Abstract
Previously we demonstrated that the heat shock transcription factor Hsf1 is indispensable for transformation of mammary epithelial cells by the Her2 oncogene. Since Hsf1 affects oncogene-induced senescence (OIS), these findings suggest that Hsf1 affects tumor initiation when OIS plays a role. Indeed, here we report that Hsf1 knockout suppressed mammary hyperplasia in Her2-expressing mice and reduced tumor emergence. On the other hand, Hsf1 expression increases with advanced breast cancer, indicating that there is an additional role of Hsf1 in tumor progression. We studied rare tumors that developed in Hsf1-knockout mice and found that these tumors grew slower than tumors in control animals and showed suppressed angiogenesis. Similarly, in the xenograft model, knockdown of Hsf1 suppressed angiogenesis, which was associated with suppression of the HIF-1 pathway. Suppression of HIF-1 was at the level of translation due to downregulation of the RNA-binding protein HuR. Importantly, besides HIF-1, HuR controls translation of other major regulators of cancer progression, many of which were suppressed in Hsf1-knockdown cells. Therefore, in addition to OIS, Hsf1 regulates the HuR–HIF-1 pathway, thus affecting both cancer initiation and progression.
INTRODUCTION
The heat shock transcription factor 1 (Hsf1) is the major regulator of the heat shock response (33) that is involved in protection of cells and organisms from heat, ischemia, inflammation, oxidative stress, and some other noxious conditions. Among proteins activated by Hsf1, the major defensive function is attributed to heat shock proteins (Hsps) such as Hsp70, Hsp90, and Hsp27, though besides Hsps, Hsf1 regulates hundreds of other targets (32). Initially, Hsps were described as molecular chaperones that prevent and repair protein damage, but later it became clear that their functions go beyond protein folding or degradation, since Hsps also play a distinct and essential role in cell signaling, for example, in suppression of apoptotic pathways or mitogen-activated protein (MAP) signaling cascades (4, 15).
Numerous studies indicate that Hsf1 and Hsps are overexpressed in a variety of human tumors (11). Furthermore, it was recently demonstrated that Hsf1 is critical for tumorigenesis by certain oncogenes. Indeed, it was demonstrated that Hsf1 knockout (KO) suppresses lymphoma development in p53-knockout mice (30). Furthermore, hsf1 knockout dramatically delayed overall development of tumors and increased survival of p53-knock-in mutant (R172H) mice (13). Similarly, Hsf1 deficiency drastically postponed RAS-induced chemical skin carcinogenesis and increased survival of mice from 30% to 90% (13).
We have recently demonstrated that the major Hsf1 target Hsp72 (HSPA1, or inducible Hsp70) plays an essential role in Her2 (NeuT)-induced tumorigenesis in mice (29). This mouse tumor model is widely used to study mechanisms of human breast cancer since it recapitulates initiation and progression of Her2-positive breast cancer, occurring in 15 to 25% of patients. Interestingly, activation of Her2 signaling by heregulin or Her2 overexpression can activate Hsf1 (22, 28).
Importantly, Hsf1 and Hsp72 are essential not only for initial transformation but also for maintaining viability and growth of a variety of fully transformed cells (13, 28, 29). On the other hand, Hsf1 and Hsp72 are dispensable for viability and growth of nontransformed human cells and whole animals (13, 20, 28, 29). The specific dependence of cancer cells on Hsf1 and chaperones was named “nononcogene addiction” (35).
In searching for the mechanism of tumor addiction to Hsf1 and Hsp72, we have found that Hsf1 and Hsp72 play an important role in evasion of oncogene-induced senescence (OIS) (16, 28, 29), which is critical for early stages of neoplastic transformation. Cellular senescence was originally described as a limit to the number of divisions that a normal cell can undergo. Senescence can be triggered by telomere shortening or stressful treatments and is associated with activation of p53 and accumulation of the cell cycle inhibitors p21 and/or p16 (5). Although tumor cells are immortal and divide indefinitely, they often retain functional senescence programs and can become senescent in response to various DNA-damaging drugs and radiation (9), which is especially relevant to solid tumors of epithelial origin. Importantly, cell senescence can be triggered upon activation of oncogenes, e.g., Ras or Raf (6, 26), which was observed in various systems, including human precancerous tissues. Therefore, a novel concept has emerged that oncogene-induced senescence (OIS) represents the major block on the path toward the neoplastic transformation, and cells in emerging tumors must acquire adaptations/mutations that allow the senescence bypass (8). We have found that Hsf1 and Hsp72 are critically involved in prevention of the oncogene-induced senescence caused by PIK3CA, Ras, and Her2 oncogenes. These effects were associated with suppression of accumulation of p21 and decrease in survivin levels (16, 28, 29).
The finding of the role of Hsf1 and Hsp72 in OIS suggests that these factors control early stages of tumorigenesis. This idea is in line with our data with Hsp72-knockout mice, which show dramatic suppression of early hyperplasia induced by the Her2 oncogene and development of senescence of epithelial cells in ducts (29). On the other hand, there are multiple reports that expression levels of Hsf1 and Hsp72 closely correlate with tumor grade, suggesting that these proteins could also be involved in later stages of tumor progression, including invasion and metastasis (see reference 11 for a review).
Here we addressed the possibility of involvement of Hsf1 in later stages of tumor progression by investigating why rare tumors that emerge in Her2-expressing Hsf1-knockout animals grow slower than tumors in control mice. We found that Hsf1 plays an additional important function in tumorigenesis by affecting hypoxia-inducible factor 1 (HIF-1)-dependent angiogenesis. HIF-1 is a master regulator not only of genes that control tumor neovascularization but also of those that control glycolysis, pH regulation, invasion, and metastasis (18, 34).
Here, we found that Hsf1 controls HIF-1 via the mRNA-binding protein HuR, which in turn controls an even wider set of cancer-related genes, including genes that regulate cell cycle, apoptosis, invasion, and angiogenesis. Therefore, besides being necessary for tumor initiation by blocking oncogene-induced senescence, Hsf1 also plays a critical role in tumor progression by regulating multiple genes controlled by HIF-1 and HuR.
MATERIALS AND METHODS
Animals.
Animal maintenance and experiments were conducted in compliance with IACUC guidelines. MMTVneu+/+ mice (FVB/N; Jackson Laboratory) were crossed with wild-type (WT) mice or hsf1−/− mice (129/BALB) (27, 36) to generate WT-MMTVneu+/−, hsf1+/− MMTVneu+/−, and hsf1−/− MMTVneu+/− mice. Mice were sacrificed at 3 months of age to study mammary gland hyperplasia or kept to monitor tumor development.
Xenografts.
For establishing tumor xenografts, MCF7 cells were trypsinized and mixed at a 1:1 ratio with Matrigel, and 1 million cells were injected subcutaneously into either left (control) or right (shHsf1-knockdown) flanks of 6-week-old female NCR nude mice (Taconic). Tumor growth was monitored weekly using caliper and calculated according to the formula L × W2 × π/6, where L is length and W is width.
Angiogenesis.
Tumors were removed from animals when they reached approximately 1 cm3 in WT mice or control xenografts; corresponding tumors in knockout (KO) animals or shHsf1 xenografts were smaller. Tumors were fixed with formalin, and CD31 staining and analysis were performed by Premier Lab using Aperio ImageScope software.
Cell cultures and reagents.
MCF10A cells were cultured in Dulbecco's modified Eagle's medium (DMEM)–F-12 medium supplemented with 5% horse serum, 20 ng/ml epidermal growth factor (EGF), 0.5 μg/ml hydrocortisone, 10 μg/ml human insulin, and 100 ng/ml cholera toxin. HEK293T cells were from ATCC and were cultivated in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS). MDA-MB453 cells were cultured in Leibovitz's L-15 medium with 10% FBS. Other cell lines were grown in DMEM with 10% FBS. All media contained penicillin and streptomycin (500 units per ml).
Retroviral vectors and infection.
RNAi-Ready pSIREN-RetroQ vector from the BD Biosciences retroviral delivery system was used for knockdown of HSF1. The sequence of the human HSF1 gene was selected as reported before (5′-TATGGACTCCAACCTGGATAA-3′) (37). shHuR lentivirus was purchased from Open Biosystems (Huntsville, AL). For HuR overexpression, HuR lentiviral open reading frames (ORFs) were purchased from Open Biosystems and HuR sequence was cloned by PCR and inserted into pQCXIN vector (BD Biosciences) at AgeI and PacI sites.
Retroviruses were produced as reported before (16). Briefly, HEK293T cells were cotransfected with plasmids expressing retroviral proteins Gag-Pol, vesicular stomatitis virus glycoprotein (VSV-G) pseudotype, and enhanced green fluorescent protein (EGFP) or our constructs using Lipofectamine 2000 (Invitrogen). Lentiviruses were produced by transfection of 293T cells with plasmids psPAX2, pMD2.G, and our constructs. Forty-eight hours after transfection, supernatants containing the retroviral particles were collected and frozen at −80°C until use. Cells were infected with diluted supernatant in the presence of 10 μg/ml Polybrene overnight and were selected with puromycin (0.75 μg/ml) 48 h after infection. Retroviral or lentiviral vectors expressing EGFP were used as an infection efficiency indicator: usually ∼90% of cells were fluorescent 2 days after infection.
qRT-PCR.
For total RNA preparation and quantitative real-time PCR (qRT-PCR), tissues from mice were harvested and preserved in RNAlater reagent (Qiagen). Total RNAs from tissues or cells were isolated using the RNeasy minikit (Qiagen) and reverse transcribed with the RetroScript kit (Ambion), according to the manufacturer's instructions. qRT-PCR was performed using SYBR green Rox Master Mix (Qiagen).
Primer sequences used in qRT-PCR are as follows: human HuR, For, 5′-CAGGAAACGCCTCCTCCGGC-3′, and Rev, 5′-ACGGCACCAAACGGCCCAAA-3′; human glyceraldehyde-3-phosphate dehydrogenase (GAPDH), For, 5′-GGCCTCCAAGGAGTAAGACC-3′, and Rev, 5′-AGGGGAGAGATTCAGTGTGGTG-3′; mouse HuR, For, 5′-GGATGACATTGGGAGAACGAAT-3′, and Rev, 5′-TGTCCTGCTACTTTATCCCGAA-3′; mouse GAPDH, For, 5′-AAATTCAACGGCACAGTCAAGG-3′, and Rev, 5′-GCCTCACCCCATTTGATGTTAGT-3′; human HIF-1, 5′-AAAAGAGGTGGATATGTCTGGGTT, and Rev, 5′-TGCTGAATAATACCACTCACAACGT.
mRNA stability study.
HuR mRNA half-life (t1/2) was measured in control (CT) and Hsf1-depleted (shHsf1) cells by incubation with 5 μg/ml of actinomycin D and collection of RNA after 1.5 h and 3 h. HuR and GAPDH mRNAs were measured by qRT-PCR and normalized by GAPDH mRNA. Data are represented as percentages of HuR mRNA measured at time zero (prior to adding actinomycin D), on a semilogarithmic scale.
Immunoblotting and translation analysis.
Cells were washed twice with phosphate-buffered saline (PBS) and lysed in lysis buffer (40 mM HEPES, pH 7.5, 50 mM KCl, 1% Triton X-100, 1 mM Na3VO4, 50 mM glycerophosphate, 50 mM NaF, 5 mM EDTA, 5 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 5 μg/ml each leupeptin, pepstatin A, and aprotinin). Mouse tissues or xenografted tumor was snap-frozen in liquid nitrogen and then homogenized in the same buffer and centrifuged for 5 min at 1,000 × g, and supernatants were collected. Protein concentration of the lysates was measured with the Bio-Rad protein assay reagent, after which they were diluted with lysis buffer to achieve equal protein concentrations. For measuring HIF-1α translation, MCF7 cells were labeled in 35-mm plates with EasyTag [35S]methionine (Perkin-Elmer) in methionine-free medium for 10 min. Lysates were prepared, and HIF-1α was immunoprecipitated.
Antibodies used for this study were anti-pS6, anti-p4E-BP1, p-eIF2α, and anti-Sirt1 from Cell Signaling; anti-HIF-1α, anti-p21, and anti-cyclin B from BD PharMingen; anti-HuR, anti-p53, and antisurvivin from Santa Cruz; anti-HSF1 from Stressgen; anti-HIF-2α, anti-CA9, and anti-Glut-1 from Novus Biolab; anti-PTBP1 from BioLegend; and anti-β-actin from Sigma.
RESULTS
Hsf1 knockout suppresses Her2-induced hyperplasia and tumor development.
We have previously found that knockdown of Hsf1 in MCF10A human mammary epithelial cells prevents neoplastic transformation by the Her2 oncogene. Indeed, while expression of Her2 in control MCF10A cells facilitated focus formation in culture and tumor appearance in nude mice, expression of this oncogene in Hsf1 knockdown MCF10A cells led to growth arrest and OIS, and tumors could not form in nude mice (28). To further dissect where in the tumorigenic process Hsf1 exerts its activity, here we used the transgenic animal model. Accordingly, we crossed Hsf1-knockout animals with mice expressing Her2/NeuT (a rodent homolog of Her2 carrying an activating mutation) under the control of the mouse mammary tumor virus (MMTV) promoter (MMTVneu) (19) to generate WT-MMTVneu+, hsf1+/− MMTVneu+, and hsf1−/− MMTVneu+ mice. Because of the mixed background, in the experiments with transgenic animals we used WT littermates as controls for the knockouts.
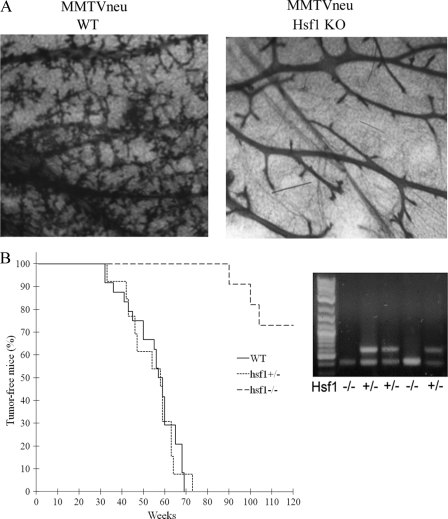
To investigate the role of Hsf1 in Her2-induced hyperplasia, mammary glands were taken from 3-month-old virgin mice to evaluate duct branching. Expression of Her2 in WT-MMTVneu+ mammary gland led to high density of ducts and extensive alveolar branching, as reported previously (31). Importantly, in hsf1−/− MMTVneu+ animals there was a low duct density and almost no alveolar branching (Fig. 1A). Therefore, Hsf1 KO prevented Her2-induced tissue hyperplasia, possibly by aggravating senescence, similar to what we have found recently with NeuT-induced mammary tumors in the Hsp72-knockout mouse model (29).
Fig 1.
Knockout of Hsf1 blocks NeuT-induced mammary duct and alveolar branching and delays tumor emergence. (A) WT-MMTVneu+ and hsf1−/−MMTVneu+ mice were sacrificed at 3 months of age, and whole mounts of their mammary glands were observed after application of Carnoy's fixative and staining with carmine. (B) Emergence of NeuT-induced tumors in WT-MMTVneu+ (n = 16), heterozygous hsf1+/− MMTVneu+ (n = 13), and hsf1−/− MMTVneu+ (n = 11) animals. Lack of Hsf1 in the knockout animals is shown by PCR.
To address whether Hsf1 KO suppresses NeuT-dependent tumorigenesis in vivo, we analyzed effects of Hsf1 knockout on NeuT-induced tumor development. Tumor incidences were similar between heterozygous hsf1+/− MMTVneu+ and WT-MMTVneu+ mice (median tumor appearance in this strain was about 55 weeks), indicating that one copy of the Hsf1 gene is sufficient to support mammary tumor emergence induced by Her2/NeuT (Fig. 1B). In contrast, the absence of Hsf1 in homozygous knockout animals markedly inhibited mammary tumor development (Fig. 1B). Indeed, tumor emergence was strongly delayed, and only three tumors of 11 animals appeared (Fig. 1B). Therefore, this model of Her2-positive breast cancer establishes that Hsf1 is critical for tumor initiation and hyperplasia.
Hsf1 knockdown suppresses tumor growth and angiogenesis.
To investigate whether Hsf1 has additional effects on later stages of tumor development, we measured growth rates of rare tumors that emerge in Hsf1-KO animals compared to control mice. Indeed, these tumors grew significantly slower than in control animals (Fig. 2A), indicating that Hsf1 may be required not only for NeuT-induced initial transformation but for tumor progression as well.
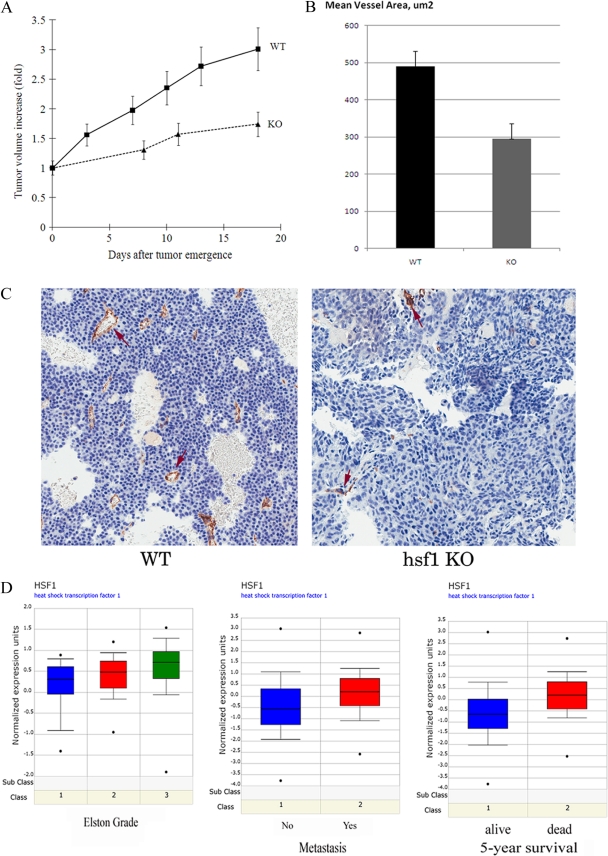
Fig 2.
Hsf1 is involved in breast tumor progression. (A) NeuT-induced tumors in Hsf1 KO mice demonstrate reduced growth rate. The data shown are means ± standard errors of the means. (B) Tumors in Hsf1-KO mice demonstrate reduced angiogenesis. Tumors from WT and KO animals were excised, fixed, stained for endothelial marker CD31, and analyzed for mean vessel area. (C) Tumor tissue staining with anti-CD31 antibody. Tumors from control and Hsf1-KO animals are shown. (D) Higher expression of Hsf1 mRNA in human breast cancer correlates with higher Elston grade, metastasis, and lower 5-year survival according to the Oncomine database (www.oncomine.org).
Since among the major factors limiting growth of solid tumors in vivo is neovascularization, we excised tumors from control and knockout animals, prepared slides, and immunostained them with a marker of angiogenesis (endothelial cells), CD31. We observed that although the numbers of blood vessels were similar in WT and KO animals (not shown), the mean vessel area in tumors from Hsf1-knockout animals was almost twice as small as in wild-type animals (Fig. 2B and C), indicating that the vessels were underdeveloped. These data were in line with microarray data deposited in Oncomine, a human cancer gene array database (www.oncomine.org), where elevated levels of Hsf1 in human breast cancer correlated with grade, metastasis, and poor prognosis (Fig. 2D), also suggesting that Hsf1 is involved in tumor progression.
Since in the transgenic mouse model Hsf1 is lacking both in mammary tumor and in surrounding stroma, to understand mechanisms by which Hsf1 can regulate angiogenesis, we decided to switch from NeuT-induced mouse mammary tumors to a simpler system, i.e., xenograft with human breast cancer cells following Hsf1 knockdown. In this system, Hsf1 can be downregulated specifically in human tumor cells but remain expressed normally in surrounding mouse stroma. As we reported previously, growth of many cancer cell lines (e.g., NeuT-expressing MCF10A cells or MDA-MB453 cells) is dependent on Hsf1, since Hsf1 knockdown causes senescence due to accumulation of p21 and downregulation of the mitotic and antiapoptotic protein survivin (28). Therefore, these cell lines cannot propagate even in vitro upon depletion of Hsf1, and accordingly, effects of Hsf1 on angiogenesis cannot be studied in this system. To avoid this problem, we screened several breast tumor cell lines and found that Hsf1 knockdown does not decrease survivin levels and does not increase p21 levels in MCF7 human breast carcinoma (Fig. 3A). Accordingly, growth of these cells in vitro was not significantly affected by Hsf1 knockdown (Fig. 3A).
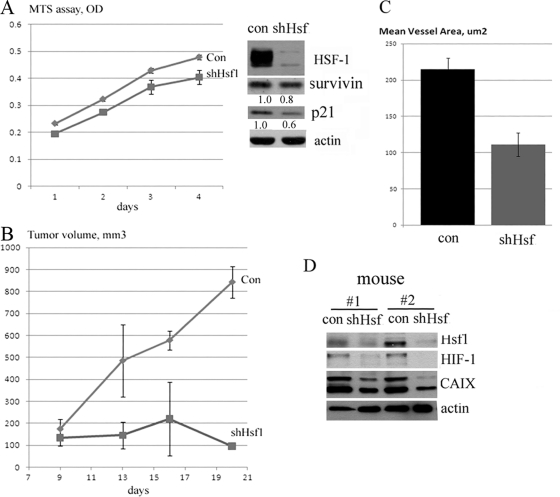
Fig 3.
Hsf1 knockdown suppresses growth and angiogenesis of tumors in the xenograft model. (A) Knockdown of Hsf1 has little effect on growth of MCF7 cells in vitro, and it does not increase p21 or decrease survivin levels. Cells were infected with shHsf1 retrovirus and selected for 4 days; their growth was assessed by formazan production MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay, and expression of p21 and survivin was assessed by immunoblotting. (B) Knockdown of Hsf1 blocks growth of MCF7 cells in vivo in xenografts. Cells infected with shHsf1 retrovirus as in panel A were injected in nude mice (106 cells per injection), and growth of tumors was monitored by caliper (see Materials and Methods for details). (C) Xenograft tumors from animals (as described in panel B) were excised, fixed, stained for CD31, and analyzed for mean vessel area as in Fig. 2B. Data are means ± standard errors. (D) Knockdown of Hsf1 reduces expression of HIF-1 and its target CAIX in xenografts. Expression of HIF-1 and CAIX tumor xenografts was analyzed by immunoblotting.
Therefore, we have chosen MCF7 cells to assess the effect of Hsf1 knockdown on tumor angiogenesis and growth in vivo in the xenograft model. MCF7 cells were infected with retroviral vector expressing shHsf1 as described before and selected with puromycin for 5 days. To avoid possible variations of host factor(s) which could affect tumor growth, control cells were injected in right flanks and shHsf1-knockdown cells were injected in left flanks of the same animals and their growth was monitored by caliper. Tumors emerged at the sites of injection of both control and Hsf1-depleted cells on day 9 after inoculation (Fig. 3B). Importantly, after tumor emergence, tumors formed by control MCF7 cells grew rapidly, while tumors formed by the Hsf1 knockdown ceased to grow soon after emergence (Fig. 3B). The strong inhibitory effect of Hsf1 knockdown on growth of MCF7 cells in xenografts was in sharp contrast with cell culture, where Hsf1 knockdown practically did not affect the growth rate (Fig. 3A). We have isolated tumors, stained them for CD31 as described above, and found that, similar to NeuT-induced mammary tumors in Hsf1-KO mice, Hsf1 knockdown markedly decreased mean vessel area in MCF7 human breast cancer xenografts (Fig. 3C) (of note, there was no significant difference in the number of vessels). These data indicate that the xenograft model recapitulates effects of Hsf1 seen in the transgenic model and suggest that control of tumor angiogenesis may represent an important factor regulated by Hsf1.
Hsf1 controls expression of HIF-1.
Hypoxia-inducible factor 1 (HIF-1) is considered to be the major regulator of tumor angiogenesis (18, 34), and therefore, we assessed its expression in xenografts formed by MCF7 cells with Hsf1 knockdown. We found high levels of HIF-1α in control tumors (which indicated hypoxic conditions in xenografts), but in tumors with Hsf1 knockdown, there were much lower levels of HIF-1α (Fig. 3D). Similarly, in Hsf1 knockdown tumors we observed downregulation of HIF-1α target CAIX (carbonic anhydrase 9) (Fig. 3D). Accordingly, Hsf1 appears to control angiogenesis in xenografts via regulating accumulation of HIF-1α.
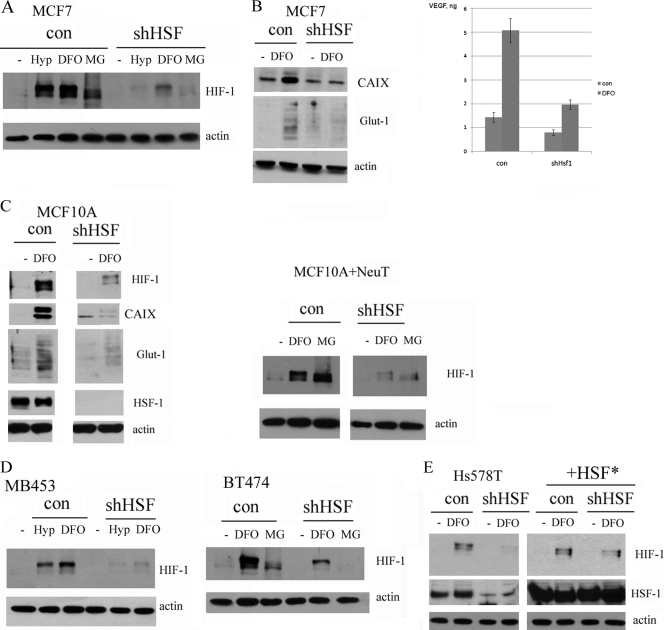
To elucidate mechanisms by which Hsf1 regulates HIF-1α expression, we studied effects of Hsf1 knockdown on HIF-1α expression in cell culture. Control and shHsf1 MCF7 cells were exposed to hypoxia (1% oxygen for 16 h) or the hypoxia mimetic deferoxamine (DFO) (100 μM), and levels of HIF-1α were monitored by immunoblotting. Knockdown of Hsf1 markedly suppressed accumulation of HIF-1α in response to these stimuli (Fig. 4A), similar to suppression of HIF-1α accumulation in xenografts formed by shHsf1 MCF7 cells (Fig. 3D). Importantly, Hsf1 knockdown also strongly inhibited secretion of vascular endothelial growth factor (VEGF), the major growth factor responsible for neovascularization, as well as other targets of HIF-1α, CAIX, and Glut-1 (Fig. 4B).
Fig 4.
Knockdown of Hsf1 suppresses HIF-1 signaling in cell lines. (A) Knockdown of Hsf1 in MCF7 cells inhibits HIF-1 accumulation after cells were subjected to hypoxia (Hyp, 1% O2, 16 h) or treated with the hypoxia mimetic DFO (100 μM, 4 h) or proteasome inhibitor MG132 (5 μM, 4 h). Cells were infected with shHsf1 retrovirus as in Fig. 3A, and HIF-1 expression was analyzed by immunoblotting. (B) Hsf1 knockdown reduces induction of HIF-1α targets VEGF, CAIX, and Glut-1 in MCF7 cells. Cells were infected with Hsf1 retrovirus as in panel A and treated with DFO for 48 h, and medium was collected and analyzed by enzyme-linked immunosorbent assay for VEGF by Quansys Biosciences or lysates were blotted with anti-CAIX or anti-Glut-1 antibodies. (C) Knockdown of Hsf1 inhibits accumulation of HIF-1 and its targets in MCF10A (left panel) and NeuT-infected MCF10A (right panel) cells. Cells were treated with DFO (100 μM, 4 h) or MG132 (5 μM, 4 h), and accumulation of HIF-1 and its targets CAIX and Glut-1 was analyzed by immunoblotting. (D) Her2-positive MB453 (left panel) and BT474 (right panel) cells were infected with Hsf1 retrovirus as in panel A and treated with hypoxia for 16 h or DFO (100 μM, 4 h) or MG132 (5 μM, 4 h), and accumulation of HIF-1 was analyzed by immunoblotting. (E) Expression of the shRNA-resistant Hsf1 mutant reverses the effect of shRNA on HIF-1α. Hs578T cells were infected with retrovirus containing Hsf1* or control retrovirus and selected. Then Hsf1 was depleted by shRNA, and HIF-1α expression was measured by immunoblotting in naïve cells or following DFO treatments.
To assess whether the effect of Hsf1 knockdown on HIF-1α has a general significance, we used other breast cell lines, including normal untransformed cell line MCF10A, NeuT-transformed MCF10A, Her2-positive cancer lines MB453 and BT474, and triple-negative Hs578T. In all these cell lines, Hsf1 knockdown strongly inhibited accumulation of HIF-1α and its targets CAIX and Glut-1 in response to the hypoxia mimetic DFO (Fig. 4C to E and data not shown). This phenomenon was not an off-target effect of Hsf1 knockdown, since expression of a short hairpin RNA (shRNA)-resistant mutant of Hsf1 (Hsf1*) prevented downregulation of HIF-1α. Therefore, impairment of angiogenesis in NeuT-induced mammary tumors in hsf1-knockout animals can be associated with inhibition of HIF-1α expression in tumors. Of note, these effects were not limited to Her2-positive breast cancer cells, since they were seen in Her2-negative, estrogen receptor (ER)-positive MCF7 cells and in triple-negative cancer line Hs578T (Fig. 4E).
Hsf1 is involved in HIF-1 translation.
HIF-1α expression can be regulated at multiple levels, including protein degradation (e.g., following hypoxia or hypoxia mimetics), transcription (e.g., upon exposure to lipopolysaccharide or cytokines), or translation (e.g., upon exposure to autocrine growth factors, like epidermal growth factor or insulin-like growth factor) (see reference 7 for a review). Hsf1 knockdown may block HIF-1α accumulation by affecting any of these mechanisms, and therefore, we addressed what mechanisms are targeted by Hsf1.
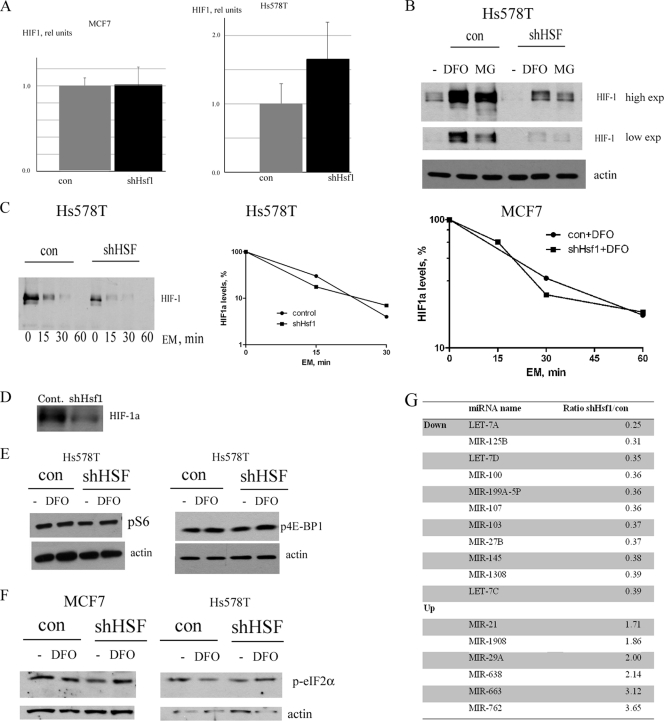
To test for transcription and mRNA degradation, we analyzed expression of HIF-1α mRNA by qRT-PCR. For these experiments, we used MCF7 cells which have undetectable levels of HIF-1α under normal conditions but accumulate this protein under hypoxia. We also used Hs578T breast tumor cells, which have elevated basal levels of HIF-1α, but these levels can be further increased by hypoxia mimetics (Fig. 4E and 5B). Although in both lines Hsf1 knockdown markedly reduced expression of HIF-1α protein (Fig. 4A and E and Fig. 5B), there was no downregulation of HIF-1α mRNA levels (Fig. 5A), indicating that Hsf1 regulates HIF-1 by affecting a step in the pathway downstream of HIF-1α mRNA transcription or degradation. To assess stability of HIF-1α protein, we treated cells with a proteasome inhibitor, MG132. If Hsf1 knockdown enhances degradation of HIF-1, inhibition of proteasome should lead to restoration of HIF-1α in Hsf1-depleted cells to levels seen in control cells treated with MG132. In the experiment, MG132 treatment caused robust accumulation of HIF-1α in control cells, but much weaker buildup of HIF-1α was seen in Hsf1-depleted cells (Fig. 4A, C, and D and Fig. 5B). Therefore, inhibition of protein degradation was insufficient to restore HIF-1 levels, suggesting that the Hsf1-dependent regulation of HIF-1 is at the level not of protein stability but rather of protein synthesis. (Of note, in MG132-treated cells HIF-1 migrated on gels faster than hypoxia-induced HIF-1, apparently due to the absence of phosphorylation [21].) To directly assess the effects of Hsf1 knockdown on HIF-1 degradation, we measured the half-lives of HIF-1 in control and Hsf1 knockdown MCF7 cells treated with DFO. Samples were taken at different time points following addition of the protein synthesis inhibitor emetine, and HIF-1 levels were analyzed by immunoblotting. In naïve MCF7 cells, degradation of HIF-1α was very fast (half-life [t1/2] of about 10 min), while in DFO-treated cells HIF-1α was more stable (t1/2 of about 25 min) (Fig. 5C), which is in line with previous reports. While strongly reducing HIF-1α expression levels (Fig. 4A), depletion of Hsf1 did not significantly affect stability of HIF-1 in either control or DFO-treated cells (Fig. 5C). Similarly, Hsf1 knockdown did not affect the rates of HIF-1α degradation in naïve Hs578T cells, while it decreased HIF-1 levels (Fig. 5C).
Fig 5.
Effect of Hsf1 knockdown on various levels of HIF-1 expression. (A) Effect of Hsf1 knockdown on HIF-1α mRNA levels in MCF7 or Hs578T cells; DFO treatment did not change mRNA levels (not shown). qRT-PCR was done as described in Materials and Methods. (B) shHsf1 suppresses both basal and DFO (100 μM, 4 h)- or MG132 (5 μM, 4 h)-induced levels of HIF-1 in Hs578T cells. (C) Knockdown of Hsf1 does not affect stability of HIF-1. Hs578T or MCF7 cells were treated with the protein synthesis inhibitor emetine (EM; 10 μM) in the absence or presence of DFO (100 μM) for the time intervals indicated, and the levels of HIF-1 were determined. The middle panel shows quantification of the left panel. (D) Hsf1 knockdown reduces translation of HIF-1α. MCF7 cells were labeled with [35S]methionine for 10 min, and HIF-1 was immunoprecipitated, run on SDS-PAGE gels, and exposed to film. The initial material was normalized by trichloroacetic acid-precipitable count (label incorporated into proteins). (E) Hsf1 knockdown does not affect activity of mTOR pathway. Activity of the mTOR pathway was assayed by antibody to phosphorylated (Ser235/236) S6 ribosomal protein (left panel) or phosphorylated (Ser65) 4E-BP1 (right panel). (F) Hsf1 knockdown does not affect phosphorylation of eIF2α, assayed by antibody to phosphorylated (Ser51) eIF2α. (G) Effect of shHsf1 knockdown on microRNA profile of Hs578T cells. MicroRNA was isolated and assayed by Miltenyi Biotec. See text for further explanation.
To assess translation of HIF-1α directly, cells were pulse-labeled with [35S]methionine for 10 min, and HIF-1α was immunoprecipitated with the corresponding antibody. Hsf1 knockdown significantly reduced incorporation of label into HIF-1α (Fig. 5D), which, together with the data that mRNA levels were similar, indicated that the main regulation of HIF-1 by Hsf1 is at the level of translation.
Effects of Hsf1 on major pathways regulating HIF-1α translation and microRNA (miRNA).
We further explored effects of Hsf1 on translation of HIF-1α. One possibility involves the mTOR pathway, since HIF-1α translation is dependent on mTOR activity (7, 21, 34). Accordingly, we tested whether Hsf1 knockdown downregulates components of the mTOR pathway, including phospho-S6 ribosomal protein and phospho-4E-BP1, and found no effects of Hsf1 knockdown on either p-S6 or p-4E-BP1 (Fig. 5E). Therefore, the mTOR pathway apparently is not involved in regulation of HIF-1 by Hsf1 depletion.
Another potential mechanism which can regulate HIF-1 translation is associated with downregulation of eIF2α by its phosphorylation on Ser51. We explored this possibility but did not find any difference in eIF2α phosphorylation upon Hsf1 knockdown (Fig. 5F).
Next we used microarrays to assess how Hsf1 knockdown affects levels of microRNAs which may potentially regulate HIF-1α translation. Hsf1 knockdown had profound effects on expression of a number of microRNAs in Hs578T cells, but none of the upregulated microRNAs had binding sites in the untranslated regions (UTRs) of HIF-1α mRNA. Therefore, these miRNAs cannot be directly involved in regulation of HIF-1α (Fig. 5G). On the other hand, interestingly, 7 out of 11 microRNAs reduced upon Hsf1 knockdown (i.e., Let-7A, Let-7C, Let-7D, MiR-199A-5P, MiR-210, MiR-125B, and MiR-107) were previously shown to be upregulated by HIF-1 or hypoxia (23) (Fig. 5F). Therefore, downregulation of these microRNAs upon Hsf1 knockdown most likely reflects suppression of HIF-1. Importantly, some of these miRNAs, e.g., Let-7 and MiR-125, regulate multiple targets involved in cancer, e.g., RAS, MYC, and p53 (12). Accordingly, effects of Hsf1 on HIF-1 not only result in suppression of angiogenesis but stimulate a variety of cancer-related pathways.
Hsf1 knockdown downregulates HuR and its targets associated with cancer traits.
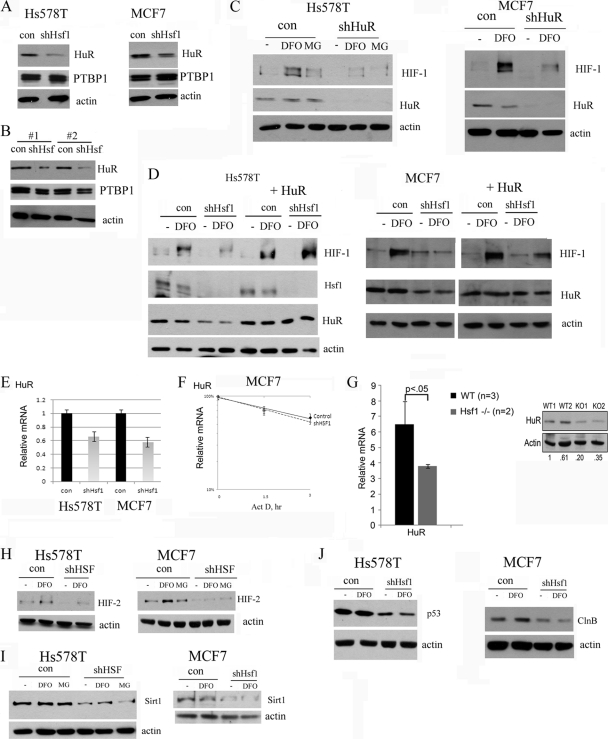
Among regulators of HIF-1α translation, there are mRNA-binding proteins PTBP1 and HuR. Since the mTOR pathway, eIF2α, or microRNA was not involved in HIF-1 regulation by Hsf1, we assessed expression of PTBP1 and HuR upon Hsf1 knockdown. By immunoblotting, we did not find any decrease in PTBP1 levels (Fig. 6A). On the other hand, HuR levels decreased by about 70% upon Hsf1 knockdown in both MCF7 and Hs578 cells (Fig. 6A). Furthermore, we found 70 to 80% downregulation of HuR levels in MCF7 xenografts as well (Fig. 6B), suggesting that this factor may be involved in HIF-1 regulation by Hsf1. With many targets, HuR regulates stability of mRNA; however, specifically with HIF-1, it was demonstrated that HuR regulates its translation without affecting mRNA stability (17). Therefore, effects of HuR on HIF-1 are consistent with our results with qRT-PCR (Fig. 5A).
Fig 6.
Suppression of HIF-1 accumulation by shHsf1 is associated with HuR downregulation. (A) Hsf1 knockdown decreases HuR but not PTBP1 levels in Hs578T (left panel) and MCF7 (right panel) cells. (B) Hsf1 knockdown decreases HuR but not PTBP1 levels in MCF7 xenografts (Fig. 3B to D). (C) Knockdown of HuR suppresses HIF-1 accumulation in Hs578T (left panel) and MCF7 (right panel) cells. Cells were infected with shHuR retrovirus, selected, and treated with DFO (100 μM) or MG132 (5 μM) for 4 h. (D) HuR overexpression prevents inhibition of HIF-1 accumulation by Hsf1 knockdown. Hs578T and MCF7 cells were first infected with HuR retrovirus and selected. HuR-expressing cells were infected with shHsf1 retrovirus as described in Materials and Methods and treated with 100 μM DFO for 4 h. (E) Hsf1 knockdown leads to reduction of HuR mRNA levels in Hs579T and MCF7 cells. (F) Hsf1 knockdown does not affect stability of HuR mRNA. Levels of HuR mRNA were measured by qRT-PCR after inhibition of transcription by actinomycin at the indicated time points. (G) Levels of HuR mRNA and protein were assessed in liver of control and Hsf1-KO mice. (H to J) Hs578T and MCF7 cells were infected with shHsf1 retrovirus as in Fig. 4A and 5B and treated with DFO (100 μM) or MG132 (5 μM) for 4 h, and the levels of HIF-2α (Η), Sirt1 (I), p53 (mutant) (J), or cyclin B (ClnB) (J) were assayed by immunoblotting.
To test whether downregulation of HuR is responsible for suppression of HIF-1 upon Hsf1 knockdown, we first assessed whether decrease in HuR levels by shRNA can downregulate HIF-1 in cell lines which we used for the study. Indeed, as seen in Fig. 6C, downregulation of HuR markedly suppressed HIF-1 accumulation in both MCF7 and Hs578T cells. Furthermore, we assessed whether overexpression of HuR can prevent effect of Hsf1 knockdown on HIF-1 accumulation. Hs578T and MCF7 cells were first infected with retrovirus expressing HuR, selected with neomycin, and then infected with shHsf1 retrovirus and selected with puromycin. HuR overexpression completely prevented an effect of Hsf1 knockdown on HIF-1 accumulation in both Hs578T and MCF7 cells (Fig. 6D). These results demonstrate that downregulation of HuR by Hsf1 knockdown appears to be the major mechanism responsible for HIF-1 inhibition.
We next addressed whether Hsf1 affects HuR transcription, translation, or degradation. Hsf1 apparently did not affect HuR protein stability, since addition of the proteasome inhibitor MG132 did not restore HuR levels in shHsf1 cells (data not shown). At the same time, shHsf1 decreased levels of HuR mRNA (Fig. 6E). This effect was apparently at the level of transcription, since we did not observe any effect on the HuR mRNA stability (Fig. 6F). Furthermore, we measured levels of HuR mRNA and protein in Hsf1-KO animals. In line with results of the cell culture experiments, Hsf1-KO mice had significantly lower levels of HuR than did control mice (Fig. 6G). These data emphasize the relevance of the role of Hsf1 in regulation of HuR to the mammalian organism.
Along with overexpression of Hsf1 and HIF-1, tumor progression in various human cancers is associated with higher expression of HuR (24, 25). HuR controls mRNA stability and/or translation of many proteins involved in cancer, including proteins involved in angiogenesis (e.g., HIF-1, HIF-2, and vascular endothelial growth factor [VEGF]), cell survival (e.g., p53 and Sirt1), proliferation (e.g., cyclins, Cdc2, and p21), and others (1, 14, 24, 25). Since Hsf1 knockdown leads to HuR decrease, we assessed whether some of the known targets of HuR are also downregulated in these cells. Indeed, along with HIF-1 and VEGFA, knockdown of Hsf1 in Hs578 or MCF7 cells decreased levels of other known targets of HuR such as HIF-2, Sirt1, p53, and cyclin B (Fig. 6H to J). Therefore, by regulating HuR, Hsf1 controls a multitude of genes and pathways involved in various stages of tumorigenesis, including initiation, invasion, and angiogenesis.
DISCUSSION
Previous studies demonstrated that human cancers often have higher levels of expression of Hsf1 and heat shock proteins, which are associated with tumor progression and resistance to anticancer therapy, thus indicating that Hsf1 and Hsps play an important role in cancer. Indeed, recent works using knockout mice demonstrated that Hsf1 is essential for tumorigenesis induced by RAS or p53 knock-in mutant (R172H). Following these developments, we attempted to dissect the mechanism of Hsf1 effects on tumorigenesis and found that Hsf1 interferes with senescence signaling, thus playing a critical role in prevention of oncogene-induced senescence (OIS). We demonstrated that when Hsf1 or Hsp72 is downregulated, the Her2/NeuT oncogene can no longer transform mammary epithelial cells but instead causes senescence which is accompanied by accumulation of a cell cycle inhibitor, p21, and decrease in levels of the cell cycle promoter and apoptosis inhibitor survivin (28, 29). Furthermore, in established Her2-positive human breast cancer lines, knockdown of Hsf1 led to an increase in p21, a decrease in survivin, and inhibition of their growth (28). Due to this mechanism, Hsf1 is essential for transformation of human mammary epithelial cells by the Her2/NeuT oncogene and maintenance of growth.
These findings suggested that Hsf1 should mainly function at early stages of cancer development when the Her2 oncogene is activated and OIS blocks cell proliferation and transformation. Indeed, here we demonstrate that in Hsf1-KO mice emergence of tumors driven by the Her2/NeuT oncogene is dramatically suppressed. There was no NeuT-induced hyperplasia of mammary tissue and development of alveoli was strongly reduced (Fig. 1), further supporting the notion that Hsf1 is essential early in tumor development.
These effects of Hsf1 KO were almost indistinguishable from effects of Hsp72 KO, which, as we recently demonstrated, also inhibits mammary tissue hyperplasia and alveolar development, causes senescence of epithelial cells, and overall blocks tumor initiation (29). These data are consistent with our prior suggestion that the major effect of Hsf1 depletion on OIS may be mediated by downregulation of Hsp72. Indeed, we found that in cell lines where depletion of Hsf1 causes downregulation of Hsp72, it also causes OIS, while in cell lines where depletion of Hsf1 does not cause Hsp72 downregulation, it does not trigger OIS (unpublished data). Nevertheless, the mechanism of OIS remains in these latter lines, since depletion of Hsp72 in them does trigger OIS (16).
Effects of Hsf1 on cancer are not limited to the control of OIS, and literature data suggest that Hsf1 plays a more general role in tumor development. For example, it was demonstrated that in a series of prostate cancer clones that developed invasiveness and metastasis upon passage through nude mice, expression of Hsf1 was further increased in more aggressive tumorigenic clones (2). Furthermore, data from the cancer microarray archive Oncomine also indicate that levels of Hsf1 correlate with the tumor grade, ultimately indicating that besides tumor initiation Hsf1 should be involved in tumor progression (Fig. 2C).
Based on these considerations, we addressed the role of Hsf1 in tumor progression, focusing on a mouse model of Her2-positive breast cancer. Initially, we looked at rare tumors that emerged in Hsf1-KO animals that express NeuT. Of note, these tumors emerged much later than did tumors in control NeuT-expressing mice (Fig. 1B). Importantly, tumors in Hsf1-knockout mice grew much slower than in control mice (Fig. 2A), further confirming that in addition to tumor initiation, Hsf1 plays a role in tumor growth.
In these rare tumors emerging in Hsf1-KO animals, we observed that angiogenesis is impaired, which may be a factor in slow growth. In order to investigate mechanisms of these effects, we had to develop xenograft and cell culture models. Testing several breast cancer mammary epithelium lines in culture demonstrated that levels of the major regulator of angiogenesis HIF-1 are dramatically reduced upon depletion of Hsf1. The xenograft model was more difficult to apply, since cancer cell lines usually develop senescence or apoptosis following Hsf1 depletion (13, 28), and lack of growth in xenografts could be due to OIS or apoptosis, rather than insufficient angiogenesis. After screening of a collection of breast cancer lines, we found that in two lines, MCF7 and Hs578T, depletion of Hsf1 does not downregulate Hsp72 and does not cause senescence, while it does cause downregulation of HIF-1α. Accordingly, MCF7 cells with depleted Hsf1 could form tumors in xenografts, but after emergence tumors grew very slowly and had reduced angiogenesis. This model therefore recapitulates suppression of tumor progression seen in the knockout animals. An interesting conclusion from these experiments is that while Hsf1 effects on OIS and tumor initiation are mediated by Hsp72, effects on HIF-1 and tumor progression are independent of Hsp72 and must involve distinct Hsf1 transcription targets.
Interestingly, there are very complex bidirectional relations between heat shock transcription factors and HIF-1. For example, it has been demonstrated in Drosophila that hypoxia induces heat shock response via HIF-1 (3). On the other hand, in mammalian cells Hsf2 and Hsf4 were shown to regulate HIF-1 transcription (10).
In addition to angiogenesis, HIF-1 regulates expression of genes involved in other aspects of tumorigenesis, including survival of hypoxia and other harmful conditions, invasion, etc. We have found that beyond angiogenesis-related targets of HIF-1 (e.g., VEGF), Hsf1 also controls expression of Glut-1 and CAIX and most likely other HIF-regulated genes involved in tumor progression. For example, Hsf1 regulates a set of cancer-related miRNAs (e.g., Let-7, MiR-199A, or MiR-125B), which are known to be controlled by HIF-1 or hypoxia (23).
In the search for a molecular mechanism of the effect of Hsf1 on HIF-1, we found that Hsf1 controls another major regulator of tumor progression, HuR. HIF-1 accumulation under hypoxia is mediated by inhibition of constitutive ubiquitination by the E3 ligase VHL, resulting in suppression of HIF-1α degradation by the proteasome. Originally, we suggested that Hsf1 deficiency increases the rate of HIF-1α degradation. However, direct measurement of the rate of HIF-1α degradation showed that Hsf1 knockdown does not increase the half-life of HIF-1α. Similarly, we found that there is no difference in the levels of HIF-1α mRNA in Hsf1-depleted cells. Therefore, Hsf1 knockdown does not affect either degradation of HIF-1α protein or transcription of degradation of HIF-1α mRNA, and indeed we found that Hsf1 regulates HIF-1α translation (Fig. 5). Assessing major mechanisms known to regulate HIF-1 translation (miRNAs, mTOR, and eIF2α pathways), we could not implicate any of these mechanisms in regulation of HIF-1 translation following Hsf1 knockdown. On the other hand, there was a marked decrease in the levels of the mRNA-binding protein HuR, a major regulator of translation which is known to promote translation of HIF-1α (Fig. 6). This decrease was seen both in cell culture and in xenograft tumors as well in Hsf1-knockout animals (Fig. 6). Effects of Hsf1 on HIF-1 were mediated by HuR, since knockdown of HuR suppressed HIF-1 accumulation in our models, while restoration of HuR levels in Hsf1-depleted cells prevented HIF-1 downregulation (Fig. 6). These data demonstrate that downregulation of HuR is the main mechanism by which Hsf1 regulates HIF-1 translation.
HuR is overexpressed in various human cancers, and its expression correlates with cancer progression (1, 14, 24, 25). Besides HIF-1, HuR controls mRNA stability and translation of a plethora of proteins associated with cancer, and we report that several known cancer-related targets of HuR, including HIF-2, Sirt1, p53, and cyclin B, are also downregulated upon Hsf1 knockdown (Fig. 6). Therefore, we have established an important link between Hsf1 and two major players in cancer progression, HIF-1 and HuR. This association between Hsf1, HuR, and HIF-1 demonstrates that Hsf1 is essential not only for initial stages of tumorigenesis by preventing oncogene-induced senescence but also for progression of established tumors.
ACKNOWLEDGMENT
This work was supported by a Department of Defense Breast Cancer Grant (M.Y.S.).
Footnotes
Published ahead of print 3 January 2012
REFERENCES
- 1. Abdelmohsen K, Lal A, Kim HH, Gorospe M. 2007. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle 6:1288–1292 [DOI] [PubMed] [Google Scholar]
- 2. Akalin A, et al. 2001. A novel mechanism for chaperone-mediated telomerase regulation during prostate cancer progression. Cancer Res. 61:4791–4796 [PubMed] [Google Scholar]
- 3. Baird NA, Turnbull DW, Johnson EA. 2006. Induction of the heat shock pathway during hypoxia requires regulation of heat shock factor by hypoxia-inducible factor-1. J. Biol. Chem. 281:38675–38681 [DOI] [PubMed] [Google Scholar]
- 4. Beere HM. 2005. Death versus survival: functional interaction between the apoptotic and stress-inducible heat shock protein pathways. J. Clin. Invest. 115:2633–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ben-Porath I, Weinberg RA. 2005. The signals and pathways activating cellular senescence. Int. J. Biochem. Cell Biol. 37:961–976 [DOI] [PubMed] [Google Scholar]
- 6. Bihani T, et al. 2004. Differential oncogenic Ras signaling and senescence in tumor cells. Cell Cycle 3:1201–1207 [PubMed] [Google Scholar]
- 7. Brahimi-Horn MC, Pouyssegur J. 2009. HIF at a glance. J. Cell Sci. 122:1055–1057 [DOI] [PubMed] [Google Scholar]
- 8. Braig M, Schmitt CA. 2006. Oncogene-induced senescence: putting the brakes on tumor development. Cancer Res. 66:2881–2884 [DOI] [PubMed] [Google Scholar]
- 9. Chang B-D, et al. 2002. Molecular determinants of terminal growth arrest induced in tumor cells by a chemotherapeutic agent. Proc. Natl. Acad. Sci. U. S. A. 99:389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen R, Liliental JE, Kowalski PE, Lu Q, Cohen SN. 2011. Regulation of transcription of hypoxia-inducible factor-1alpha (HIF-1alpha) by heat shock factors HSF2 and HSF4. Oncogene 30:2570–2580 [DOI] [PubMed] [Google Scholar]
- 11. Ciocca DR, Calderwood SK. 2005. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 10:86–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Croce CM. 2009. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10:704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dai C, Whitesell L, Rogers AB, Lindquist S. 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130:1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Danilin S, et al. 2010. Role of the RNA-binding protein HuR in human renal cell carcinoma. Carcinogenesis 31:1018–1026 [DOI] [PubMed] [Google Scholar]
- 15. Gabai VL, Mabuchi K, Mosser DD, Sherman MY. 2002. Hsp72 and stress kinase c-jun N-terminal kinase regulate the Bid-dependent pathway in tumor necrosis factor-induced apoptosis. Mol. Cell. Biol. 22:3415–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gabai VL, Yaglom JA, Waldman T, Sherman MY. 2009. Heat shock protein Hsp72 controls oncogene-induced senescence pathways in cancer cells. Mol. Cell. Biol. 29:559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galban S, et al. 2008. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol. Cell. Biol. 28:93–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gordan JD, Simon MC. 2007. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr. Opin. Genet. Dev. 17:71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guy CT, et al. 1992. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc. Natl. Acad. Sci. U. S. A. 89:10578–10582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hunt CR, et al. 2004. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol. Cell. Biol. 24:899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ke Q, Costa M. 2006. Hypoxia-inducible factor-1 (HIF-1). Mol. Pharmacol. 70:1469–1480 [DOI] [PubMed] [Google Scholar]
- 22. Khaleque M, et al. 2005. Induction of heat shock proteins by heregulin beta1 leads to protection from apoptosis and anchorage-independent growth. Oncogene 24:6564–6573 [DOI] [PubMed] [Google Scholar]
- 23. Kulshreshtha R, Davuluri RV, Calin GA, Ivan M. 2008. A microRNA component of the hypoxic response. Cell Death Differ. 15:667–671 [DOI] [PubMed] [Google Scholar]
- 24. Lopez de Silanes I, et al. 2003. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 22:7146–7154 [DOI] [PubMed] [Google Scholar]
- 25. Lopez de Silanes I, Lal A, Gorospe M. 2005. HuR: post-transcriptional paths to malignancy. RNA Biol. 2:11–13 [DOI] [PubMed] [Google Scholar]
- 26. Lowe SW, Cepero E, Evan G. 2004. Intrinsic tumour suppression. Nature 432:307–315 [DOI] [PubMed] [Google Scholar]
- 27. McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. 1998. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J. Biol. Chem. 273:7523–7528 [DOI] [PubMed] [Google Scholar]
- 28. Meng L, Gabai VL, Sherman MY. 2010. Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene 29:5204–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meng L, Hunt C, Yaglom JA, Gabai VL, Sherman MY. 2011. Heat shock protein Hsp72 plays an essential role in Her2-induced mammary tumorigenesis. Oncogene 30:2836–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Min J-N, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF. 2007. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene 26:5086–5097 [DOI] [PubMed] [Google Scholar]
- 31. Muller WJ, et al. 1996. Synergistic interaction of the Neu proto-oncogene product and transforming growth factor alpha in the mammary epithelium of transgenic mice. Mol. Cell. Biol. 16:5726–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Page TJ, et al. 2006. Genome-wide analysis of human HSF1 signaling reveals a transcriptional program linked to cellular adaptation and survival. Mol. Biosyst. 2:627–639 [DOI] [PubMed] [Google Scholar]
- 33. Pirkkala L, Nykanen P, Sistonen L. 2001. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15:1118–1131 [DOI] [PubMed] [Google Scholar]
- 34. Semenza GL. 2010. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 29:625–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Solimini NL, Luo J, Elledge SJ. 2007. Non-oncogene addiction and the stress phenotype of cancer cells. Cell 130:986–988 [DOI] [PubMed] [Google Scholar]
- 36. Xiao X, et al. 1999. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 18:5943–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zaarur NG, Porco VLJ, Calderwood S, Sherman M. 2006. Targeting heat shock response to sensitize cancer cell to proteasome and Hsp90 inhibitors. Cancer Res. 66:1783–1791 [DOI] [PubMed] [Google Scholar]