Abstract
Human hemochromatosis (HC) has been associated with the common C282Y polymorphism of HFE or rare pathogenic mutations of TfR2, HJV, FPN and HAMP. All forms of human HC seem to be caused by low or inadequate levels of hepcidin, the iron hormone. We and others have recently shown that Hfe−/−mice exhibit an impairment in the bone morphogenetic protein (BMP) signaling pathway controlling hepcidin. However, all data indicating the central role of BMPs in hepcidin regulation and an impaired BMP/SMAD signaling in HC have been collected in mice. In this study we investigated whether also in humans the expression of BMP signaling targets, SMAD7 and Id1, are associated with liver iron concentration (LIC) and whether such regulation is disrupted in HFE-HC. We correlated the mRNA expression, assessed by RT-PCR, of HAMP, SMAD7 and Id1 with LIC in liver biopsies from patients with normal iron status, HFE-HC or non-HC hepatic iron overload. We found that in human liver, not only HAMP, but also SMAD7 and Id1 mRNA significantly correlate with the extent of hepatic iron burden. However, this correlation is lost in patients with HFE-HC, but maintained in subjects with non-hemochromatotic iron overload. These data indicate that in human HFE-HC a disrupted BMP/SMAD signaling in the liver is key in the pathogenesis of the disease.
Keywords: iron overload, bone morphogenetic proteins, hepcidin, SMAD proteins
Iron is an important nutrient that is continuously recycled by the body; its uptake, storage, and utilization must be carefully coordinated at the cellular and systemic level. Signals from iron storage compartments (mainly hepatocytes and macrophages) and iron utilization sites (primarily the bone marrow)–historically referred to as “store” and “erythroid” regulatory signals [1] –are transmitted to a central control site in the liver, where they regulate expression of the iron hormone hepcidin. Pigeon et al. [2] were the first to link hepcidin to iron, and shortly thereafter, transgenic mouse studies revealed that hepcidin is in fact the principal down-regulator of iron traffic directed toward the bloodstream from the external environment (i.e., the intestinal lumen or, for the fetus, maternal blood) and from the body storage sites. Hepcidin is believed to regulate iron homeostasis by binding to FPN, the main cellular iron exporter in mammals. As a result of its interaction with circulating hepcidin, FPN is internalized and degraded [3], thereby diminishing the cells’ ability to transfer iron to the plasma compartment. Later, if iron is needed in the bone marrow for hemoglobin synthesis, hepcidin production is reduced, FPN is re-expressed at the cell surface, and iron export to the bloodstream resumes. This negative-feedback mechanism keeps circulating iron at the proper level for erythropoiesis without causing oxidative damage to cells.
Hepcidin responds to a variety of signals, both inhibitory and stimulatory, but a main stimulus for hepcidin synthesis is plasma iron, and recent studies highlight a central regulatory role for bone morphogenic proteins (BMPs). BMPs are members of the TGF-β superfamily [4] that signal through transmembrane receptors (BMPRs) to Smad proteins, which translocate to the nucleus and activate the expression of target genes, such as ID1, SMAD6, SMAD7, and HAMP[5–7]. The timing, location, and specific downstream effects of BMP signaling are controlled by a network of regulatory proteins, which includes a family of BMP co-receptors known as the repulsive guidance molecules (RGMs). The RGM that provides specificity to the iron signal in the liver, RGMc, is HJV. [7] In 2004 mutations involving the HJV gene were identified as the most common cause of juvenile hemochromatosis (HC). [8] SMAD6 and SMAD7, known as inhibitory SMADs, are able to modulate the BMP signal at the intracellular level. They interact with BMPRs, promoting their dephosphorylation or degradation or interfering with Smad proteins phosphorylation. SMAD6 also interferes with the formation of SMAD1/SMAD4 complex. [4]
In mice, iron administration has been shown to increase hepatic BMP signaling [9] while the administration of BMP increases hepcidin expression and reduces serum iron. [7; 10; 11] Various BMPs have proved to be capable of stimulating hepcidin synthesis in vitro, but a particularly important role is emerging for BMP6. [12; 13], which can physically interact with soluble HJV and increases hepcidin expression and reduces serum iron levels in mice. [12] Notably, Bmp6-null mice exhibit an hemochromatosis-like phenotype characterized by reduced hepcidin expression and tissue iron overload. [12; 13] These data point to BMP6 as an endogenous regulator of hepcidin expression and iron metabolism in vivo. Dietary iron has also been shown to induce Smad7 trascription in mice in a Smad4-dependent fashion, concordantly with Id1 and Hamp. [14] Moreover SMAD7 has been described as a specific inhibitor of hepcidin expression in vitro [6]. However the physiologic role of SMAD7 in human iron homeostasis is unknown.
Human hemochromatosis (HC) has been associated with the common C282Y polymorphism of HFE [15] or rare pathogenic mutations of TfR2,[16] HJV [8], FPN [17] and HAMP itself [18]. With a prevalence of approximately 5 in 1000 individuals of Northern European descent, HFE hemochromatosis is the most common inherited metabolic disorder in Caucasians. [19] HFE hemochromatosis is characterized by a failure to prevent excess iron release into the circulation from absorptive enterocytes and from reticuloendothelial cell stores, leading to progressive iron accumulation in other tissues with the potential for multi-organ damage and disease, including cirrhosis, diabetes, cardiomyopathy, hypogonadism, arthritis, skin pigmentation, and increased risk of cancer. [20] All forms of human HC are characterized by low or inadequate levels of hepcidin [21–24]. In fact, HFE, TfR2, and HJV all appear to be independent but complementary regulators of hepcidin synthesis in the liver. [20] In particular, we and others have recently shown that Hfe−/− mice exhibit an impairment in the bone morphogenetic protein (BMP)-SMAD signaling pathway. [25] [26] In mice, loss of the genes encoding the ligand BMP6 [12–13], the BMP co-receptor hemojuvelin (HJV) [27; 28], and the intracellular signaling molecule SMAD4 [10] all result in inappropriately suppressed hepcidin expression and tissue iron overload, supporting the central importance of the BMP6-HJV-SMAD signaling pathway in hepcidin regulation and hemochromatosis.
However, all data indicating the central role of BMPs in hepcidin regulation and showing an impaired BMP/SMAD signaling in HC, have been collected in mice. Since BMPs have pleiotropic activities which depend on a number of variables including the cellular context, different receptor-coreceptor combination or species specificity, [4] we investigated if also in humans expression of post-receptor BMP targets is associated with liver iron concentration (LIC) and such regulation is disrupted in HC, particularly HFE-HC.
MATERIAL AND METHODS
Patients
We investigated 30 subjects undergoing liver biopsy at the Center for Hemochromatosis at the University Hospital of Modena and the Department of Medicine II, Medical University of Innsbruck. The investigated population included: 14 subjects (8 M/6 F; age 51 ± 14)without iron abnormalities (LIC 424 ± 307μg/gram dry wt): n. 4 steatosis; n. 2 chronic hepatitis B; n. 2 chronic hepatitis C; n. 2 Wilson disease; n. 2 alcoholic liver disease; 10 patients (8 M/2 F; age 48 ±17) with untreated HFE HC (LIC 6179 ± 4111 μg/gram dry wt): n. 7 C282Y homozygotes; n. 2 C282Y/H63D compound heterozygotes; n.1 H63D/H63D homozygote); 6 patients (3 M/3 F; age 42 ±8) with non-HC iron overload (9467 ± 7152 μg/gram dry wt): n.3 A77D ferroportin disease; 2 aceruloplasminemia; n.1 parenteral iron overload. Informed consent was obtained by all patients and the study was approved by the local ethical committees. All patients underwent full clinical evaluation and iron assessment.
RNA analysis
RNA was extracted by the biopsy specimen using RNeasy Kit (Qiagen, Hilden, GmbH-Germany) according to the manufacturer’s instructions.
Quantitative RT-PCR
The cDNA was generated by reverse transcription of 400 ng of total human liver RNA with 0,5 μg random examer, 500 μM dNTPs, 3 mM MgCl2, 1U RNasin Ribonuclease Inhibitor and 1μl ImProm-II Reverse Transcriptase (Promega Corp., Madison, WI) in 20 μl of 1X reverse transcriptase buffer for 1 hour at 42°C. The primers for HAMP, BMP6, SMAD7, ID1 and GAPDH were as in Table 1. The real-time quantification of genes of interest were performed using two-step quantitative real-time RT-PCR. 1 μl of cDNA was amplified using SsoFast EvaGreen Supermix (Biorad, Hercules, CA) at 60°C for 40 cycles in iCycler Thermal Cycler (Bio-Rad Hercules, CA), and data were analyzed using iCycler iQ Optical System Software. The relative expression in each sample was calculated by a mathematical method based on the real-time PCR efficiencies using as reference GAPDH mRNA. All samples were assayed in triplicate. After 40 amplification cycles, threshold cycle values were automatically calculated and relative quantification was perform using standard curves constructed from serial dilution of PCR products covering a range of four orders of magnitude. Expression levels were normalized to the housekeeping gene GAPDH.
Table 1.
RT-PCR primers
| FORWARD | REVERSE | |
|---|---|---|
| hHAMP | 5′-TGTTTTCCCACAACAGACGGG-3′ | 5′-CGCAGCAGAAAATGCAGATGG-3′ |
| hID1 | 5′-ACGATCGCATCTTGTGTCGCTGAA-3′ | 5′-AGACCCACAGAGCACGTAATTCCT-3′ |
| hSMAD7 | 5′-CGATGGATTTTCTCAAACCAA-3′ | 5′-ATTCGTTCCCCCTGTTTCA-3′ |
| hGAPDH | 5′-TAGCCCAGGATGCCCTTGAG-3′ | 5′-GGACCTGACCTGCCGTCTAG-3′ |
Tissue Iron Content
Tissue specimens were analyzed for non-heme iron content as describe by Torrance and Bothwell [29] with slight modifications. Briefly, tissue samples were dry weighted, digested over-night at room temperature in nitric acid. Subsequently samples were digested for 2 hours at 100°C in Nitric Acid solution (Nitric Acid: H2O2; 1:0,25). Determination of iron content was performed using Mass Spectrometer ICP-MS X Series II (ThermoFischer Scientific, Waltham, MA).
Statistical analyses
Correlation between gene expression and LIC was assessed by linear regression analysis. Difference of gene expression among the three patient groups was evaluated by Kruskall-Wallis non-parametric test and differences between individual groups by Dunn’s Multiple Comparisons post-test analysis. P < 0.05 was used to determine statistical significance.
RESULTS AND DISCUSSION
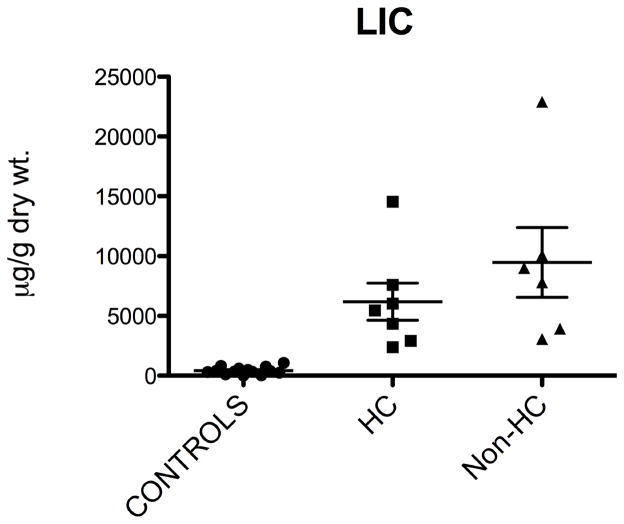
We have investigated patients with different iron status and correlated the expression of genes involved in the BMP/SMAD pathway controlling hepcidin with LIC. Figure 1 shows LIC values of our population: patients with HC and non-HC iron overload presented with a significant increase of LIC as compared to the control population. LIC was not significantly different between HC and non-HC iron overload patients.
Figure 1. Liver iron concentration in different patient groups.
Liver iron concentration (LIC) was assessed in liver biopsy specimens as specified in the Methods. Statistical difference between groups is reported (Dunn’s Multiple Comparisons post-test analysis)
Hamp expression is inappropriate to iron burden in HC
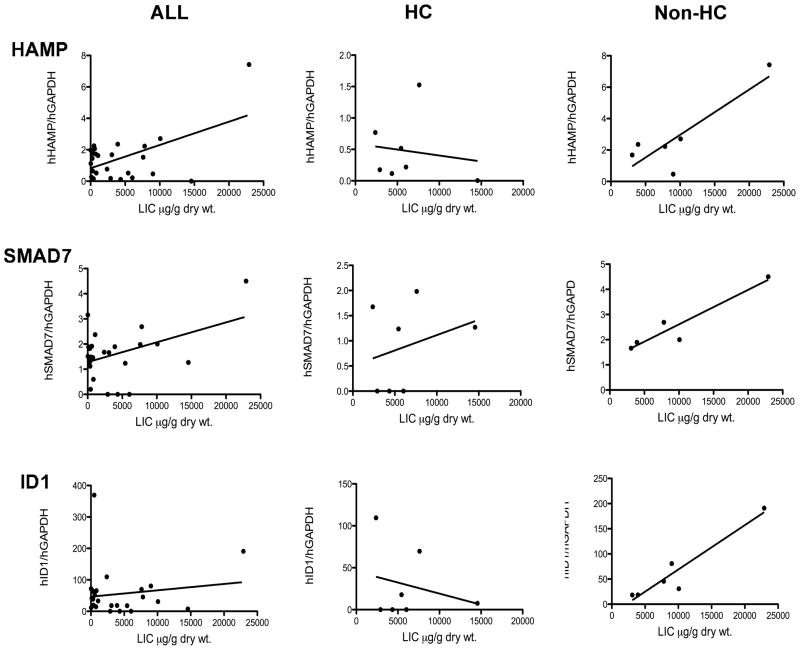
Figure 2 (upper panels) shows the relationship between HAMP mRNA expression and LIC in the different populations based on a linear regression analysis. It has already been established that in humans hepcidin expression in the liver is directly related to liver iron content [21] [22; 30]. In order to validate our population in terms of iron gene expression (particularly HAMP) as related to LIC, we first investigated whether HAMP expression correlated with LIC. We found a significant linear relationship between HAMP and LIC in the whole population (Figure 2, HAMP panel ALL). Then we wondered whether HAMP expression was inappropriate to LIC in our HC patients as already described by others in human HC [21] [22; 30]. The direct correlation between LIC and HAMP expression was lost in the sub-population of HC patients whereas it was still present in patients with non-HC iron overload (Figure 2, HAMP panel, HC vs. non-HC).
Figure 2. Correlation between LIC, HAMP and BMP target genes in human HC.
HAMP, SMAD7 and Id1 mRNA expression was assessed in liver biopsies as specified in the Methods. Linear regression analysis was performed and p value is reported for each assay.
Expression of BMP/SMAD target genes is inappropriate in human HC
In order to verify whether in human HC the BMP post-receptor signaling pathway is impaired as recently reported in mouse HC [25; 26], we investigated the expression of two known intracellular targets of BMPs, SMAD7 and Id1 [5] [7] in our HC patients and control groups.
In mice, SMAD7, a putative negative regulator of hepcidin transcription, is readily responsive to iron [7; 14]. In this study we found that in humans SMAD7 mRNA expression is strictly related to LIC, as shown by the significant correlation found in the whole population (Figure 2, SMAD7, panel ALL). However, this direct relationship was lost in HC patients, while still maintained in non-HC iron overload subjects (Figure 2, SMAD7, panel HC vs non-HC). Similar results were obtained when analyzing the expression of Id1 mRNA, another BMP target gene (Figure 2, Id1). Although the linear correlation between LIC and Id1 did not reach statistical significance in the whole studied population (likely due to the presence of the HC patients), there was a clear strong correlation between Id1 mRNA and LIC in non-HC patients; this correlation was negative, and almost inverse, in the HC group (Figure 2, Id1 panel HC vs non-HC).
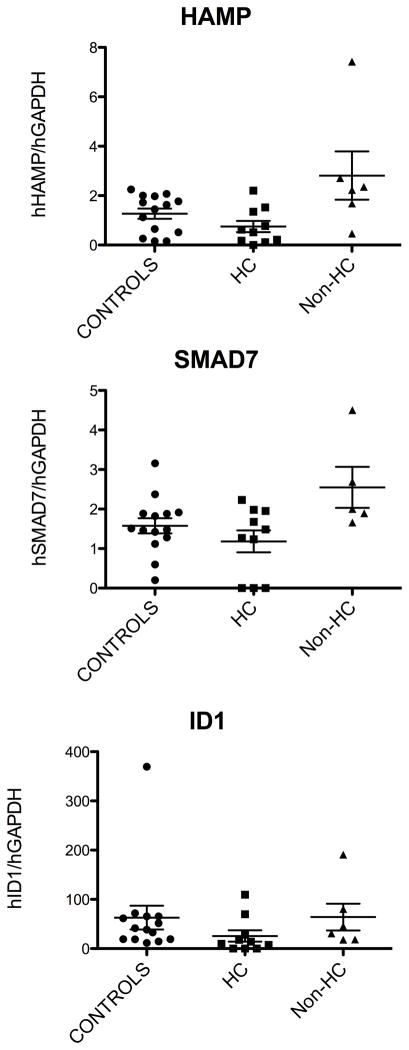
Even when we performed data sub-analysis and compared the mean value of iron gene expression between different groups, we found that neither SMAD7 nor Id1 mRNA expression were appropriately increased in patients with HC as compared to control subjects (Figure 3). In fact, Kruskall-Wallis non-parametric test showed that a significant change of the relevant gene expression occurred among the three groups for each tested gene (HAMP: p= 0.01; SMAD7: p=0.05; Id1: p=0.04). When comparing the individual groups by Dunn’s Multiple Comparisons post-test analysis, we found a significant difference only for HAMP expression in HC vs. non-HC patients (Figure 3, HAMP panel). This may be due to the small sample. Nevertheless, there was a constant trend for lower expression of all tested genes in the HC groups as compared to the control group or, more importantly, expression the tested genes did not increase in the HC-group as much as in the non-HC iron overloaded subjects, in spite of comparable liver iron burden.
Figure 3. Expression of HAMP and BMP target genes in human HC.
HAMP, SMAD7 and Id1 mRNA expression was assessed in liver biopsies as specified in the Methods. Difference of gene expression among the three different patient groups was assessed by Kruskall-Wallis non-parametric test and differences between individual groups by Dunn’s Multiple Comparisons post-test analysis.
While our manuscript was in preparation, Ryan et al. [31] have reported a lack of hepatic Id1 mRNA activation in human HC as compared to control, supporting our results. Surprisingly, in contrast with our study, SMAD7, which, like Id1 is a target gene of the BMP signaling, was increased in HC as compared to control. Smad7 is also induced by non-BMP signals, especially cytokines, such as interferongamma and TNFα [32]. Therefore, our ‘control’ population may have increased Smad7, because of upregulation of Smad7 by non-BMP signals arising from their disease state (e.g. chronic infections). This could explain the discrepancy with Ryan et al as regards to Smad7 [31]. However, in our non-HC patients, hepatic SMAD7 mRNA was consistently higher than in control subjects or HC patients indicating that hepatic iron burden per se may increase SMAD7 expression (as reported in mice [14]), and this does not occur in our HC patients with marked hepatic iron overload, Unfortunately, no data on expression of the tested genes in non-HC hemochromatosis were presented in the Ryan study and this may affect proper interpretation of the results and comparison with our data. In addition, not only is Smad7 responsive to BMP and non-BMP signals, but Smad7 also negatively regulates BMP-transduced signals and hepcidin expression [6]. Therefore, increased Smad7 might be expected to cause reduced hepcidin, as postulated by Ryan et al. [31]. However, when the BMP signaling pathway is activated by iron [14] or overexpression of a HFE transgene [33], SMAD7 mRNA is readily upregulated in the liver, concordantly with hepcidin. We hypothesize that SMAD7 operates in vivo by limiting hepcidin overexpression, and not by preventing any hepcidin activation. Timing and effects of this inhibitory activity in vivo are yet to be clarified.
Our study, although limited to a small number of patients, clearly indicates that in human hemochromatosis, as already demonstrated in mice, the inappropriate expression of hepcidin is associated with an impairment of the post-receptor BMP signaling pathway. While this is clearly expected in HJV-HC where a key protein of the BMP/SMAD regulatory pathway is missing, it is less obvious in HFE- HC. How in human HFE- HC a mutant HFE interferes with the BMP/SMAD pathway controlling HAMP expression in the liver remains unknown.
Acknowledgments
This study was supported by the Italian University and Research Council grant PRIN-08 and the Telethon 2010 grant to AP, the Austrian Science Fund FWF Project P195579 to HZ, NIH grants K08 DK075846 and RO1 DK087727 to JLB and RO1 DK-069533 and RO1 DK-071837 grants to HYL.
References
- 1.Finch CA. Iron Balance in Man. Nutr Rev. 1965;23:129–31. doi: 10.1111/j.1753-4887.1965.tb02108.x. [DOI] [PubMed] [Google Scholar]
- 2.Pigeon C, Ilyin G, Courselaud B, et al. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811–9. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- 3.Nemeth E, Tuttle MS, Powelson J, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–3. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 4.Corradini E, Babitt JL, Lin HY. The RGM/DRAGON family of BMP co-receptors. Cytokine Growth Factor Rev. 2009;20:389–98. doi: 10.1016/j.cytogfr.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazawa K, Shinozaki M, Hara T, Furuya T, Miyazono K. Two major Smad pathways in TGF-beta superfamily signalling. Genes Cells. 2002;7:1191–204. doi: 10.1046/j.1365-2443.2002.00599.x. [DOI] [PubMed] [Google Scholar]
- 6.Mleczko-Sanecka K, Casanovas G, Ragab A, et al. SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression. Blood. 2009 doi: 10.1182/blood-2009-09-238105. [DOI] [PubMed] [Google Scholar]
- 7.Babitt JL, Huang FW, Wrighting DM, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–9. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 8.Papanikolaou G, Samuels ME, Ludwig EH, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 9.Yu PB, Hong CC, Sachidanandan C, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang RH, Li C, Xu X, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Babitt JL, Huang FW, Xia Y, et al. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest. 2007;117:1933–1939. doi: 10.1172/JCI31342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andriopoulos B, Jr, Corradini E, Xia Y, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41:482–7. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meynard D, Kautz L, Darnaud V, et al. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet. 2009;41:478–81. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- 14.Kautz L, Meynard D, Monnier A, et al. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood. 2008;112:1503–9. doi: 10.1182/blood-2008-03-143354. [DOI] [PubMed] [Google Scholar]
- 15.Feder JN, Gnirke A, Thomas W, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 16.Camaschella C, Roetto A, Cali A, et al. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25:14–5. doi: 10.1038/75534. [DOI] [PubMed] [Google Scholar]
- 17.Njajou OT, Vaessen N, Joosse M, et al. A mutation in SLC11A3 is associated with autosomal dominant hemochromatosis. Nat Genet. 2001;28:213–4. doi: 10.1038/90038. [DOI] [PubMed] [Google Scholar]
- 18.Roetto A, Papanikolaou G, Politou M, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21–2. doi: 10.1038/ng1053. [DOI] [PubMed] [Google Scholar]
- 19.Merryweather-Clarke AT, Pointon JJ, Shearman JD, Robson KJ. Global prevalence of putative haemochromatosis mutations. J Med Genet. 1997;34:275–8. doi: 10.1136/jmg.34.4.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietrangelo A. Hereditary Hemochromatosis: Pathogenesis, Diagnosis and Treatment. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Bridle KR, Frazer DM, Wilkins SJ, et al. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361:669–73. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- 22.Gehrke SG, Kulaksiz H, Herrmann T, et al. Expression of hepcidin in hereditary hemochromatosis: evidence for a regulation in response to the serum transferrin saturation and to non-transferrin-bound iron. Blood. 2003;102:371–6. doi: 10.1182/blood-2002-11-3610. [DOI] [PubMed] [Google Scholar]
- 23.Nemeth E, Roetto A, Garozzo G, Ganz T, Camaschella C. Hepcidin is decreased in TFR2 hemochromatosis. Blood. 2005;105:1803–6. doi: 10.1182/blood-2004-08-3042. [DOI] [PubMed] [Google Scholar]
- 24.Kattamis A, Papassotiriou I, Palaiologou D, et al. The effects of erythropoetic activity and iron burden on hepcidin expression in patients with thalassemia major. Haematologica. 2006;91:809–12. [PubMed] [Google Scholar]
- 25.Corradini E, Garuti C, Montosi G, et al. Bone morphogenetic protein signaling is impaired in an HFE knockout mouse model of hemochromatosis. Gastroenterology. 2009;137:1489–97. doi: 10.1053/j.gastro.2009.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kautz L, Meynard D, Besson-Fournier C, et al. BMP/Smad signaling is not enhanced in Hfe-deficient mice despite increased Bmp6 expression. Blood. 2009;114:2515–20. doi: 10.1182/blood-2009-02-206771. [DOI] [PubMed] [Google Scholar]
- 27.Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. A mouse model of juvenile hemochromatosis. J Clin Invest. 2005;115:2187–91. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest. 2005;115:2180–6. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torrance JD, Bothwell TH. Tissue iron stores. In: Cook JD, editor. Methods in Hematology. Churchill Livingstone; New York, NY: 1980. p. 104. [Google Scholar]
- 30.Gehrke SG, Herrmann T, Kulaksiz H, et al. Iron stores modulate hepatic hepcidin expression by an HFE-independent pathway. Digestion. 2005;72:25–32. doi: 10.1159/000087400. [DOI] [PubMed] [Google Scholar]
- 31.Ryan JD, Ryan E, Fabre A, Lawless MW, Crowe J. Defective bone morphogenic protein signaling underlies hepcidin deficiency in HFE hereditary hemochromatosis. Hepatology. 2010 doi: 10.1002/hep.23814. [DOI] [PubMed] [Google Scholar]
- 32.Monteleone G, Del Vecchio Blanco G, Palmieri G, et al. Induction and regulation of Smad7 in the gastric mucosa of patients with Helicobacter pylori infection. Gastroenterology. 2004;126:674–682. doi: 10.1053/j.gastro.2003.11.048. [DOI] [PubMed] [Google Scholar]
- 33.Corradini E, Schmidt PJ, Meynard D, et al. BMP6 treatment compensates for the molecular defect and ameliorates hemochromatosis in Hfe knockout mice. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]