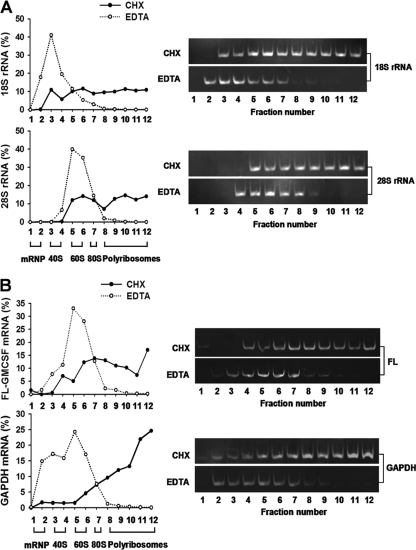
Fig 2.
Polyribosome profile analysis of FL-GM-CSF reporter mRNA. 293T cells were transfected with the FL-GM-CSF reporter plasmid. Two days later, cell lysates were prepared in the presence of cycloheximide (CHX) or EDTA. These were fractionated by sucrose density gradient centrifugation, and RNA was purified from each collected fraction. (A) Distributions of 18S and 28S ribosomal RNAs. Ribosomal RNAs were quantified in each fraction by qRT-PCR (left) or semiquantitative PCR (right) and plotted as a percentage of the total of 18S or 28S rRNA. Based on the distributions of 18S and 28S RNAs, gradient fractions represent the following: 1 and 2, free mRNP; 3 and 4, 40S subunits; 5 and 6, 60S subunits; 7, 80S ribosomes; and 8 to 12, polyribosomes. EDTA shifts the sedimentation of ribosomal subunits to lighter fractions, indicative of disrupted polyribosomes. (B) Polyribosome distribution of FL-GM-CSF reporter mRNA. FL-GM-CSF reporter mRNA and GAPDH mRNA were quantified by qRT-PCR (left) or semiquantitative PCR (right). EDTA shifted the sedimentation of both reporter and GAPDH mRNAs to lighter fractions, indicative of disrupted polyribosomes.
Coimmunoprecipitation (co-IP) and Western blotting.
For co-IP experiments, 293T cells in 6-well plates were transfected with plasmids as follows: 100 ng of Flag-TTP or Myc-TTP, 100 ng of HA-hnRNP-A1, or 200 ng of Myc-Ago2 or Myc-RCK. pcDNA3.0 was added to make a total of 1 μg of plasmid in each experiment. 293T cells expressing the indicated plasmids were lysed 48 h after transfection by incubation for 10 min at 4°C in 600 μl of lysis buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM sodium orthovanadate, 1 mM sodium pyrophosphate, and 1 mM NaF supplemented with a protease inhibitor cocktail [Roche]). Five microliters of RNase A (10 mg/ml) was added to the 600 μl of lysis buffer. Ten microliters of cell lysate was preserved as input, and the remainder was preincubated with protein A/G agarose for 1 h. The supernatant was incubated at 4°C for 4 h with a monoclonal anti-Myc agarose conjugate (Santa Cruz). The beads were washed five times with lysis buffer. For endogenous co-IP, cell lysates of RAW264.7 were prepared in lysis buffer as described above and incubated with 5 μg of RCK-specific antibody (Santa Cruz) for 2 h at 4°C, followed by the addition of protein A-Sepharose beads for another 2 h. Coprecipitated proteins were detected by Western blotting with the indicated antibodies at the following dilutions: horseradish peroxidase (HRP)-conjugated monoclonal anti-Myc antibody at 1:5,000 (Santa Cruz); mouse monoclonal anti-Flag antibody at 1:5,000 (Sigma-Aldrich); mouse monoclonal anti-HA antibody at 1:5,000 (Abmart); mouse monoclonal anti-RCK antibody at 1:1,000 (Santa Cruz); rabbit polyclonal anti-Dcp1a antibody at 1:10,000 (gift from Jens Lykke-Andersen); and goat polyclonal anti-TTP antibody at 1:500 (Santa Cruz). HRP-conjugated goat anti-mouse IgG antibodies at 1:5,000 (Abmart), rabbit anti-goat IgG at 1:5,000 (Abmart), or goat anti-rabbit at 1:5,000 (Abmart) was used as a secondary antibody for detection with a chemiluminescence reagent (peroxide/luminol enhancer; Millipore).
Pulse-labeling cells.
For pulse-labeling, the culture medium of RAW264.7 cells was replaced with methionine-free Dulbecco's modified Eagle medium (DMEM) and incubated for 30 min to deplete intracellular methionine, and then [35S] methionine (100 μCi/ml) was added to the medium. Cells then were cultured for an additional 2 h at 37°C. Then the culture medium was removed, and cells were lysed with radioimmunoprecipitation assay buffer (RIPA) (50 mM Tris [pH 7.5], 150 mM NaCl, 0.5% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]) supplemented with protease inhibitor and sedimented at 12,000 rpm for 10 min. The cell lysates were incubated with 5 μg of rat anti-mouse TNF-α antibody (Santa Cruz) or normal rat IgG at 4°C for 5 h; then 50 μl of protein A-agarose was added to each lysate, and the mixtures were incubated overnight at 4°C with rocking. The protein A-agarose then was washed four times with washing buffer (150 mM triethanolamine [pH 8.0] containing 150 mM NaCl, 5 mM EDTA, and 0.1% SDS), suspended in 30 μl of sample buffer, and heated for 5 min. Immunoprecipitates were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE). After the gels had been dried, autoradiography was performed.
Indirect immunofluorescence (IF).
293T cells in DMEM–10% fetal bovine serum (FBS) at 50% confluence in 6-well plates were transfected using Lipofectamine 2000 according to the manufacturer's protocols (Invitrogen). Cells were divided and applied to chamber slides 24 h later. For indirect immunofluorescence experiments, cells were transfected with 50 ng of TTP-EGFP or 100 ng of Myc-Ago2. pcDNA3.0 was added to make a total of 1 μg of plasmid in each experiment. Forty-eight hours after transfection, cells were fixed in 4% paraformaldehyde for 15 min and permeabilized and blocked with PBS–1% goat serum–0.1% Triton X-100 for 30 min. For indirect immunofluorescence microscopy, all the primary and secondary antibodies were diluted 1:1,000 with 1% (wt/vol) bovine serum albumin (BSA) in PBS. Endogenous Dcp1a was detected using rabbit polyclonal anti-Dcp1a antibody and Alexa Fluor 555-labeled goat anti-rabbit IgG. Myc-Ago2 was detected with rhodamine-labeled mouse anti-Myc antibody (Myc-TRITC; Santa Cruz).
Statistical analysis.
Data are reported as means and standard deviations (SD) from three independent experiments. For luciferase assays, transfection control plasmid RL (or FL) was included and the FL (or RL) activity, mRNA level, and translation efficiency were normalized to that of RL (or FL) in all experiments. The statistical significance of the differences between the two groups was determined using Student's t test. P values of <0.05 were considered significant.
RESULTS
ARE-mRNA is translationally repressed, and the ARE is required for translation inhibition.
We adopted the system described by Vasudevan and Steitz (46) to construct a firefly luciferase reporter containing the 3′ UTR of GM-CSF (FL-GM-CSF) (Fig. 1A). A mutant reporter was also constructed by deleting the core ARE nucleotides (46 nt) from its 3′ UTR (FL-GM-CSFΔARE) (Fig. 1A). The same 3′ UTR (i.e., antisense orientation) of GM-CSF was reversely inserted following the luciferase-coding region as a control (FL-Control in Fig. 1A). For all firefly luciferase assays, a plasmid expressing Renilla luciferase (RL) (Fig. 1A) was cotransfected as an internal control.
We first established a correlation between protein levels of the reporter genes and their cognate mRNA levels. To do this, different amounts of FL-Control plasmid were individually transfected into 293T cells, and then firefly/Renilla luciferase (FL/RL) activities were assayed and mRNA levels were quantified by real-time PCR (qRT-PCR) 48 h later. In order to minimize detection of mRNA fragments resulting from incomplete mRNA processing or degradation, we used oligo(dT) as a primer to initiate reverse transcription in the poly(A) tail, and the primer for real-time PCR of FL was designed near the 5′ end of the mRNA. All RNAs were treated with DNase I before reverse transcription to eliminate residual reporter plasmid DNA that could lead to high background levels in qRT-PCRs. Figure 1B shows that FL/RL activity is strictly proportional to the FL/RL mRNA level in the dosage range (5 to 160 ng/well, in 24-well plates) of reporter plasmid transfected. Therefore, translational efficiency was calculated with the equation in Fig. 1A.
Next, we examined whether ARE-mRNA was translationally repressed in 293T cells. Cells were transfected with reporter FL-GM-CSF or FL-Control, together with RL as an internal control. As shown in Fig. 1C, luciferase activity from FL-GM-CSF declined 8-fold compared to that from FL-Control, while its mRNA level was about 50% that of FL-Control (Fig. 1C). The quantitation of the mRNA level was validated by qRT-PCR with a different set of FL- or RL-specific primers, with similar results (data not shown). With the formula mentioned above, translation efficiency of FL-GM-CSF was calculated as 25% that of FL-Control. This result indicates that the ARE exerts a greater effect on translation inhibition than on mRNA decay with this reporter system.
To validate the function of the core ARE sequence in translation inhibition, the 46-nt core ARE was deleted from the GM-CSF 3′ UTR, and translation efficiency was assessed. The FL-GM-CSFΔARE activity level increased 5-fold but its mRNA level increased only 1.3-fold compared to FL-GM-CSF (Fig. 1C). Therefore, deletion of the ARE increased FL-GM-CSF translation efficiency from 30% to 80% of the control value (Fig. 1C). We note that FL-GM-CSFΔARE still displayed 20% less translation efficiency than FL-Control, indicating that sequences outside the core ARE may make a modest contribution to translation repression. These results show that the core ARE elicits the major effect of the GM-CSF 3′ UTR on translation inhibition.
To further define the effect of the core ARE upon translation regulation, we performed polyribosome profile experiments. 293T cells were transfected with either FL-GM-CSF or FL-GM-CSFΔARE reporter plasmids, and then cytoplasmic lysates were used for polyribosome profile assays. The presence of the ARE significantly shifted the GM-CSF reporter mRNA to the top, lighter fractions, thus reducing the proportion of mRNAs associated with the heavier fractions as determined by qRT-PCR and semiquantitative RT-PCR assay (Fig. 1D, top). In contrast, sedimentation of GAPDH mRNA was unaffected (Fig. 1D, bottom).
To ascertain that FL-GM-CSF mRNA was indeed associated with polyribosomes, we performed control experiments employing EDTA-mediated disruption of polyribosomes. 293T cells were transfected with the FL-GM-CSF reporter plasmid, and after 48 h, cell lysates were prepared in the presence of cycloheximide or EDTA and fractionated by sucrose density gradient centrifugation. Analyses of 18S and 28S ribosomal RNAs permitted estimates of sedimentation positions for free mRNP, 40S/60S subunits, monosomes, and polyribosomes (Fig. 2A). EDTA shifted the sedimentation of ribosomal RNAs to lighter fractions, indicative of disrupted polyribosomes. Approximately 60% of FL-GM-CSF reporter mRNA sedimented with polyribosomes in control extracts (i.e., prepared in cycloheximide; fractions 8 to 12) (Fig. 2B, top). In contrast, EDTA reduced the sedimentation of FL-GM-CSF mRNA in fractions 8 to 12 to less than 5% compared to control extracts; EDTA resulted in a concomitant increase in the reporter mRNA to 65% in fractions 5 to 6. Likewise, EDTA shifted sedimentation of GAPDH mRNA to lighter fractions (Fig. 2B, bottom). We conclude that FL-GM-CSF reporter mRNA sediments with polyribosomes in the fractionation conditions utilized throughout our study. Taken together, the data in Fig. 1 and 2 indicate that the core ARE exerts its effects at the translational level.
TTP is essential for ARE-mRNA translation repression.
TTP binds to a number of AREs (4, 30). To test whether TTP is involved in translational regulation, we silenced the expression of endogenous TTP by transfecting 293T cells with two siRNAs, siTTP-1 and siTTP-2, which target distinct sequences in human TTP mRNA. Transfection of siTTP-1 and siTTP-2 decreased the abundance of TTP protein to about 40% and 20%, respectively, of control siRNA (Fig. 3A). Since siTTP-2 was more efficient than siTTP-1, it was used in subsequent experiments. The GM-CSF reporter mRNA level was unchanged upon transfection of 293T cells with siRNA against TTP (siTTP-2) (Fig. 3B, left). However, reducing TTP expression increased luciferase activity from the GM-CSF reporter 3-fold (Fig. 3B, middle), which elevated FL-GM-CSF translation efficiency from 19% to 58% compared to FL-Control (Fig. 3B, right). Importantly, introduction of the siRNA-resistant form of HA-TTP (HA-TTPr) restored translation inhibition in cells depleted of endogenous TTP (Fig. 3B, right), indicating that the effect of TTP knockdown was specific. To further examine the effect of TTP, we constructed a reporter, FL-TNF, in which the entire human TNF-α (NM_000594.2) The 3′ UTR was inserted downstream of the FL coding sequence. Knockdown of TTP led to increased translation of the FL-TNF reporter. Also, translation repression was restored by reintroduction of HA-TTPr (Fig. 3C). Consistent with results from the GM-CSF reporter, knockdown of TTP mainly increased protein levels from the TNF reporter without affecting mRNA levels (Fig. 3C). Thus, the knockdown experiments provide evidence that TTP represses ARE-mRNA translation.
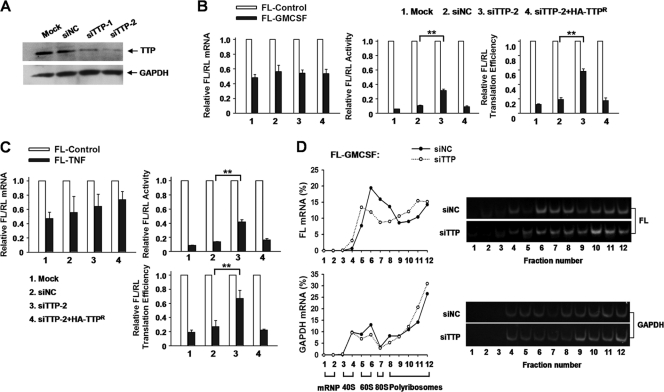
Fig 3.
TTP knockdown increases ARE-mRNA reporter mRNA translation in 293T cells. (A) Knockdown effect of TTP siRNAs. 293T cells were transfected twice with the indicated siRNAs. Endogenous TTP was analyzed by Western blotting with TTP polyclonal antibody. GAPDH served as a loading control. (B) TTP depletion increases FL-GM-CSF reporter mRNA translation efficiency. 293T cells transfected with control siRNA (siNC) or siTTP-2 or left untransfected (Mock) were also transfected with the indicated reporter plasmids and overexpression plasmids. The normalized values of FL mRNA level, activity, and translation efficiency were set to 1 for cells transfected with FL-Control in each knockdown condition. SiTTP-2-resistant, HA-tagged TTP (HA-TTPr) was also included in the transfection mixture where indicated. Means and SD from three independent experiments are shown. **, P < 0.01. (C) Results of an experiment similar to that for panel B, except that the reporter FL-GM-CSF was replaced with FL-TNF. (D) Polyribosome distribution of FL and GAPDH mRNAs in extracts prepared from 293T cells subjected to TTP knockdown. 293T cells were transfected with FL-GM-CSF together with siTTP or siNC. Forty-eight hours later, cytoplasmic lysates were prepared and loaded onto 10 to 50% sucrose gradients. Subsequent procedures and analyses are the same as for Fig. 1D. Representative data from three independent experiments are shown, with ribosome subunit distributions indicated below the bottom graph.
To further substantiate the inhibitory effect of TTP, we examined the polyribosome profiles of the GM-CSF reporter mRNA after silencing TTP in 293T cells. As shown in Fig. 3D, depletion of TTP reduced GM-CSF reporter mRNA accumulation in middensity fractions and increased its distribution in high-density fractions as determined by qRT-PCR and semiquantitative RT-PCR assay (Fig. 3D, top). GAPDH mRNA was unaffected by TTP knockdown (Fig. 3D, bottom). Also, EDTA treatment disrupted polyribosomes and shifted the bulk of GM-CSF reporter mRNA and GAPDH mRNA to lighter gradient fractions (data not shown), indicating that TTP specifically represses target mRNA loading onto polyribosomes.
To determine whether overexpression of TTP could affect ARE-mRNA translation, 293T cells were transfected with various reporters and a plasmid expressing HA-tagged TTP or septin 1 (a cell mitosis-related cytoskeletal protein which served as a control). septin 1 transfection did not affect FL-GM-CSF reporter expression at either the protein or the mRNA level (data not shown). The normalized values of FL activity, mRNA level, and translation efficiency were set to 1 in cells transfected with FL-Control and the septin 1-encoding plasmid. Overexpression of TTP decreased both FL-GM-CSF reporter mRNA levels and activity (Fig. 4A, left and middle). FL-GM-CSF reporter translation efficiency was significantly reduced by overexpression of TTP (Fig. 4A, right). In contrast, in cells transfected with FL-Control or FL-GM-CSFΔARE, the effect of TTP on translation repression was not observed (Fig. 4A, right), indicating that the ARE is required for TTP to inhibit translation. However, overexpression of HA-tagged TTP did not obviously affect the distribution of GM-CSF reporter mRNA on sucrose gradients (Fig. 4B), implying that TTP alone is not sufficient to affect mRNA loading with polyribosomes.
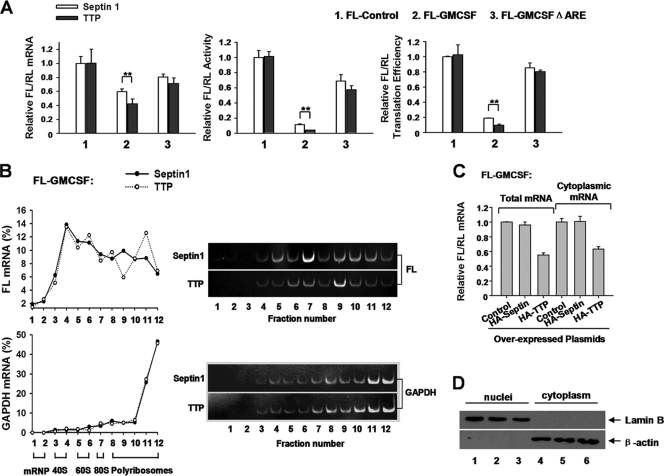
Fig 4.
Overexpression of TTP represses FL-GM-CSF reporter mRNA translation via the ARE. (A) 293T cells were transfected with the indicated reporter plasmids (50 ng/well) and plasmids expressing HA-tagged TTP or septin 1 (10 ng/well). The samples were analyzed as described for Fig. 3B. The normalized values of FL mRNA level, activity, and translation efficiency were set to 1 for cells transfected with FL-Control and the plasmid encoding septin 1. Means and SD from three independent experiments are shown. **, P < 0.01. (B) Relative distributions of FL and GAPDH mRNAs in polyribosome gradients after TTP overexpression. 293T cells in 100-mm dishes were transfected with 4 μg of FL-GM-CSF reporter, together with 2 μg of HA-TTP-expressing plasmid. The plasmid expressing HA-septin 1 served as a control. Forty-eight hours later, cytoplasmic lysates were prepared and loaded onto 10 to 50% sucrose gradients as described for Fig. 1D. Representative data from three independent experiments are shown, with ribosome subunit distributions indicated below the bottom graph. (C and D) TTP does not alter the relative distribution of FL-GM-CSF reporter RNA between the nucleus and cytoplasm. (C) Relative intracellular distributions of FL-GM-CSF reporter mRNA. Cells were transfected with the FL-GM-CSF reporter plasmid and the indicated plasmids for protein overexpression. RL served as an internal control. Nuclear and cytoplasmic fractionation was performed as described previously (51). For cytoplasmic mRNA preparation, 250 μl (of 400 μl total) of the supernatant was added to 750 μl TRIzol LS (Invitrogen). Detailed procedures for RNA extraction and qRT-PCR are described in Materials and Methods. Means and SD from three independent experiments are shown. (D) 293T cells were cotransfected with the FL-GM-CSF reporter plasmid and a plasmid expressing HA-tagged TTP (lanes 3 and 6) or septin 1 (lanes 2 and 5), or the corresponding empty vector (lanes 1 and 4). RL served as an internal control. Nuclear and cytoplasmic fractionation was performed as described previously (51). Five percent of the cytoplasmic protein extracts and the nuclear protein extracts were loaded for Western blotting. Lamin B (Santa Cruz) and β-actin (Abmart) were used as markers indicative of nuclear and cytoplasmic fractions, respectively.
It is possible that the observed effect of TTP on decreased translation efficiency is derived from interference with mRNA trafficking from the nucleus to cytoplasm. To examine this possibility, we determined whether TTP influences the distribution of GM-CSF reporter mRNA in cellular compartments. It is a reasonable assumption that TTP should not influence the export from the nucleus of RL mRNA bearing control sequence (plasmid vector sequence) in its 3′ UTR. If TTP inhibits ARE-mRNA transport, then TTP overexpression should give rise to a decrease in the cytoplasmic fraction of FL-GM-CSF reporter in the total mRNA pool. However, we found a similar reduction in FL-GM-CSF reporter mRNA level in both the total and cytoplasmic RNA pools upon TTP overexpression in comparison with empty vector (control) or septin 1 cotransfection (Fig. 4C). Western blot analyses showed that lamin B was exclusively detected in the nuclear fraction, while β-actin was detected mainly in the cytoplasmic fraction (Fig. 4D), indicating that the nucleus and cytoplasm were effectively separated during fractionation. Thus, we conclude that TTP does not alter the relative distribution of GM-CSF reporter mRNA between nucleus and cytoplasm. Taken together, the TTP knockdown, overexpression, and polyribosome profile experiments suggest that TTP is involved in ARE-mRNA translation repression via the ARE.
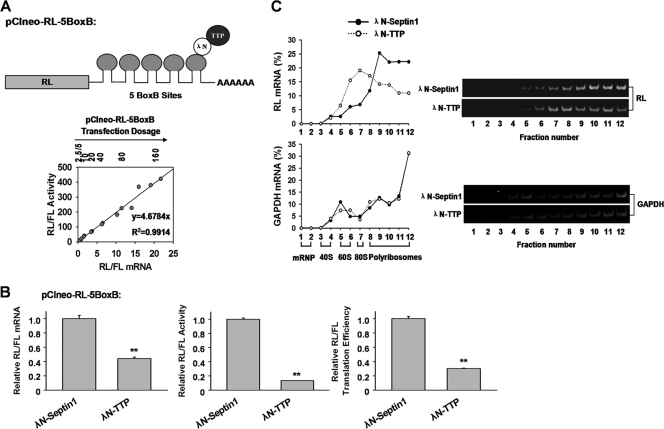
Tethering TTP to mRNA inhibits translation.
The requirement of the ARE suggests that 3′ UTR binding is essential for TTP-mediated translation repression. We asked whether binding of mRNA by TTP is sufficient to repress translation. To this end, a previously described tethering assay was performed (15). The assay involves expression of TTP fused with N-peptide derived from bacteriophage λ (λN-TTP), which binds to five BoxB sites with high affinity in the 3′ UTR of RL mRNA (pCIneo-RL-5BoxB) (Fig. 5A). Again, there was a linear relation between mRNA level and luciferase activity for all tested transfection dosages of RL-5BoxB reporter plasmid (Fig. 5A). As seen in Fig. 5B, the λN-TTP fusion protein reduced translation efficiency of pCIneo-RL-5BoxB by 75% by decreasing mRNA and protein levels 2- and 8-fold, respectively, compared with the λN-septin 1 fusion protein (Fig. 5B). The inhibitory effect of λN-TTP was not observed with plasmid pCIneo-RL, which lacks 5BoxB elements (data not shown), suggesting specificity of the λN-TTP tethering assay. Furthermore, polyribosome profile assays showed that in cells transfected with λN-septin 1, about 70% of RL-5BoxB mRNA was distributed in high-density sucrose fractions (Fig. 5C, left, fractions 10 to 12). Upon λN-TTP expression, only 40% of RL-5BoxB mRNA sedimented in high-density sucrose fractions (Fig. 5C, left), while the remaining 60% of mRNA was primarily distributed in the middensity sucrose fractions (Fig. 5C, left). The distributions of endogenous GAPDH (Fig. 5C, left) remained unchanged, indicating specificity of the observed change in polyribosome distribution. Semiquantitative RT-PCR confirmed that λN-TTP shifted the target RL-5BoxB mRNA, but not GAPDH mRNA, to the middensity sucrose fractions (Fig. 5C, right), similar to results obtained in the presence of the core ARE sequence in the FL-GM-CSF reporter (Fig. 1D). EDTA addition to extracts and sucrose gradients eliminated the RL mRNA shift induced by λN-TTP (data not shown). Thus, the tethering assay provides further evidence that TTP affects target mRNA translation.
Fig 5.
TTP inhibits target mRNA translation in tethering assays. (A) A diagram of the pCIneo-RL-5BoxB reporter containing five BoxB hairpins that bind λN-TTP, and a standard curve for pCIneo-RL-5BoxB reporter expression for mRNA and corresponding protein levels. Different dosages of pCIneo-RL-5BoxB (2.5 to 160 ng/well, 2-fold dilution), together with 50 ng of control pCIneo-FL, were transfected into 293T cells. Plasmid pcDNA3.0 was added to make a total of 200 ng of DNA. Relative RL/FL activity and mRNA levels were determined as above. Three dosage experiments were performed. The equation of RL/FL activity (y) versus the corresponding mRNA level (x) is y = 4.6784x (R2 = 0.9914). (B) Ten nanograms of plasmid expressing λN-TTP or λN-septin 1 was cotransfected with 50 ng of plasmid pCIneo-RL-5BoxB into 293T cells. Plasmid pCIneo-FL (50 ng) was a transfection control. pcDNA3.0 was added to make a total of 200 ng of plasmid in each experiment. RL mRNA level, activity, and translation efficiency were normalized to those of the FL transfection control, respectively. The values were set to 1 for cells transfected with the λN-septin 1 plasmid. Means and SD from three independent experiments are shown. **, P < 0.01. (C) TTP decreases target mRNA loading onto polyribosomes in tethering assays. Five hundred nanograms of plasmid encoding λN-TTP or λN-septin 1 was cotransfected with 2 μg of plasmid pCIneo-RL-5BoxB into 293T cells. Forty-eight hours later, cytoplasmic lysates were prepared and subjected to sucrose density gradient fractionation as in Fig. 1D. Representative data from three independent experiments are shown, with ribosome subunit distributions indicated below the bottom graph.
TTP represses endogenous TNF-α mRNA translation in macrophages.
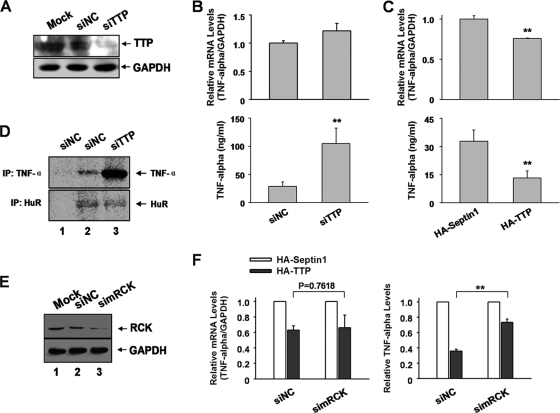
To determine whether TTP affects endogenous ARE-mRNA translation, RAW264.7 macrophages were transfected with murine TTP-specific siRNA (simTTP). Transfection of simTTP decreased the abundance of TTP protein to about 30% of control 2 days after transfection (Fig. 6A). The expression levels of cytokines in the supernatant and their intracellular mRNA levels were detected by ELISA and qRT-PCR, respectively. The GM-CSF level in the supernatant was below the detection limit by ELISA; hence, we focused on TNF-α. As shown in Fig. 6B, the production of TNF-α in the supernatant increased about 3-fold after downregulation of TTP in RAW264.7 cells (Fig. 6B, bottom). In contrast, TTP knockdown slightly increased the corresponding mRNA level about 1.3-fold (Fig. 6B, top). The observation that TTP affects TNF-α expression more strongly at the protein level indicates that TTP has an additional function in translational control besides regulation of mRNA stability.
Fig 6.
TTP represses endogenous TNF-α mRNA translation in murine macrophages. (A) RAW264.7 macrophages seeded in 6-well plates were transfected with murine TTP-specific siRNA and control siRNA at a final concentration 50 μM. Forty-eight hours later, cells were harvested and analyzed for TTP expression by Western blotting. GAPDH served as a loading control. (B) TTP depletion increases TNF-α mRNA translation. Cells seeded in 12-well plates were transfected with TTP-specific siRNA or control siRNA at a final concentration of 50 μM. Forty-eight hours later, supernatants were collected, and the TNF-α level was determined by ELISA. The corresponding mRNA level of TNF-α was determined by qRT-PCR. The relative TNF-α/GAPDH mRNA level was set as 1 in siNC-transfected cells. Means and SD from three independent experiments are shown. **, P < 0.01. (C) Overexpressed TTP represses TNF-α mRNA translation. Cells seeded in 12-well plates were transfected with HA-tagged TTP plasmid. The plasmid expressing HA-septin 1 served as a control. Forty-eight hours later, cells were collected for protein and mRNA analyses as for panel B. Means and SD from three independent experiments are shown. **, P < 0.01. (D) SDS-PAGE of [35S]methionine-labeled proteins immunoprecipitated with anti-mouse TNF-α antibody. RAW264.7 cells transfected with TTP-specific siRNA or control siRNA were labeled for 2 h with [35S]methionine. Cells were lysed and immunoprecipitated as described in Materials and Methods. Lanes 1 and 2, lysates of siNC-treated RAW264.7 cells; lane 3, lysate of siTTP-transfected RAW264.7 cells. (Top) Lane 1, normal rat IgG; lanes 2 and 3, rat anti-mouse TNF-α antibody. (Bottom) Lane 1, normal mouse IgG; lanes 2 and 3, mouse anti-HuR antibody. (E) The effectiveness of siRNA against murine RCK in RAW264.7 cells was analyzed by Western blotting with anti-RCK antibody; GAPDH served as a loading control. (F) RAW264.7 cells transfected with RCK-specific siRNA or control siRNA were subsequently transfected with plasmids encoding HA-tagged septin 1 or HA-TTP. Forty-eight hours later, protein and mRNA levels of TNF-α were analyzed as described for panel B. Means and SD from three independent experiments are shown. **, P < 0.01.
To determine if overexpression of TTP leads to TNF-α translational repression, RAW264.7 cells were transfected with plasmids encoding HA-tagged TTP or HA-septin 1 as a control. Overexpression of TTP decreased TNF-α protein and mRNA levels by 60% and 25%, respectively, as determined by ELISA and qRT-PCR (Fig. 6C). Together, the TTP knockdown and overexpression experiments suggest that TTP affects endogenous TNF-α mRNA translation just as it does the ARE-reporter mRNAs.
To further ascertain the effect of TTP upon TNF-α translation, we performed a pulse-labeling experiment to assess the biosynthesis rate of TNF-α. Newly synthesized proteins in RAW264.7 cells were metabolically radiolabeled with methionine-free DMEM supplemented with [35S]methionine, and cells were lysed shortly thereafter. Cell lysates were immunoprecipitated with TNF-α-specific antibody and analyzed by SDS-PAGE and autoradiography. TNF-α immunoprecipitated with TNF-α-specific antibody (Fig. 6D, lane 2, top) but not with normal IgG (negative control) (Fig. 6D, top, lane 1). Upon TTP knockdown, the TNF-α band density significantly increased (Fig. 6D, top, lane 3), suggesting an increased rate of TNF-α biosynthesis after TTP silencing. In contrast, the band density immunoprecipitated by the anti-HuR antibody remained unchanged after TTP knockdown (Fig. 6D, bottom, compare lane 2 with lane 3). This band was not seen upon immunoprecipitation with normal IgG (negative control) (Fig. 6D, bottom, lane 1). As TTP depletion only modestly affected TNF-α mRNA levels, our data suggest that TTP inhibits endogenous TNF-α translation in macrophages.
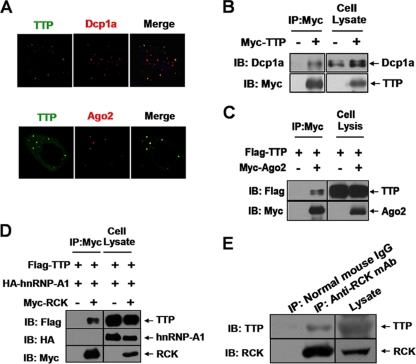
TTP localizes to P bodies and interacts with the DEAD box member RCK/P54.
Previous studies showed that TTP localizes to P bodies and associates with multiple P-body components (5, 17, 23, 25–27, 45). We confirmed that localization of TTP overlaps with P-body markers Dcp1a and Ago2 (a functional partner for TTP in ARE-mRNA decay), suggesting their interactions (Fig. 7A to C). We also cotransfected Flag-tagged TTP and Myc-tagged RCK (a conserved helicase member) and performed immunoprecipitations to determine if they copurify. Indeed, Flag-tagged TTP was specifically detected in a complex with Myc-tagged RCK (Fig. 7D), while the negative control HA-tagged hnRNP-A1 (a general RNA binding protein) was not detected (Fig. 7D). The interaction was further confirmed in the natural cellular context of RAW264.7 cells (Fig. 7E). As RNase A was added to these immunoprecipitation mixtures, an indirect interaction by ARE-RNA can be ruled out. These results indicate that RCK associates with protein complexes containing TTP.
Fig 7.
TTP associates with P bodies. (A) Ectopically expressed TTP colocalizes with Dcp1a and Ago2. 293T cells were transfected with plasmid TTP-EGFP or Myc-Ago2. Immunofluorescence staining was performed with anti-Dcp1a or anti-Myc antibody. (B and C) TTP interacts with multiple P-body components. The expression vectors Myc-TTP (100 ng) (B), Flag-TTP (100 ng) (C), and Myc-Ago2 (200 ng) (C) were cotransfected into 293T cells in the indicated combinations. Cytoplasmic lysates were immunoprecipitated (IP) with anti-Myc agarose beads, followed by immunoblotting (IB) with anti-Flag, anti-Myc, or anti-Dcp1a. Cell lysates were also immunoblotted with anti-Flag, anti-Myc, or anti-Dcp1a to show inputs. (D) Co-IP assay in HEK 293 cells to show that TTP associates with RCK. 293T cells were transfected with the combined plasmids as indicated. Cytoplasmic lysates were immunoprecipitated (IP) with anti-Myc, followed by immunoblotting (IB) with anti-Flag, anti-Myc, or anti-HA. Cell lysates were also immunoblotted with anti-Flag, anti-Myc, or anti-HA to show inputs. (E) Association of endogenous TTP with RCK in RAW264.7 cells. RAW264.7 cells were stimulated with LPS (10 ng/ml) for 2 h and then cytoplasmic lysates were subjected to immunoprecipitation (IP) with anti-RCK antibody or control normal mouse IgG, followed by immunoblotting (IB) with anti-RCK or anti-TTP. Cell lysates were also immunoblotted with anti-RCK or anti-TTP to show inputs.
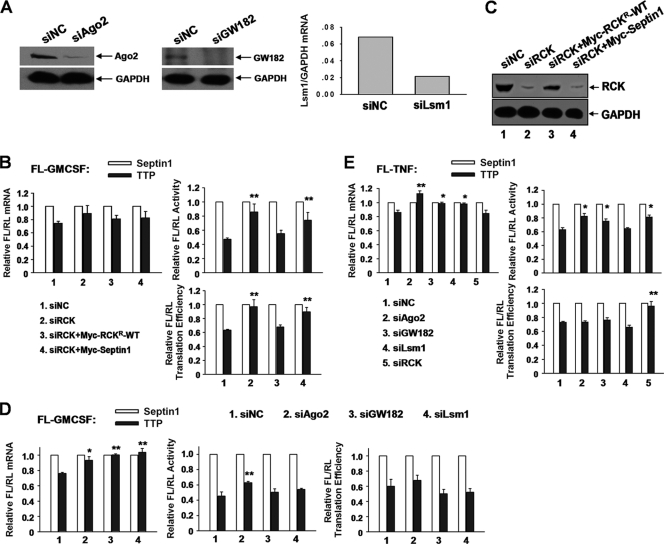
RCK is required for ARE-mRNA translation inhibition mediated by TTP.
We asked whether P bodies are necessary for translational inhibition of ARE-mRNA mediated by TTP, since TTP colocalized with P bodies, as shown earlier. To this end, several P-body proteins that repress translation were screened, including Ago2, GW182, Lsm1, and RCK. To compensate for possible nonspecific effects of knockdown on general protein or mRNA levels, the normalized values of FL activity, mRNA level, and translation efficiency in the presence of Myc-TTP were divided by those obtained in the presence of Myc-septin 1 for each knockdown. Knockdowns of Ago2, GW182, and RCK were analyzed by Western blotting (Fig. 8A and C); knockdown of Lsm1 was analyzed with real-time PCR (Fig. 8A). As expected, overexpression of TTP reduced translation efficiency of FL-GM-CSF reporter mRNA (Fig. 8B and D). Silencing RCK increased relative translation efficiency in the presence of TTP, and this change was reversed by expressing siRNA-resistant RCK (Myc-RCKr-WT) (Fig. 8B). Western blotting showed that the RCK protein level was significantly reduced by siRCK and restored effectively upon adding back of Myc-RCKr-WT (Fig. 8C). RCK depletion increased relative FL/RL activity of the FL-GM-CSF reporter but not the abundance of cognate mRNA (Fig. 8B), implying its potential function in translation repression. In contrast, knockdown of Ago2, GW182, or Lsm1 had little or no influence on FL-GM-CSF reporter translation efficiency mediated by TTP (Fig. 8D). The same assay for the FL-TNF reporter was performed, and similar results were obtained (Fig. 8E). These data provide evidence that RCK is involved in TTP-mediated translational repression of ARE-mRNAs.
Fig 8.
RCK is involved in TTP-mediated translational repression. (A) Knockdown effects of P-body components. The effectiveness of siRNAs against Ago2 and GW182 was analyzed by Western blotting with anti-Ago2 (left) and anti-GW182 (middle). GAPDH served as a loading control. The effectiveness of Lsm1 siRNA was analyzed by qRT-PCR (right). GAPDH mRNA served as an internal control. (B) 293T cells were transfected with the indicated siRNAs and then transfected with reporter FL-GM-CSF, RL, and the indicated plasmids expressing HA-tagged TTP or HA-septin 1. Wild-type RCK resistant to siRCK (Myc-RCKr-WT) was also included in the transfections where indicated. Myc-tagged septin served as a negative control for adding back of Myc-RCKr-WT. Normalized values of FL mRNA level, activity and translation efficiency were set to 1 for cells transfected with the HA-septin 1 plasmid in each knockdown condition. Means and SD from three independent experiments are shown. **, P < 0.01. (C) Effects of RCK silencing and adding back with plasmid Myc-RCKr-WT, as determined by Western blotting with anti-RCK antibody. GAPDH served as a loading control. (D) An experiment similar to that whose results are shown in panel C was done to screen P-body components that are involved in TTP-mediated translational repression. The siRNAs used are indicated. Means and SD from three independent experiments are shown. *, P < 0.05; **, P < 0.01. (E) An experiment similar to that whose results are shown in panel D was done to screen P-body components that are involved in TTP-mediated FL-TNF reporter mRNA translational repression. Means and SD from three independent experiments are shown. *, P < 0.05; **, P < 0.01.
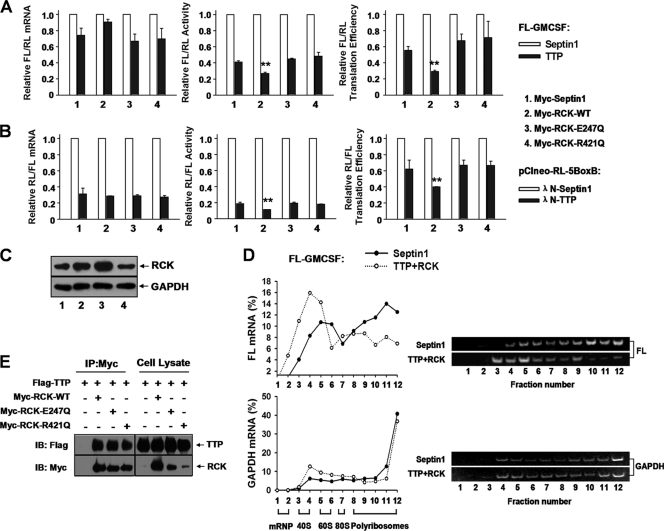
Next, we examined whether overexpressed RCK reinforces the repression effect of TTP upon ARE reporters. Without RCK coexpression, TTP decreased translation efficiency of the FL-GM-CSF reporter to 60% compared to control HA-septin 1 (Fig. 9A, right, first two bars). When RCK was coexpressed, TTP decreased FL-GM-CSF reporter translation efficiency to 30% in contrast to septin 1 (Fig. 9A, right, second black bar). The presence of RCK mainly decreased the FL-GM-CSF reporter protein level (Fig. 9A, middle) with a marginal increase in mRNA level (Fig. 9A, left) mediated by TTP. This effect was also observed with pCIneo-RL-5BoxB (Fig. 9B) and the FL-TNF reporter (data not shown). Western blots showed that RCK protein levels increased about 2-fold upon Myc-tagged RCK transfection (Fig. 9C, compare lane 2 with lane 1, top). Polyribosome profiles showed that coexpression of both TTP and RCK significantly shifted FL-GM-CSF reporter mRNA, but not GAPDH mRNA, toward the top, lighter fractions of the gradients (Fig. 9D), and the effect on the FL-GM-CSF reporter mRNA could be eliminated by addition of EDTA to the sucrose gradients (data not shown). Since overexpression of either TTP or RCK alone did not significantly alter the polyribosome profiles of the FL-GM-CSF reporter mRNA (Fig. 4B and 10A), RCK must act in concert with TTP to repress ARE-mRNA translation.
Fig 9.
RCK promotes TTP-mediated translation repression and is dependent on its helicase activity. (A) Wild-type RCK, but not its mutants RCKE247Q and RCKR421Q, acts together with TTP to repress FL-GM-CSF reporter translation. 293T cells were transfected with the FL-GM-CSF reporter and RL plasmids, together with two plasmids, one expressing either HA-tagged TTP or HA-septin 1 and another expressing either Myc-tagged RCK or its mutated forms, as indicated. The relative values of FL activity, mRNA level, and translation efficiency were set to 1 for cells transfected with the plasmid expressing HA-tagged septin 1 in each condition. Means and SD from three independent experiments are shown. **, P < 0.01. (B) Wild-type RCK promotes TTP-mediated translational repression in tethering assays, but its mutants RCKE247Q and RCKR421Q do not. 293T cells in 24-well plates were transfected with 40 ng of pCIneo-RL-5BoxB and 50 ng of FL-Control, together with two plasmids, one expressing either λN-TTP or septin 1 (10 ng/well) and another expressing either Myc-tagged RCK or its mutated forms (50 ng/well), as indicated. The relative values of RL activity, mRNA level, and translation efficiency were set to 1 for cells transfected with the λN-septin 1 plasmid in each condition. Means and SD from three independent experiments are shown. **, P < 0.01. (C) Expression levels of RCK were analyzed by Western blotting using anti-RCK antibody. GAPDH served as a loading control. Results show that transfection with plasmids expressing Myc-tagged RCK-WT (lane 2) or its mutants RCKE247Q (lane 3) and RCKR421Q (lane 4) increased RCK protein level about 2-fold compared with control septin 1 transfection (lane 1). (D) Accumulation of FL-GM-CSF reporter mRNA in polyribosomes is significantly reduced upon cotransfection of TTP and RCK. 293T cells in 100-mm dishes were transfected with 4 μg of FL-GM-CSF reporter, together with plasmids expressing HA-TTP (2 μg) and Myc-RCK (2 μg). The HA-septin 1 plasmid served as a control. Forty-eight hours later, cytoplasmic lysates were prepared and loaded onto 10-to-50% sucrose gradients as described for Fig. 1D. Representative data from three independent experiments are shown, with ribosome subunit distributions indicated below the bottom graph. (E) The helicase domain mutant of RCK does not affect its association with TTP. The indicated plasmids were transfected into 293T cells, and cytoplasmic lysates were immunoprecipitated with anti-Myc antibody immobilized on agarose beads. Inputs and immunoprecipitates were analyzed by Western blotting with anti-Myc or anti-Flag antibody.
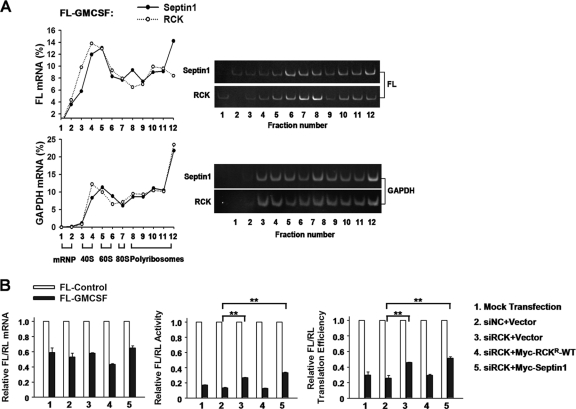
Fig 10.
RCK represses FL-GM-CSF reporter mRNA translation. (A) RCK overexpression alone does not affect FL-GM-CSF reporter mRNA distribution in polyribosomes. 293T cells in 100-mm dishes were transfected with 4 μg of FL-GM-CSF reporter, together with 2 μg of the plasmid expressing Myc-RCK. The septin 1 plasmid served as a control. Forty-eight hours later, cytoplasmic lysates were prepared and loaded onto 10-to-50% sucrose gradients as described for Fig. 1D. Representative data from three independent experiments are shown, with ribosome subunit distributions indicated below the bottom graph. (B) 293T cells were transfected with the indicated siRNAs and then transfected with the reporter FL-Control or FL-GM-CSF, together with plasmids expressing Myc-RCKr-WT (the wild-type Myc-tagged RCK which is resistant to siRCK) or Myc-septin 1 or the corresponding empty vector. The normalized values of FL mRNA level, activity, and translation efficiency were set to 1 for cells transfected with FL-Control in each knockdown condition. Means and standard errors from three independent experiments are shown. **, P < 0.01.
To examine the contribution of RCK to repression of ARE-mRNA translation, we silenced RCK in 293T cells and then evaluated the relative translation efficiency of ARE reporters. RCK knockdown elevated relative translation efficiency of the FL-GM-CSF reporter mRNA about 1.5-fold without significantly affecting mRNA level (Fig. 10B). This phenotype was reversed by addition of Myc-RCKr-WT (Fig. 10B). The same effect was found with the TNF reporter (data not shown). By comparison, knockdown of TTP resulted in a 3-fold increase in reporter translation efficiency (Fig. 3B). Thus, RCK contributes to repression of ARE-mRNA translation, but the magnitude of repression is smaller than that conferred by TTP.
To determine whether RCK affects translation of endogenous TNF-α mRNA, we transfected RAW264.7 cells with siRNA against RCK and then examined the effects of overexpressed TTP upon TNF-α expression at both protein and mRNA levels. As shown in Fig. 6E, RCK protein level was reduced by RCK-specific siRNA (simRCK). TTP expression led to reduced TNF-α expression, with a stronger effect at the protein level than at the mRNA level (Fig. 6F, siNC). However, when RCK was depleted, the change in protein and mRNA levels of TNF-α were comparable (Fig. 6F), suggesting that RCK depletion abolished the effect of TTP upon TNF-α translation. Thus, our results indicate that RCK is involved in the translational regulation of endogenous TNF-α mRNA mediated by TTP.
Next, we wished to explore whether the helicase activity of RCK is involved in repression of ARE-mRNA translation. RCK is a DEAD box helicase family member. The conserved domains DEAD and HRIGQ are required for its helicase function. The DQAD mutation (in the DEAD domain, RCKE247Q) and the HRIGQ mutation (in the HRIGR domain, RCKR421Q) each abolish its helicase activity (37). As shown in Fig. 9A, neither RCKE247Q nor RCKR421Q could promote TTP-dependent translational suppression. This result was confirmed with the pCIneo-RL-5BoxB reporter (Fig. 9B). We note that the Myc-tagged RCK mutant proteins were expressed effectively (Fig. 9C). To determine whether these mutations diminish interaction between TTP and RCK, we performed coimmunoprecipitation assays. TTP was similarly detected in the complex of immunoprecipitated Myc-RCK-WT, RCKE247Q, and RCKR421Q (Fig. 9E). Thus, the helicase domain mutations did not appear to affect RCK interaction with protein complexes containing TTP. Together, these data indicate that RCK promotes TTP-mediated translation repression in a helicase-dependent fashion.
DISCUSSION
We have identified TTP as a novel translational repressor of ARE-mRNAs. Besides the evidence obtained from knockdown experiments and tethering assays, the polyribosome profile experiments strongly support the notion that TTP inhibits ARE-mRNA translation, possibly through interfering with the loading of polyribosomes on mRNAs. Furthermore, the conserved helicase RCK acts to facilitate translational inhibition directed by TTP, and it achieves this effect by its helicase activity. These results provide novel insights into the unresolved issue of molecular mechanisms by which AREs trigger a translational block.
TTP represses ARE-mRNA translation.
The mechanism by which AREs repress translation is not well understood. A previous study showed that macrophages from TTP-deficient mice secreted approximately 5-fold more TNF-α, accompanied by a 2-fold increase in cellular levels of TNF-α mRNA, than wild-type macrophages (2). This result pointed to a potential function of TTP in TNF-α mRNA translational silencing. Likewise, in the present work with the luciferase-based system, we determined that TTP represses ARE reporter mRNA translation (Fig. 3B, 3C, 4A, and 5B). Additionally, our results suggest that TTP serves as a translational repressor of TNF-α mRNA in macrophages with assays utilizing knockdowns, overexpression, and pulse-labeling (Fig. 6B, C, and D). Furthermore, sucrose density gradient fractionation assays demonstrated TTP decreases target mRNA association with polyribosomes (Fig. 3D and 5C), confirming its function in translation. We noticed that TTP exhibited stronger translation repression in tethering assays than ARE 3′ UTR-containing reporters (compare Fig. 5B with Fig. 4A and 8E) and that TTP more strongly shifts the target mRNA to the top, lighter gradient fractions in tethering assays by polyribosome profile analyses (compare Fig. 5C with Fig. 3D). This may be due to the synergistic effect of multiple binding sites for λN-TTP in the tethering assay. This suggests that the affinity of TTP for target mRNAs might determine the magnitude of translational repression by TTP. It is also possible that TTP oligomers formed between mRNAs or on an mRNA molecule is a prerequisite to repress ARE-mRNA translation. Indeed, by UV cross-linking assay, we have observed that TTP effectively binds the GM-CSF ARE (46-nt) probe at 42 kDa (the molecular mass of TTP is 42 kDa) when incubating in vitro-transcribed GM-CSF-ARE-probe with a cytoplasmic lysate of 293T cells in which TTP was overexpressed (data not shown). However, a large complex was simultaneously observed upon addition of TTP (data not shown). We propose that the large complex may be the mRNP organized by TTP and the GM-CSF ARE. This mRNP might contribute to translational silencing by sequestrating RNA away from the translation machinery. Nonetheless, it remains to be determined whether TTP oligomerization is essential for formation of this putative mRNP.
We also noticed that TTP silencing predominantly increases protein abundance from the target gene (Fig. 3B, 3C, and 6B). These results suggest that TTP mainly acts to repress target mRNA translation rather than degrade mRNAs, at least in the systems utilized here. TTP deficiency in mice results in a systemic inflammatory syndrome, and this syndrome results from increased production of TNF-α and GM-CSF (2, 3). Translational repression of TNF-α exerted by TTP might be an essential mechanism for controlling cytokine expression, thereby likely contributing to maintaining normal physiological function.
Depletion of TTP did not obviously influence mRNA levels of the target gene. This was not unexpected. We considered the possibility that a relatively low TTP protein level is not sufficient to effectively exert its mRNA destabilizing activity. TTP is a low-abundance protein in mouse tissues (1). Indeed, the TTP mRNA level is 200-fold less than the GAPDH mRNA level in 293T cells, as determined by qRT-PCR (data not shown). Previous reports showed that TTP is involved in degradation of ARE-mRNAs. However, this evidence is largely based on TTP overexpression (3, 10, 35). For example, Carballo et al. (3) demonstrated that TTP deficiency resulted in increased stability of GM-CSF mRNA in bone marrow stromal cells (BMSCs) when BMSCs were stimulated with LPS for 4 h. This condition induced overproduction of TTP (1). Thus, its mRNA degradation activity could be easily seen in wild-type BMSCs. Nevertheless, there was no evidence that low basal levels of TTP influence decay of ARE-mRNAs. Combined with our results here, we believe that TTP protein level is proportional to its destabilizing activity. Thus, our results suggest that TTP mainly serves as a translational repressor in unstimulated conditions. This may be a fundamental mechanism for limiting ARE-mRNA expression in cells such as macrophages.
RCK is essential for TTP-mediated translational repression.
Through an RNAi-based screen, we determined that RCK is required for TTP-mediated translation repression. Depleting RCK relieved TTP-dependent repression of the FL-GM-CSF and FL-TNF reporters and endogenous TNF-α mRNA (Fig. 6F, 8B, and 8E). Conversely, overexpression of RCK further enhanced translation repression directed by TTP (Fig. 9A and B). Previous studies showed that RCK is a general translational repressor. Here, we present evidence that RCK is specifically involved in TTP-mediated translation repression as well. Furthermore, overexpression of RCK greatly reduced polyribosome loading of FL-GM-CSF reporter mRNA induced by TTP (Fig. 9D), suggesting that RCK is an important partner for TTP to interfere with loading of the translation machinery on mRNAs. Interactions with translation-regulatory factors located at the 5′ and 3′ ends of mRNAs are considered a mechanism by which RCK triggers large mRNP complex formation and subsequent translation repression (21, 37, 39). The translation initiation factor eIF4E and poly(A)-binding protein (PABP) are necessary for effective translation initiation through circularization of mRNA (48). TTP and PABP are found in the same complexes (24). We consider the possibility that interactions of TTP and PABP, as well as RCK and eIF4E, will block assembly of translation initiation complexes, thus sequestering mRNA away from the translation machinery. As mentioned above, a putative large mRNP complex formed in vitro by incubation of TTP and GM-CSF ARE-RNA was diminished when either RCKE247Q or RCKR421Q was added as well (data not shown). Based on the result that either of the two helicase domain mutants impaired RCK's function in promoting TTP-mediated translational repression (Fig. 9A and B), we speculate that the large RNP complex represents translation silencing complexes formed by ARE-mRNA, TTP, RCK, and possibly translation regulatory factors. Indeed, we detected GM-CSF reporter mRNAs associated with RCK, as determined by mRNP-immunoprecipitation and qRT-PCR (data not shown). Association was impaired by mutation of the HRIGQ domain (RCKR421Q) (data not shown), consistent with a report that the HRIGQ domain is essential for RNA binding (37). However, the 2-fold enrichment in mRNP with wild-type RCK (compared with control septin 1) (data not shown) suggests to us that RCK may not directly bind to ARE-mRNA but rather requires TTP, which in turn recruits translation regulatory factors. Alternatively, the helicase activity of RCK may act to unwind ARE-mRNA, which in turn could unmask the ARE, exposing it to ARE-binding proteins. We favor the first possibility because of the stronger translational repression activity of TTP (Fig. 3B) than RCK (Fig. 10B). Helicase activity may promote mRNA remodeling to subsequently allow mRNP formation. As such, translation repression promoted by the concerted actions of TTP and RCK might be through inhibition of translation initiation together with subsequent formation of translationally silenced mRNP complexes (49).
Additionally, our data show that the miRISC subunit Ago2 is involved in ARE-mRNA instability mediated by TTP (Fig. 8D and E), which is consistent with a previous study (23). Knockdown of GW182 also partially increased ARE reporter mRNA levels (Fig. 8D and E). In contrast, depletion of RCK did not significantly affect ARE reporter mRNA levels (Fig. 8B and E), suggesting its specific function in translation repression. Thus, it seems there are two independent pathways for TTP to promote ARE-mRNA degradation and translation repression by cooperating with miRISC members (Ago2 and GW182) and RCK, respectively. Finally, the integrity of P bodies is not likely required for TTP-mediated translation repression: GW182 and Lsm1 are required for P-body integrity (9, 53), but depletion of either protein did not reverse translational repression of ARE reporter mRNA mediated by TTP (Fig. 8D and E). Hence, delivery of ARE-mRNAs to P bodies may be a consequence of TTP-mediated translational repression rather than the cause.
In summary, we have discovered a novel function for TTP as a translational repressor of ARE-mRNAs by acting in collaboration with RCK. These results provide an increasingly clear picture of how AREs trigger translation inhibition. Future work will be required to fully understand the mechanisms in detail.
ACKNOWLEDGMENTS
We are grateful to Jens Lykke-Andersen (University of California, San Diego) for providing anti-Dcp1a antibody, Elisa Izaurralde (Max Planck Institute for Developmental Biology, Tübingen, Germany) and Witold Filipowicz (Friedrich Miescher Institute, Basel, Switzerland) for providing plasmids for tethering assays, and Gunter Meister (University of Regensburg, Germany) for providing anti-Ago2 antibody. We thank Mingjuan Shen for reviewing the manuscript. We thank all members of the laboratory for helpful discussions and comments on the manuscript.
This work was supported in part by the National Natural Science Foundation of China (no. 81130005 and 30828006), Ministry of Science and Technology of China (no. 2010CB945600, 2011CB811304, and 2007CB947002), Chinese Academy of Sciences (grants XDA01040306, KSCX2-YW-R-233, and KSCX2-YW-R-096), and Shanghai Pujiang Program (grant 05PJ14105). G.B. was supported by NIH grant CA052443 and a visiting professorship at the Institute of Health Sciences, Shanghai Institutes of Biological Sciences, Chinese Academy of Sciences.
Footnotes
Published ahead of print 27 December 2011
REFERENCES
- 1. Cao H, Tuttle JS, Blackshear PJ. 2004. Immunological characterization of tristetraprolin as a low abundance, inducible, stable cytosolic protein. J. Biol. Chem. 279:21489–21499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carballo E, Gilkeson GS, Blackshear PJ. 1997. Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 (-/-) mice. Evidence that monocyte/macrophage progenitors may be responsible for TNFα overproduction. J. Clin. Invest. 100:986–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carballo E, Lai WS, Blackshear PJ. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95:1891–1899 [PubMed] [Google Scholar]
- 4. Carballo E, Lai WS, Blackshear PJ. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001–1005 [DOI] [PubMed] [Google Scholar]
- 5. Chang WL, Tarn WY. 2009. A role for transportin in deposition of TTP to cytoplasmic RNA granules and mRNA decay. Nucleic Acids Res. 37:6600–6612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen CY, et al. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451–464 [DOI] [PubMed] [Google Scholar]
- 7. Chen CY, Shyu AB. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465–470 [DOI] [PubMed] [Google Scholar]
- 8. Chen CY, Zheng D, Xia Z, Shyu AB. 2009. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 16:1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chu CY, Rana TM. 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 4:e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. 2011. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol. Cell. Biol. 31:256–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coller J, Parker R. 2005. General translational repression by activators of mRNA decapping. Cell 122:875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Datta S, et al. 2008. Tristetraprolin regulates CXCL1 (KC) mRNA stability. J. Immunol. 180:2545–2552 [DOI] [PubMed] [Google Scholar]
- 13. Dean JL, et al. 2001. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol. Cell. Biol. 21:721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dhamija S, et al. 2011. Interleukin-1 activates synthesis of interleukin-6 by interfering with a KH-type splicing regulatory protein (KSRP)-dependent translational silencing mechanism. J. Biol. Chem. 286:33279–33288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eulalio A, Huntzinger E, Izaurralde E. 2008. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 15:346–353 [DOI] [PubMed] [Google Scholar]
- 16. Fabian MR, et al. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell 35:868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20:905–915 [DOI] [PubMed] [Google Scholar]
- 18. Franks TM, Lykke-Andersen J. 2007. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 21:719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fukao A, et al. 2009. The ELAV protein HuD stimulates cap-dependent translation in a poly(A)- and eIF4A-dependent manner. Mol. Cell 36:1007–1017 [DOI] [PubMed] [Google Scholar]
- 20. Gray NK, Wickens M. 1998. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 14:399–458 [DOI] [PubMed] [Google Scholar]
- 21. Groisman I, et al. 2000. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell 103:435–447 [DOI] [PubMed] [Google Scholar]
- 22. Han J, Brown T, Beutler B. 1990. Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level. J. Exp. Med. 171:465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jing Q, et al. 2005. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120:623–634 [DOI] [PubMed] [Google Scholar]
- 24. Kedar VP, Darby MK, Williams JG, Blackshear PJ. 2010. Phosphorylation of human tristetraprolin in response to its interaction with the Cbl interacting protein CIN85. PLoS One 5:e9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kedersha N, Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81 [DOI] [PubMed] [Google Scholar]
- 26. Kedersha N, et al. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kedersha N, Tisdale S, Hickman T, Anderson P. 2008. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 448:521–552 [DOI] [PubMed] [Google Scholar]
- 28. Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10:387–398 [DOI] [PubMed] [Google Scholar]
- 29. Kruys V, Marinx O, Shaw G, Deschamps J, Huez G. 1989. Translational blockade imposed by cytokine-derived UA-rich sequences. Science 245:852–855 [DOI] [PubMed] [Google Scholar]
- 30. Lai WS, et al. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19:4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. 2006. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol. Cell. Biol. 26:9196–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liao B, Hu Y, Brewer G. 2007. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 14:511–518 [DOI] [PubMed] [Google Scholar]
- 33. Lin WJ, et al. 2011. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Mol. Cell. Biol. 31:3196–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Linder P. 2006. Dead-box proteins: a family affair–active and passive players in RNP-remodeling. Nucleic Acids Res. 34:4168–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lykke-Andersen J, Wagner E. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19:351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M. 2006. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 26:2716–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Minshall N, Thom G, Standart N. 2001. A conserved role of a DEAD box helicase in mRNA masking. RNA 7:1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mukherjee D, et al. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21:165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nakahata S, et al. 2001. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 276:20945–20953 [DOI] [PubMed] [Google Scholar]
- 40. Piecyk M, et al. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19:4154–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qi M, et al. 2005. septin 1, a new interaction partner for human serine/threonine kinase aurora-B. Biochem. Biophys. Res. Commun. 336:994–1000 [DOI] [PubMed] [Google Scholar]
- 42. Shim J, Karin M. 2002. The control of mRNA stability in response to extracellular stimuli. Mol. Cells 14:323–331 [PubMed] [Google Scholar]
- 43. Taylor GA, et al. 1996. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4:445–454 [DOI] [PubMed] [Google Scholar]
- 44. Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20:515–524 [DOI] [PubMed] [Google Scholar]
- 45. van Dijk E, et al. 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21:6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vasudevan S, Steitz JA. 2007. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128:1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vasudevan S, Tong Y, Steitz JA. 2007. Switching from repression to activation: microRNAs can up-regulate translation. Science 318:1931–1934 [DOI] [PubMed] [Google Scholar]
- 48. Wells SE, Hillner PE, Vale RD, Sachs AB. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2:135–140 [DOI] [PubMed] [Google Scholar]
- 49. Weston A, Sommerville J. 2006. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 34:3082–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wilson GM, et al. 2003. Regulation of A + U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J. Biol. Chem. 278:33029–33038 [DOI] [PubMed] [Google Scholar]
- 51. Wu L, Belasco JG. 2008. Examining the influence of microRNAs on translation efficiency and on mRNA deadenylation and decay. Methods Enzymol. 449:373–393 [DOI] [PubMed] [Google Scholar]
- 52. Xu YZ, Di Marco S, Gallouzi I, Rola-Pleszczynski M, Radzioch D. 2005. RNA-binding protein HuR is required for stabilization of SLC11A1 mRNA and SLC11A1 protein expression. Mol. Cell. Biol. 25:8139–8149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang Z, et al. 2004. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 117:5567–5578 [DOI] [PubMed] [Google Scholar]
- 54. Zhang T, Kruys V, Huez G, Gueydan C. 2002. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem. Soc. Trans. 30:952–958 [DOI] [PubMed] [Google Scholar]
- 55. Zhao KW, et al. 2004. Protein kinase Cδ mediates retinoic acid and phorbol myristate acetate-induced phospholipid scramblase 1 gene expression: its role in leukemic cell differentiation. Blood 104:3731–3738 [DOI] [PubMed] [Google Scholar]