Abstract
Germline mutations in BRCA2 gene predispose women to early-onset familial breast and ovarian cancer. BRCA2 is a protein of multiple functions. In addition to its role in DNA double-strand breaks repair, BRCA2 also plays a role in stabilization of stalled DNA replication forks, cytokinesis, transcription regulation, mammalian gametogenesis, centrosome duplication, and suppression of cell proliferation. However, how BRCA2 mutations predispose women specifically to breast and ovarian cancer remains undefined. Here we found that BRCA2 binds and stabilizes MAGE-D1, a member of the MAGE gene family of proteins. Expression of BRCA2 and MAGE-D1 synergistically suppresses cell proliferation independently of the p53 pathway. Using two MAGE-D1 RNAis and two cell lines expressing low or undetectable levels of MAGE-D1, we further demonstrated that the expression of MAGE-D1 is required for BRCA2-mediated suppression of cell proliferation, indicating that MAGE-D1 is a downstream target of BRCA2 and that BRCA2 suppresses cell proliferation via stabilizing MAGE-D1. Importantly, MAGE-D1 protein expression was reduced in 6 out of 16 breast carcinoma cell lines tested as compared with untransformed immortal mammary epithelial cell lines, suggesting that suppression of MAGE-D1 expression may be involved in the tumorigenesis of a subset of sporadic breast cancers.
Introduction
Germ-line mutations in the breast cancer susceptibility gene BRCA2 confer susceptibility to familial early-onset breast and ovarian cancers (1-3). However, mutation of the BRCA2 gene in sporadic breast cancer rarely occurs (4). BRCA2 is a large protein of multiple functions. Extensive studies indicate that BRCA2 plays an important role in DNA damage repair by homologous recombination (5-13). BRCA2 also plays an important role in transcription regulation (13), centrosome duplication (14), cytokinesis (15), mammalian gametogenesis (16), and stabilization of stalled DNA replication forks (17). Importantly, BRCA2 inhibits tumor cell proliferation in vitro and tumor growth in vivo (18). However, how BRCA2 mediates the negative regulation of cell proliferation remains undefined.
The MAGE (MAGE for melanoma antigen) family contains a large group of proteins that harbor the MAGE homology domain, a well-conserved domain of about 200 amino acids (19). Members of the hMAGE-A, -B, and -C subfamilies are completely silent in most normal tissues, the exceptions being male germ cells, and, for some members of the subfamilies, the placenta (19). However, many members of the MAGE family are expressed in tumor cells of various histological types, and thus are of particular interest for anti-tumor immunotherapy. Members of the hMAGE-D, hMAGE-E, hMAGE-F, hMAGE-G, and hMAGE-H subfamilies exhibit a “ubiquitous” expression pattern; that is, they are expressed in many normal tissues at various levels (19). Recent studies indicate that MAGE-D1 is a regulator of apoptosis, transcription, and cell cycle progression (19). MAGE-D1 has been found to interact with Dlx/Msx homedomain proteins and is required for Dlx5-dependent transcription (20). Recently it was reported that MAGE-D1 cooperated with Necdin, another MAGE family protein, to regulate the function of Msx homeodomain proteins in cellular differentiation (21). MAGE-D1 also interacts with inhibitors of apoptosis proteins (IAPs), ubiquitin ligase praja1, axon guidance receptor UNC5H, and Ror receptor kinases (22-25). MAGE-D1 has also been reported to bind to p75 neurotrophin receptor and to mediate nerve growth-factor-dependent apoptosis (26).
In this study, using a conserved fragment of BRCA2 as bait, we identified MAGE-D1 as a BRCA2 binding protein and demonstrated that BRCA2 and MAGE-D1 interacted in vivo. Moreover, BRCA2 can stabilize MAGE-D1 and expression of BRCA2 and MAGE-D1 can suppress cell proliferation synergistically independently of p53 pathway. We also found that BRCA2 mediated-growth suppression is dependent on the expression of MAGE-D1, suggesting that MAGE-D1 is likely a downstream target of BRCA2. Importantly, MAGE-D1 expression was reduced in 6 out of 16 breast carcinoma cell lines tested as compared with untransformed immortal mammary epithelial cells, indicating that suppression of MAGE-D1 expression may disrupt the function of BRCA2 and play a role in the tumorigenesis of a subset of sporadic breast cancers.
Materials and Methods
Yeast two-hybrid constructs and screening
The cDNA encoding the residues 2392-2952 of BRCA2 was cloned into pGBT9 (Clontech, CA) to generate the bait plasmid, pGBT9-BRCA2-C2. Yeast two-hybrid screening was performed as described previously (27, 28).
Cell culture
Human embryonic kidney cell line 293T was cultured in DMEM medium supplemented with 10% fetal calf serum (Hyclone). 76NTert, 76R30, 184A1, 184B5, MCF-10A, 21MT-1, and 21MT-2 cells were grown in DFCI medium (29). HCC2157 cells were cultured in DFCI medium supplemented with 10% fetal calf serum. MCF-7, T47D, ZR-75-30, UACC812, MDAMB134, MDAMB361, MDAMB415, MDAMB453, and BT549 cells were cultured in α-MEM medium (Life Technologies) supplemented with 10% fetal calf serum. HCC70, HCC1143, HCC 1806, and 21NT cells were cultured in α-MEM medium supplemented with 10% fetal calf serum, 1μg/ml hydrocortisone (Sigma) and 12.5ng/ml EGF (Sigma).
Plasmid constructs and RNAi
The pCDEF3-Myc-MAGE-D1 construct was generated by cloning Myc-tagged full-length human MAGE-D1 coding sequences into pCDEF3 vector (30). Myc-tagged full-length human MAGE-D1 was generated by polymerase chain reaction (PCR) using MAGE-D1 EST clone (ATCC number 6555672) as a template and verified by sequencing. The pCR3. 1-4xMyc-MAGE-D1 construct was generated by cloning full-length human MAGE-D1 coding sequences fused with 4 copies of myc tags at its 5′ end into pCR3.1 vector. Two pSuper-MAGE-D1-RNAi constructs were generated to express two MAGE-D1 specific RNAi target sequences, ACCTGCGCCCTTCTCCCAA and CTACTAAAGTGGGCCCAAA (synthesized by TriLink BioTechnologies, Inc.). The Flag-tagged BRCA2 and its mutants were generated by PCR and restriction splicing, then cloned into pEF6/HisB vector. The details of construction are available upon request.
Polyclonal antibody generation
A DNA fragment representing amino acids 36-272 of human MAGE-D1 was cloned into pPROEX to produce a His-MAGE-D1 fusion protein. This protein was purified and used to immunize rabbits. The anti-MAGE-D1 antibody was affinity purified using His-tagged MAGE-D1 (36-272 aa).
In vivo binding of MAGE-D1 and BRCA2 or its mutants
The 293T cells were plated at 1×106 per 100-mm dish and transfected with 20 μg of pEF6-FLAG-tagged BRCA2 or its mutants and 10 μg of pCDEF3-Myc-MAGE-D1, either alone or in combination, using the calcium phosphate method. Forty-eight hours later, cell lysates were prepared in LSAB buffer (100mM Tris-HCl, pH. 7.5, 150 mM NaCl, 1% NP-40). Lysates were subjected to immunoprecipitation using anti-FLAG antibody (M2, Sigma, St. Louis, MO), followed by immunoblotting using anti-Myc (9E10) or anti-FLAG monoclonal antibodies. Enhanced chemiluminescence was used for detection (Amersham, Piscataway, NJ). To examine the binding of endogenous MAGE-D1 with the endogenous BRCA2, 293T cells and Capan-1 cells were harvested in RIPA buffer without SDS (100mM Tris-HCl, pH. 7.5, 150 mM NaCl, 1% Triton, 1% DOC). The cell lysate was subjected to immunoprecipitation using preimmune, anti-BRCA2-C2 (generated by immunizing rabbits with GST-BRCA2-C2), and anti-MAGE-D1 antisera, followed by Western blotting using anti-BRCA2 (BRCA2 ab1, Oncogene Science) and anti-MAGE-D1 antibodies.
Immunofluorescence and BrdU labeling
Cells grown on coverslips (Fisher) were transfected with pCDEF3, pEF6, pCDEF3-myc-MAGE-D1, and pEF6-Flag-BRCA2, alone or in combination, together with pBB14-GFP (a membrane-localized GFP fusion protein that can be retained after permeabilization (31)), using Fugene 6 (Roche) according to the manufacturer's instructions. Two days after transfection, the cells were incubated with BrdU for 1hr, then stained with anti-BrdU antibody (Roche) according to the manufacturer's instructions.
Results
Identification of MAGE-D1 as a BRCA2 binding protein and binding of MAGE-D1 to BRCA2 in vivo
In an attempt to identify novel BRCA2 binding proteins, BRCA2 cDNA encoding the most conserved region of BRCA2 (residues 2393-2952) was used as bait to screen a yeast two hybrid library prepared from normal mammary epithelial cells (strain 76N) as described previously (27, 28). Of the positive clones identified, two identical clones encoded the C terminal 116 aa of MAGE-D1, a member of the MAGE family of proteins.
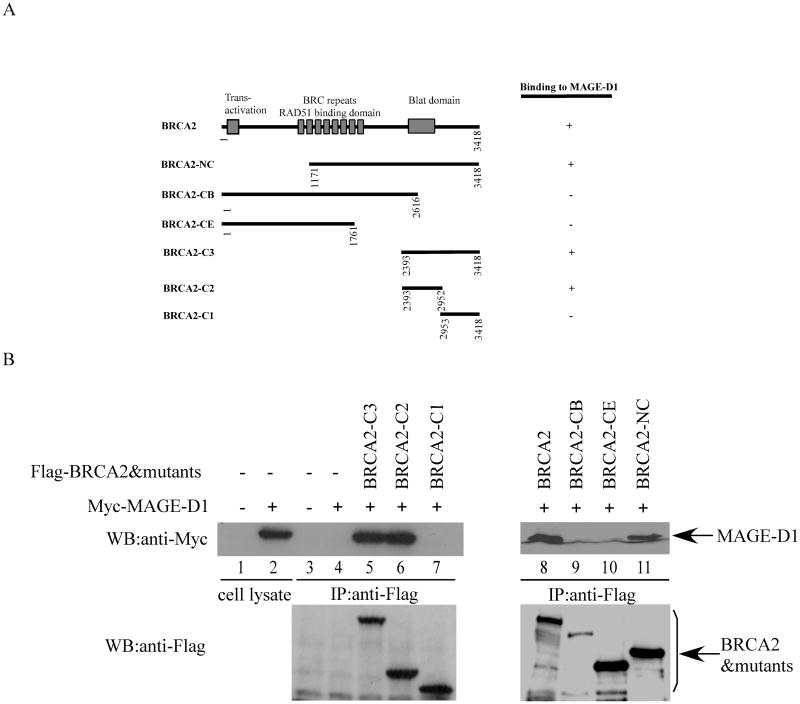
To confirm the interaction between BRCA2 and MAGE-D1 in vivo, full-length Myc-tagged-MAGE-D1 was transfected into 293T cells alone or together with full-length Flag-tagged-BRCA2. Forty-eight hours after transfection, the transfected cells were harvested in LSAB buffer and the BRCA2-MAGE-D1 complex was immunoprecipitated using anti-Flag monoclonal antibody. As shown in Figure 1B, MAGE-D1 can be specifically co-immunoprecipitated with BRCA2 using anti-Flag antibody (Fig. 1B, lane 8). Importantly, MAGE-D1 can also be co-immunoprecipitated with BRCA2 in RIPA buffer (without SDS), indicating that the interaction of MAGE-D1 and BRCA2 is unusually strong (data not shown). To define the binding domain of MAGE-D1 on BRCA2, one set of Flag-tagged BRCA2 deletion mutants as summarized in Fig.1A was co-transfected into 293T cells with full-length myc-tagged MAGE-D1 and co-immunoprecipitation was carried out using anti-Flag monoclonal antibody. We found that the only MAGE-D1-binding domain on BRCA2 localizes to a region between residues 2393 and 2952 in the BRCA2 protein (Fig. 1A, B).
Fig. 1. Binding of MAGE-D1 and BRCA2.
(A). Diagram of BRCA2 mutants. The binding of BRCA2 and its mutants to MAGE-D1 is summarized.
(B). In vivo binding of BRCA2 and MAGE-D1. 293T cells were transfected with Myc-tagged full-length MAGE-D1 and Flag-tagged BRCA2 and its mutants. The cells were harvested 48 hours after transfection in LSAB. BRCA2 and its mutants were immunoprecipitated using anti-Flag antibody. The immunoprecipitated protein complexes were fractionated and immunoblotted using anti-Myc antibody (9E10) to detect Myc-MAGE-D1. The blot was re-probed using anti-Flag antibody to detect BRCA2 and its mutants. A 4% (right panel) and a 7.5% (left panel) PAGE gels were used to fractionate the full-length BRCA2 and its mutants of varied molecular weight (from 58kd-380kd).
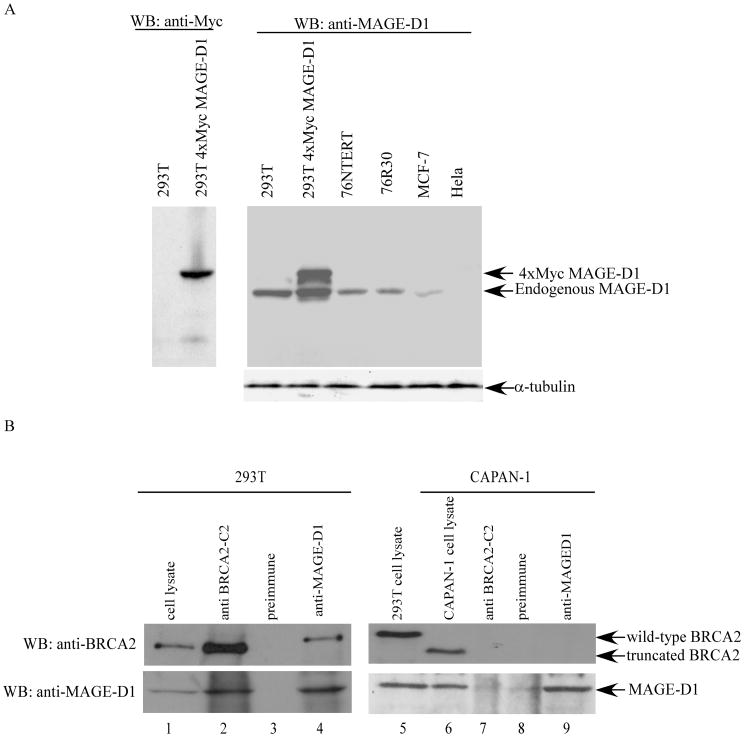
In order to detect the endogenous MAGE-D1 protein, an anti-MAGE-D1 antiserum was generated against a His-tagged fusion protein encoding the N-terminal 36-272 aa of MAGE-D1. To determine if this antiserum specifically recognizes the recombinant or endogenous MAGE-D1, cell lysates from 76NTert, 76R30, MCF-7, and HeLa cells, as well as 293T cells transfected with vector or 4×Myc-tagged MAGE-D1 were probed with affinity-purified anti-MAGE-D1 antibody. As shown in Fig. 2A, anti-MAGE-D1 antibody recognized the endogenous MAGE-D1 as an 85 kDa protein in 293T, 76NTert, 76R30, and MCF-7 cells. In the 4xMyc-tagged MAGE-D1 transfected 293T cells, anti-MAGE-D1 detected an additional 90 kDa protein corresponding to the 4×Myc-tagged-MAGE-D1 and anti-Myc antibody specifically detected the 90 kDa band at the same position (Fig. 2A). It is noteworthy that the expression of MAGE-D1 is undetectable in HeLa cells and is significantly lower in MCF-7 cells than in 293T, 76NTert, and 76R30 cells. These data clearly demonstrate that this anti-MAGE-D1 antibody can specifically detect endogenous MAGE-D1.
Fig. 2. Endogenous binding of MAGE-D1 and BRCA2 in vivo.
(A). Characterization of anti-MAGE-D1 antibody. Cell lysates (50μg of total protein) from 76NTert, 76R30, MCF-7, HeLa, 293T, and 293T cells transfected with 5 μg of pCR3.1 or 4xMyc-MAGE-D1 were fractionated by SDS-PAGE and immunoblotted using affinity-purified anti-MAGE-D1 or anti-Myc antibodies.
(B). In vivo binding of endogenous MAGE-D1 and endogenous BRCA2. 293T cells and CAPAN-1 cells were harvested in RIPA buffer (without SDS). The cell lysates were immunoprecipitated using preimmune, anti-BRCA2-C2 and anti-MAGE-D1 antisera. The immunoprecipitated protein complexes were fractionated and immunoblotted using anti-BRCA2 (BRCA2 ab1, Oncogene Science) and anti-MAGE-D1 antibodies.
Next, to determine the binding of endogenous BRCA2 and MAGE-D1, 293T and Capan-1 cells were harvested in RIPA (no SDS) buffer, and subjected to immunoprecipitation using anti-MAGE-D1, anti-BRCA2-C2 (raised against BRCA2-C2), or pre-immune antisera (as negative control). The immunoprecipitated proteins were analyzed by Western blotting using anti-BRCA2 (BRCA2 Ab1, Oncogene Science) and anti-MAGE-D1 antibodies. In 293T cells, anti-BRCA2-C2 immunoprecipitated both endogenous BRCA2 and endogenous MAGE-D1 (Fig. 2B, Lane 2). The anti-BRCA2-C2 was raised against aa 2393-2952 of BRCA2, a portion of BRCA2 deleted in Capan-1 cells (32-34). Therefore, the anti-BRCA2-C2 antibody was unable to immunoprecipitate the truncated BRCA2 in Capan-1 cells and consequently failed to co-immunoprecipitate the endogenous MAGE-D1 (Fig. 2B, Lane 7). These data also demonstrate that anti-BRCA2-C2 does not directly recognize MAGE-D1, validating that the co-immunoprecipitation of MAGE-D1 by anti-BRCA2-C2 is due to their specific interaction. Reciprocally, anti-MAGE-D1 immunoprecipitated the endogenous MAGE-D1, and consequently co-immunoprecipitated the endogenous BRCA2 in 293T cells, but not Capan-1 cells (Fig. 2B, lanes 4 and 9). In Capan-1 cells, anti-MAGE-D1 pulled down only MAGE-D1, not BRCA2 (Fig. 2B, lane 9), because the truncated BRCA2 in Capan-1 cells is devoid of the MAGE-D1 binding domain. It should be emphasized that all binding experiments were carried out in RIPA buffer (no SDS), indicating that the binding of BRCA2 and MAGE-D1 is very strong. These data conclusively demonstrate that endogenous BRCA2 can bind to endogenous MAGE-D1 in vivo.
BRCA2 stabilizes MAGE-D1
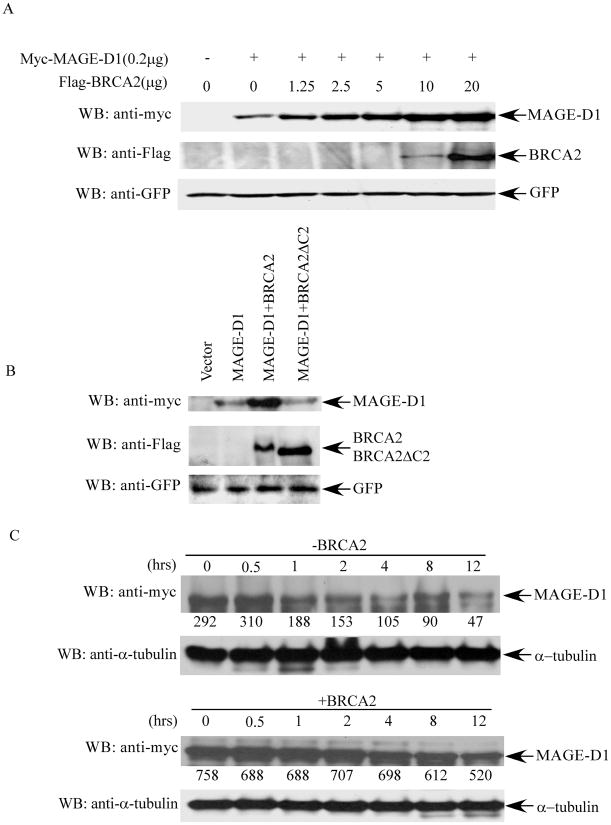
We found that the MAGE-D1 protein level was always much higher when MAGE-D1 was co-expressed with BRCA2 than when it was expressed alone. To investigate this observation further, a constant amount of MAGE-D1 was co-transfected with increasing amounts of BRCA2 into 293T cells, the MAGE-D1 protein level was examined by Western blotting using anti-Myc antibody. As shown in Fig. 3A, the MAGE-D1 protein level increased significantly and dose-dependently with increased amounts of transfected BRCA2, although BRCA2 was detectable only when more than 10μg of DNA was transfected. Furthermore, the level of MAGE-D1 did not increase when it was co-expressed with a BRCA2 mutant devoid of the MAGE-D1 binding domain (BRCA2ΔC2), indicating that the effect of BRCA2 on the MAGE-D1 protein level is due to their specific interaction (Fig. 3B). To delineate the mechanism of the elevated MAGE-D1 protein level upon BRCA2 co-expression, we examined the half-life of MAGE-D1 in 293T cells transfected with MAGE-D1 alone or with MAGE-D1 and BRCA2 together, using cycloheximide. As shown in Figure 3C, the half-life of MAGE-D1 was significantly increased up to more than 12 hours in the presence of BRCA2 co-expression, compared to about 2 hours in the absence of BRCA2 co-expression. This result clearly indicates that BRCA2 stabilizes MAGE-D1 protein.
Fig. 3. BRCA2 expression stabilizes MAGE-D1.
(A). MAGE-D1 protein levels when co-transfected with increasing amounts of BRCA2. 293T cells were transfected with 0.2 μg pBB14-GFP and 0.2 μg Myc-tagged MAGE-D1 alone or with increasing amounts of Flag-tagged BRCA2, as indicated. Cell lysates were prepared 48 hr after transfection and analyzed for Myc-MAGE-D1 and Flag-BRCA2 protein levels by immunoblotting using anti-Myc or anti-Flag antibodies. Immunoblotting using anti-GFP antibody was used to control transfection efficiency. (B). MAGE-D1 protein level when co-transfected with wild-type BRCA2 or BRCA2δC2 (BRCA2 mutant devoid of the MAGE-D1 binding domain). (C). Half-life of MAGE-D1. 293T cells were transfected with Myc-tagged MAGE-D1 alone (upper panel) or with Flag-tagged BRCA2 (lower panel). Cycloheximide (final concentration 10 μg/ml) was added 48 hr after transfection to inhibit nascent protein synthesis. Cell lysates were prepared at the indicated time points after the addition of cycloheximide, followed by immunoblotting using anti-Myc and anti-α-tubulin antibodies to determine the turnover of MAGE-D1 protein. The numbers under the panels are quantification analyses of the signals as obtained by use of LabWorks software (UVP, Inc, Upland, CA).
Expression of BRCA2 and MAGE-D1 synergistically suppresses human mammary epithelial cell proliferation independently of the p53 pathway
MAGE-D1 overexpression has been reported to inhibit progression of the cell cycle in several cell lines (26, 35). It has also been reported that expression of BRCA2 inhibits the proliferation of a pancreatic cancer cell line, Capan-1 cells, both in culture and in animals. Our finding that BRCA2 binds to MAGE-D1 and stabilizes MAGE-D1 suggests that BRCA2 and MAGE-D1 may function synergistically in the same pathway to suppress cell proliferation.
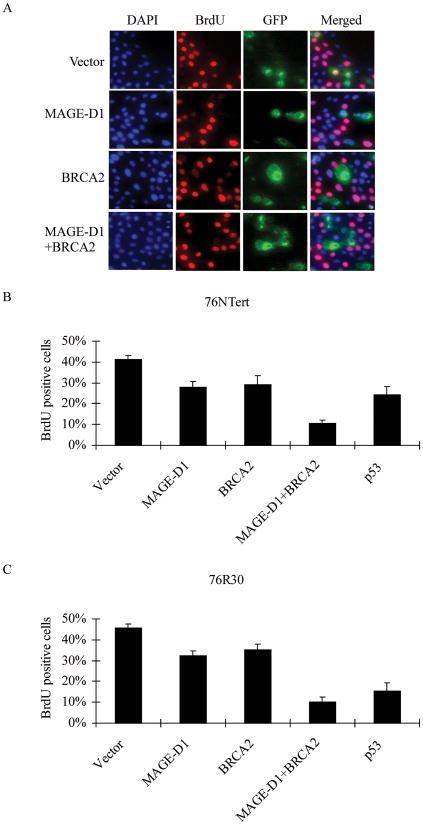
To examine the effect of co-expression of MAGE-D1 and BRCA2 on cell cycle progression, 76NTert cells (an hTERT immortalized human mammary epithelial cell line derived from 76N cells) were transfected with MAGE-D1 and BRCA2, alone or in combination, together with pBB14/GFP to indicate the transfected cells. The cells were pulse-labeled for 1 hr with bromodeoxyuridine (BrdU) at 48 hr after transfection and analyzed for BrdU incorporation by immunostaining using anti-BrdU antibody. As expected, expression of p53 suppressed 76Ntert cell proliferation, validating our cell proliferation assay. About 30% of cells transfected with MAGE-D1 or BRCA2 incorporated BrdU, while about 40% of vector-transfected cells incorporated BrdU, indicating that both MAGE-D1 and BRCA2 can inhibit progression of the cell cycle to a similar extent as can p53 (Fig. 4A, B). It is important to note that only 11% of cells overexpressing both MAGE-D1 and BRCA2 incorporated BrdU, indicating that BRCA2 and MAGE-D1 can synergistically suppress cell proliferation.
Fig. 4. BRCA2 and MAGE-D1 synergistically suppress cell proliferation.
(A) BrdU incorporation assay in 76NTert cells. 76NTert cells plated on coverslips in 24-wells plate were transfected with 0.5 μg MAGE-D1 and 1.5μg BRCA2, alone or in combination, together with pBB14-GFP (0.2 μg). The cells were incubated for 48 hr after transfection, then labeled with BrdU for 1 hr. The cells were fixed and stained using an anti-BrdU antibody. The DNA was stained with DAPI. Note that most of BRCA2 or MAGE-D1 transfected cells (GFP-positive cells) are negative for BrdU staining, while many vector- transfected cells (GFP-positive) or untransfected cells (GFP-negative) are positive for BrdU staining. (B and C) BRCA2 and MAGE-D1 synergistically suppress cell proliferation. 76NTert or 76R30 cells were transfected with MAGE-D1 and BRCA2, alone or in combination, as indicated, and assayed for BrdU incorporation as described in (A). The percentage of cells positive for BrdU staining was quantified in at least 300 transfected cells (GFP-positive cells). Each value represents the average and standard deviation from three independent experiments. The percentage of cells positive for BrdU staining was significantly lower in cells transfected with BRCA2, MAGE-D1, or MAGE-D1+BRCA2 than in cells transfected with vector alone (P < 0.05).
Wen et al. (35) recently reported that MAGE-D1 suppresses cell growth through a p53-dependent pathway in mouse embryonic fibroblasts. To test whether MAGE-D1- and BRCA2-mediated growth suppression is p53 dependent in mammary epithelial cells, we carried out the growth suppression assay by MAGE-D1 and BRCA2 in 76R30 cells, which are p53 null derivatives of 76N cells (36). As shown in Fig. 4C, the extent to which BRCA2 and MAGE-D1 suppressed proliferation of the p53 null 76R30 cells was similar to the extent as of their suppression of 76NTert cells, suggesting that BRCA2 and MAGE-D1 can retard cell cycle progression through a p53-independent pathway in human mammary epithelial cells.
BRCA2-mediated-suppression of cell proliferation is dependent on the expression of MAGE-D1
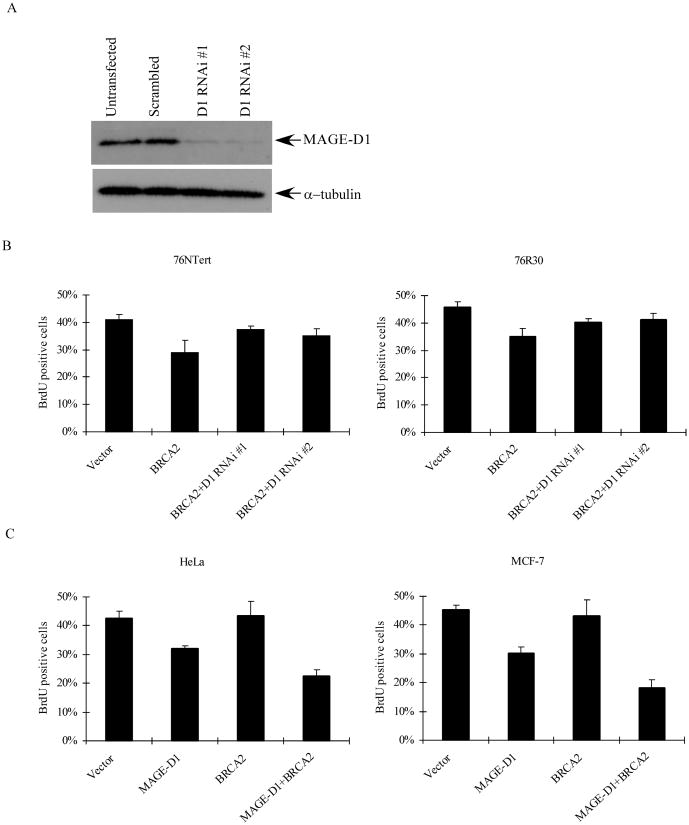
Since BRCA2 can stabilize MAGE-D1 and function synergistically with MAGE-D1 to suppress cell proliferation, it is likely that MAGE-D1 is a downstream target of BRCA2 and BRCA2 suppresses cell proliferation by stabilizing MAGE-D1. For this purpose, we used two RNAi constructs targeting the coding region of MAGE-D1 to silence the expression of MAGE-D1. As shown in Fig. 5A, the endogenous MAGE-D1 level in 293T cells was markedly reduced when these cells were transfected with both MAGE-D1 RNAi constructs (designated as D1 RNAi #1 and D1 RNAi #2). Densitometry analysis of Western blots indicated that more than 90% of MAGE-D1 was reproducibly depleted with use of these MAGE-D1 RNAi constructs. To determine whether MAGE-D1 is a downstream target of BRCA2, 76NTert and 76R30 cells were co-transfected with BRCA2 and the two MAGE-D1 RNAi constructs (Fig 5B). The proliferation of the transfected cells were analyzed by BrdU incorporation as described earlier. As shown in Fig.5B, both MAGE-D1 RNAi constructs significantly attenuated the effect of BRCA2 expression on cell proliferation (P< 0.05), indicating that BRCA2-mediated suppression of cell proliferation is dependent on the expression of MAGE-D1 and that MAGE-D1 acts downstream of BRCA2.
Fig. 5. BRCA2-mediated-suppression of cell proliferation is dependent on the expression of MAGE-D1.
(A) Western blotting analysis of 293T cells transfected with vector or MAGE-D1 RNAi constructs. 293T cells were transfected with vector or two MAGE-D1 RNAi constructs. After 48 hr, cells were harvested and fractionated through a SDS-PAGE gel, then blotted using anti-MAGE-D1 and anti-α-tubulin antibodies. (B). Silencing of MAGE-D1 expression by MAGE-D1 RNAi attenuated BRCA2 expression-mediated suppression of cell proliferation. 76NTert and 76R30 cells were transfected with 1.5μg BRCA2 alone or in combination with 0.5μg MAGE-D1 RNAi, together with pBB14-GFP (0.2μg), then BrdU incorporation was quantified as described in Fig. 4(B). The percentage of cells positive for BrdU staining was significantly lower in cells transfected with BRCA2 than in cells transfected with vector alone or BRCA2 plus MAGE-D1 RNAi (P < 0.05). (C) BRCA2 expression does not suppress cell proliferation in cells with a low level of MAGE-D1. HeLa and MCF-7 cells were transfected with BRCA2 and MAGE-D1 alone or in combination. BrdU incorporation was quantified as described in 4(B). The percentage of cells positive for BrdU staining was not lower in cells transfected with BRCA2 than in cells transfected with vector alone. However, the percentage of cells positive for BrdU staining was significantly lower in cells transfected with MAGE-D1 or BRCA2+MAGE-D1 than in cells transfected with vector alone. (P<0.05).
To further corroborate these observations, MCF-7 and HeLa cells, which express a low or undetectable level of MAGE-D1 (Fig. 2A), were transfected with BRCA2, either alone or in combination with MAGE-D1, and assayed for BrdU incorporation. We found that BRCA2 alone is unable to suppress the cell cycle progression of MCF-7 and HeLa cells (Fig. 5C), a finding that is consistent with our observation using MAGE-D1 RNAi. Co-expression of BRCA2 and MAGE-D1 significantly enhanced MAGE-D1-expression-mediated growth suppression, correlating with our observation that BRCA2 stabilizes MAGE-D1. Taken together, these data demonstrate that BRCA2 suppresses cell proliferation via stabilizing MAGE-D1.
MAGE-D1 protein expression in breast carcinoma cell lines
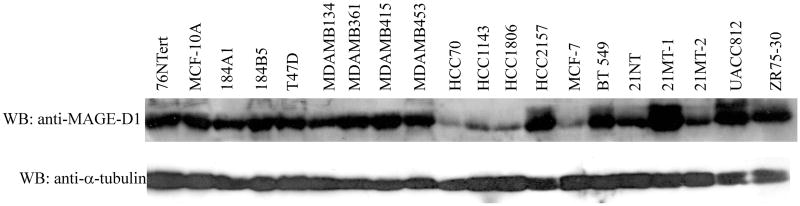
Since the expression of MAGE-D1 is required for BRCA2-mediated-suppression of cell proliferation, MAGE-D1 gene mutation or deletion, or suppression of its expression should also abrogate the function of BRCA2, which may be directly linked to tumorigenesis. To investigate if MAGE-D1 expression is altered in breast cancer, we examined the expression of MAGE-D1 in a large panel of breast cancer cell lines and four untransformed immortal mammary epithelial cell lines (76NTert, MCF-10A, 184A1, and 184B5). As shown in Fig. 6, similar levels of MAGE-D1 were readily detectable in the four untransformed mammary epithelial cell lines. However, in four breast carcinoma cell lines (HCC70, HCC1143, HCC1806, and MCF7), the MAGE-D1 protein level was significantly reduced as compared to levels in the untransformed immortal mammary epithelial cell lines (Fig 6). In 21NT and 21MT-2 cells, the MAGE-D1 level was also reduced, although to a lesser extent. This finding suggests that suppression of MAGE-D1 expression may be involved in the tumorigenesis of a subset of sporadic breast cancers.
Fig. 6. MAGE-D1 protein expression in untransformed immortal mammary epithelial cell lines and breast carcinoma cell lines.
Lysates prepared in sample buffer from the cell lines indicated were fractionated on a SDS-PAGE gel and blotted with anti-MAGE-D1 and anti–α–tubulin antibodies (loading control). 76NTert, MCF-10A, 184A1, and 184B5 are untransformed immortal mammary epithelial cell lines; the others are breast carcinoma cell lines.
Discussion
To identify molecules involved in BRCA2 functions, we performed yeast two-hybrid screening and identified MAGE-D1 as a BRCA2 binding protein. We demonstrated that endogenous BRCA2 and MAGE-D1 can interact in vivo. We also made the important observation that BRCA2 can stabilize MAGE-D1. Expression of BRCA2 and MAGE-D1 synergistically suppresses the proliferation of mammary epithelial cells independently of the p53 pathway. Furthermore, we demonstrated that MAGE-D1 acts downstream of BRCA2 and that BRCA2 suppresses cell proliferation via stabilizing MAGE-D1.
Using a set of BRCA2 mutants, we determined that the only MAGE-D1-binding domain on BRCA2 localizes to a region between residues 2393 and 2952 in the BRCA2 protein. It is notable that this region of BRCA2 encompasses almost the entire DSS1 binding domain (aa 2472-2957), the major part of BRCA2DBD (the DNA/DSS1-bindingdomain), and one of the FANCG binding domains (aa 2350–2545) (12, 37, 38). This region has been reported to play an important role in double-strand DNA breaks repair via homologous recombination and activation of the Fanconi anemia pathway (12, 37, 38). However, the functional implication of BRCA2 binding to MAGE-D1, DSS1, and FANCG at this region remain to be determined.
How BRCA2 stabilizes MAGE-D1 remains to be elucidated. It has recently been reported that BRCA2 can be co-purified with a ubiquitin E3 ligase complex, a holoenzyme complex termed BRCC that contains BRCA1, BARD1, BRCC36, and BRCC45. BRCA1 and BARD1 are proteins with a ring domain and exhibit ubiquitin ligase activities (18). Both BRCC36 and BRCC45 harbor sequence homologous to a subunit of the signalosome and proteasome complexes. A reconstituted complex containing BRCA1/BARD1/BRCC45/BRCC36 exhibits strong ubiquitin ligase activity (18). However, the way in which BRCA2 affects the ubiquitin ligase activity of BRCC holoenzyme has not been defined. Another recent study found that BRCA2 also interacts with a deubiquitinating enzyme, USP11 (39). An important BRCA2 binding protein, DSS1, was also found to be a component of the lid complex of the 26S proteasome (40, 41). The relationships among these BRCA2 containing complexes and their functions need to be elucidated. However, since all these BRCA2-containing complexes are likely to regulate protein degradation, it is reasonable to hypothesize that they regulate the stability of many target proteins. MAGE-D1 may be one of these target proteins and its stability may be regulated by these BRCA2-containing complexes.
It has been reported that expression of BRCA2 suppresses cell growth in vitro and tumor growth in vivo (18). However, the mechanism that BRCA2 suppresses cell proliferation remains undefined. Our finding that BRCA2 is able to suppress cell proliferation only in the presence of MAGE-D1 expression strongly indicates that BRCA2 exerts its function via stabilizing MAGE-D1. MAGE-D1 has been reported to suppress cell proliferation in several cell lines, including 293 cells, MEF cells, U2OS cells, and HepG2 cells (26, 35). Using p53 null MEF cells, it was found that MAGE-D1-mediated-suppression of cell proliferation occurs via the p53 pathway. However, our demonstration that MAGE-D1 can suppress cell proliferation in a p53 null mammary epithelial cell line, 76R30 cells (36), indicates that in mammary epithelial cells MAGE-D1 suppresses cell proliferation independently of the p53 pathway (Fig. 4). In corroboration of this finding, we found that MAGE-D1 is also capable of inhibiting the proliferation of HeLa cells, which express only a very low level of p53 due to the presence of HPV16 E6 (Fig. 5C). Furthermore, the previous report that MAGE-D1 can suppress the proliferation of 293 cells further supports our finding because the p53 in 293 cells is inactive due to the presence of E1B (26). This discrepancy may represent a cell-type-specific function of MAGE-D1.
At present, we can only speculate about how MAGE-D1 inhibits cell proliferation independently of the p53 pathway. Many MAGE-D1 partners have been identified recently, including homeodomain transcription factors Dlx5, Dlx7, Msx1, and Msx2, p75 neurotrophin receptor, axon guidance receptor UNC5H, inhibitors of apoptosis proteins (IAPs), ubiquitin ligase praja1, and Ror receptor kinases (19-26). Some of these MAGE-D1 binding partners may mediate the growth-suppressing function of MAGE-D1. Kuwajima T et al. recently reported that Necdin interacts with Msx homeodomain proteins via MAGE-D1 and promotes muscle differentiation by repressing cellular proliferation (21). It is likely that MAGE-D1cooperates with Necdin to suppress cell proliferation via Msx in mammary epithelial cells. Further studies are required to elucidate the proliferation-suppressing pathway used by MAGE-D1.
Multiple functions have been ascribed to BRCA2, linking it to pathways that inhibit progression to neoplasia. Heterozygous germline BRCA2 mutations predispose humans specifically to breast and ovarian cancers (1-3). However, BRCA2 is rarely mutated in sporadic breast and ovarian cancers (4). It is possible that distinct mechanisms targeting the BRCA2 pathways operate in sporadic cases. Recently, Hughes-Davies et al. demonstrated that EMSY, a BRCA2 interaction protein, was amplified in 13% of sporadic breast carcinomas and in 17% of sporadic ovarian carcinomas (42). These investigators suggested that overexpression of EMSY in sporadic breast and ovarian cancers may be functionally equivalent to BRCA2 mutation in familial cases (42). In this study, we found that BRCA2 can stabilize MAGE-D1 and suppress cell proliferation via MAGE-D1. MAGE-D1 acts downstream of BRCA2 and mediates the growth-suppressing function of BRCA2. Disruption of the function of MAGE-D1 by mutation, deletion, or suppression of expression should be functionally equivalent to BRCA2 mutations in disrupting BRCA2 function. Consistent with this hypothesis is our finding that, as compared MAGE-D1 expression in untransformed immortal cell lines, the expression level of MAGE-D1 was reduced in 6 of the 16 breast carcinoma cell lines we examined. This suggests that suppression of MAGE-D1 expression may be involved in the tumorigenesis of a subset of sporadic breast cancers. Biochemical and genetic analyses of MAGE-D1, a BRCA2 binding partner, would provide important insights into a pathway that is likely to be disrupted in human breast and ovarian carcinomas.
Acknowledgments
We thank Dr. Wen-Hwa Lee, Dr. Phang-Lang Chen, and Dr. Inder M. Verma for providing us with the full-length BRCA2 constructs. This work was supported by grants from the National Institutes of Health (1 R01 CA095221-01A1 and 1 R01 CA96986-01A1), and by the American Cancer Society (RSG-03-048-01) to Qingshen Gao.
References
- 1.Tavtigian SV, Simard J, Rommens J, et al. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet. 1996;12:333–7. doi: 10.1038/ng0396-333. [DOI] [PubMed] [Google Scholar]
- 2.Wooster R, Neuhausen SL, Mangion J, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science (Wash DC) 1994;265:2088–90. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 3.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature (Lond) 1995;378:789–92. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 4.Gayther SA, Pharoah PD, Ponder BA. The genetics of inherited breast cancer. J Mammary Gland Biol Neoplasia. 1998;3:365–76. doi: 10.1023/a:1018779830743. [DOI] [PubMed] [Google Scholar]
- 5.Sharan SK, Morimatsu M, Albrecht U, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature (Lond) 1997;386:804–10. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Silver DP, Walpita D, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–28. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 7.Connor F, Bertwistle D, Mee PJ, et al. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat Genet. 1997;17:423–30. doi: 10.1038/ng1297-423. [DOI] [PubMed] [Google Scholar]
- 8.Patel KJ, Yu VP, Lee H, et al. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–57. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 9.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–72. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 10.Xia F, Taghian DG, DeFrank JS, et al. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc Natl Acad Sci USA. 2001;98:8644–9. doi: 10.1073/pnas.151253498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellegrini L, Yu DS, Lo T, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature (Lond) 2002;420:287–93. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 12.Yang H, Jeffrey PD, Miller J, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science (Wash DC) 2002;297:1837–48. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 13.Milner J, Ponder B, Hughes-Davies L, Seltmann M, Kouzarides T. Transcriptional activation functions in BRCA2. Nature (Lond) 1997;386:772–3. doi: 10.1038/386772a0. [DOI] [PubMed] [Google Scholar]
- 14.Tutt A, Gabriel A, Bertwistle D, et al. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol. 1999;9:1107–10. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- 15.Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science (Wash DC) 2004;306:876–9. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- 16.Sharan SK, Pyle A, Coppola V, et al. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development. 2004;131:131–42. doi: 10.1242/dev.00888. [DOI] [PubMed] [Google Scholar]
- 17.Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003;17:3017–22. doi: 10.1101/gad.279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang SC, Shao R, Pao AY, Zhang S, Hung MC, Su LK. Inhibition of cancer cell growth by BRCA2. Cancer Res. 2002;62:1311–4. [PubMed] [Google Scholar]
- 19.Barker PA, Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res. 2002;67:705–12. doi: 10.1002/jnr.10160. [DOI] [PubMed] [Google Scholar]
- 20.Masuda Y, Sasaki A, Shibuya H, Ueno N, Ikeda K, Watanabe K. Dlxin-1, a novel protein that binds Dlx5 and regulates its transcriptional function. J Biol Chem. 2001;276:5331–8. doi: 10.1074/jbc.M008590200. [DOI] [PubMed] [Google Scholar]
- 21.Kuwajima T, Taniura H, Nishimura I, Yoshikawa K. Necdin interacts with the Msx2 homeodomain protein via MAGE-D1 to promote myogenic differentiation of C2C12 cells. J Biol Chem. 2004;279:40484–93. doi: 10.1074/jbc.M404143200. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki A, Masuda Y, Iwai K, Ikeda K, Watanabe K. A RING finger protein Praja1 regulates Dlx5-dependent transcription through its ubiquitin ligase activity for the Dlx/Msx-interacting MAGE/Necdin family protein Dlxin-1. J Biol Chem. 2002;277:22541–6. doi: 10.1074/jbc.M109728200. [DOI] [PubMed] [Google Scholar]
- 23.Williams ME, Strickland P, Watanabe K, Hinck L. UNC5H1 induces apoptosis via its juxtamembrane region through an interaction with NRAGE. J Biol Chem. 2003;278:17483–90. doi: 10.1074/jbc.M300415200. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda T, Suzuki H, Oishi I, et al. The receptor tyrosine kinase Ror2 associates with the melanoma-associated antigen (MAGE) family protein Dlxin-1 and regulates its intracellular distribution. J Biol Chem. 2003;278:29057–64. doi: 10.1074/jbc.M302199200. [DOI] [PubMed] [Google Scholar]
- 25.Jordan BW, Dinev D, LeMellay V, et al. Neurotrophin receptor-interacting mage homologue is an inducible inhibitor of apoptosis protein-interacting protein that augments cell death. J Biol Chem. 2001;276:39985–9. doi: 10.1074/jbc.C100171200. [DOI] [PubMed] [Google Scholar]
- 26.Salehi AH, Roux PP, Kubu CJ, et al. NRAGE, a novel MAGE protein, interacts with the p75 neurotrophin receptor and facilitates nerve growth factor-dependent apoptosis. Neuron. 2000;27:279–88. doi: 10.1016/s0896-6273(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 27.Gao Q, Srinivasan S, Boyer SN, Wazer DE, Band V. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol Cell Biol. 1999;19:733–44. doi: 10.1128/mcb.19.1.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Q, Kumar A, Srinivasan S, et al. PKN binds and phosphorylates human papillomavirus E6 oncoprotein. J Biol Chem. 2000;275:14824–30. doi: 10.1074/jbc.275.20.14824. [DOI] [PubMed] [Google Scholar]
- 29.Band V, Sager R. Distinctive traits of normal and tumor-derived human mammary epithelial cells expressed in a medium that supports long-term growth of both cell types. Proc Natl Acad Sci USA. 1989;86:1249–53. doi: 10.1073/pnas.86.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldman LA, Cutrone EC, Kotenko SV, Krause CD, Langer JA. Modifications of vectors pEF-BOS, pcDNA1 and pcDNA3 result in improved convenience and expression. Biotechniques. 1996;21:1013–5. doi: 10.2144/96216bm10. [DOI] [PubMed] [Google Scholar]
- 31.Kalejta RF, Brideau AD, Banfield BW, Beavis AJ. An integral membrane green fluorescent protein marker, Us9-GFP, is quantitatively retained in cells during propidium iodide-based cell cycle analysis by flow cytometry. Exp Cell Res. 1999;248:322–8. doi: 10.1006/excr.1999.4427. [DOI] [PubMed] [Google Scholar]
- 32.Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci USA. 1998;95:5287–92. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goggins M, Schutte M, Lu J, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–4. [PubMed] [Google Scholar]
- 34.Abbott DW, Freeman ML, Holt JT. Double-strand break repair deficiency and radiation sensitivity in BRCA2 mutant cancer cells. J Natl Cancer Inst. 1998;90:978–85. doi: 10.1093/jnci/90.13.978. [DOI] [PubMed] [Google Scholar]
- 35.Wen CJ, Xue B, Qin WX, et al. hNRAGE, a human neurotrophin receptor interacting MAGE homologue, regulates p53 transcriptional activity and inhibits cell proliferation. FEBS Lett. 2004;564:171–6. doi: 10.1016/S0014-5793(04)00353-9. [DOI] [PubMed] [Google Scholar]
- 36.Wazer DE, Chu Q, Liu XL, Gao Q, Safaii H, Band V. Loss of p53 protein during radiation transformation of primary human mammary epithelial cells. Mol Cell Biol. 1994;14:2468–78. doi: 10.1128/mcb.14.4.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol. 1999;19:4633–42. doi: 10.1128/mcb.19.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hussain S, Witt E, Huber PA, Medhurst AL, Ashworth A, Mathew CG. Direct interaction of the Fanconi anaemia protein FANCG with BRCA2/FANCD1. Hum Mol Genet. 2003;12:2503–10. doi: 10.1093/hmg/ddg266. [DOI] [PubMed] [Google Scholar]
- 39.Schoenfeld AR, Apgar S, Dolios G, Wang R, Aaronson SA. BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol Cell Biol. 2004;24:7444–55. doi: 10.1128/MCB.24.17.7444-7455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isono E, Saeki Y, Yokosawa H, Toh-e A. Rpn7 Is required for the structural integrity of the 26 S proteasome of Saccharomyces cerevisiae. J Biol Chem. 2004;279:27168–76. doi: 10.1074/jbc.M314231200. [DOI] [PubMed] [Google Scholar]
- 41.Sone T, Saeki Y, Toh-e A, Yokosawa H. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J Biol Chem. 2004;279:28807–16. doi: 10.1074/jbc.M403165200. [DOI] [PubMed] [Google Scholar]
- 42.Hughes-Davies L, Huntsman D, Ruas M, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–35. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]