Abstract
Many plants monitor day-length changes throughout the year and use the information to precisely regulate the timing of seasonal flowering for maximum reproductive success. In Arabidopsis thaliana, transcriptional regulation of the CONSTANS (CO) gene and posttranslational regulation of CO protein are crucial mechanisms for proper day-length measurement in photoperiodic flowering. Currently, the CYCLING DOF FACTOR proteins are the only transcription factors known to directly regulate CO gene expression, and the mechanisms that directly activate CO transcription have remained unknown. Here we report the identification of four CO transcriptional activators, named FLOWERING BHLH 1 (FBH1), FBH2, FBH3, and FBH4. All FBH proteins are related basic helix–loop–helix-type transcription factors that preferentially bind to the E-box cis-elements in the CO promoter. Overexpression of all FBH genes drastically elevated CO levels and caused early flowering regardless of photoperiod, whereas CO levels were reduced in the fbh quadruple mutants. In addition, FBH1 is expressed in the vascular tissue and bound near the transcription start site of the CO promoter in vivo. Furthermore, FBH homologs in poplar and rice induced CO expression in Arabidopsis. These results indicate that FBH proteins positively regulate CO transcription for photoperiodic flowering and that this mechanism may be conserved in diverse plant species. Our results suggest that the diurnal CO expression pattern is generated by a concert of redundant functions of positive and negative transcriptional regulators.
Keywords: photoperiodism, developmental transition, circadian clock
The precise alignment of flowering timing with season is crucial for successful reproduction. Various plants monitor photoperiod (day-length) changes throughout the year and use the information to regulate the timing of flowering (1). Photoperiodic flowering regulation is mediated by complex interactions between internal timekeeping mechanisms termed “circadian clocks” and “external environmental stimuli,” such as light and temperature (2). In Arabidopsis thaliana, the circadian-clock–regulated transcriptional regulation of the CONSTANS (CO) gene and the light-dependent posttranslational regulation of CO protein are the most crucial mechanisms for day-length measurement in photoperiodic flowering (3–6). In this mechanism, expression of the floral integrator gene FLOWERING LOCUS T (FT) is induced only when the CO protein expression coincides with the presence of light. FT protein synthesized in the leaf vasculature that moves to the shoot apical meristem (SAM) is thought to be the long-sought mobile floral induction signal “florigen” (7). At the SAM, FT binds to the bZIP transcription factor FD to initiate the expression of the floral meristem identity genes (8, 9). In addition, the CO/FT functional modules, as well as the daily expression patterns of CO homologs in flowering regulation, are widely conserved in many plant species (10, 11). Thus, to understand general seasonal flowering mechanisms, it is important to understand the regulatory mechanisms of the CO/FT module.
To induce FT under specific day-length conditions, the timing of daily CO transcription needs to be precisely regulated. Arabidopsis possesses a number of factors that regulate CO transcription, such as GIGANTEA (GI), FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1), RED AND FAR-RED INSENSITIVE 2 (RFI2), LONG VEGETATIVE PHASE 1 (LOV1), FIONA1 (FIO1), LIGHT-REGULATED WD1 (LWD1)/2, and CYCLING DOF FACTOR (CDF) proteins (12–21). The timing of the expression of all these genes is precisely regulated throughout the day by the circadian clock. Except for GI and FKF1, all of them are negative regulators of CO, and the mechanisms by which these proteins regulate CO transcription are largely unknown (12–21). Among these transcriptional regulators of CO, CDF1 is the only transcription factor known to directly bind to the CO promoter (15, 22), although LOV1 and FIO1 also contain DNA-binding motifs (18, 19). Overexpression of all CDF genes led to a decrease of CO transcripts and delayed flowering in long days (15, 21, 22). CDF1 was originally identified as an interacting protein of the FKF1 Kelch-repeat domain where a potential substrate for protein degradation binds (15). FKF1 absorbs blue light through its Light, Oxygen, or Voltage (LOV) domain (14, 22), and after light absorption, FKF1 binds to GI and functions as an SCF E3 ubiquitin ligase complex to target CDF proteins for degradation on the CO promoter (15, 21, 22). This mechanism enables plants to induce CO during late afternoon under long-day (LD) conditions. All CDF proteins are CO transcriptional repressors, and no transcriptional activators have been yet identified. To elucidate the mechanisms by which daily CO expression is controlled in combination with the CDF repressors, we attempted to identify additional CO regulators. Here we report a set of transcriptional activators of CO.
Results
FBH1 and FBH2 Bind to the CO Promoter.
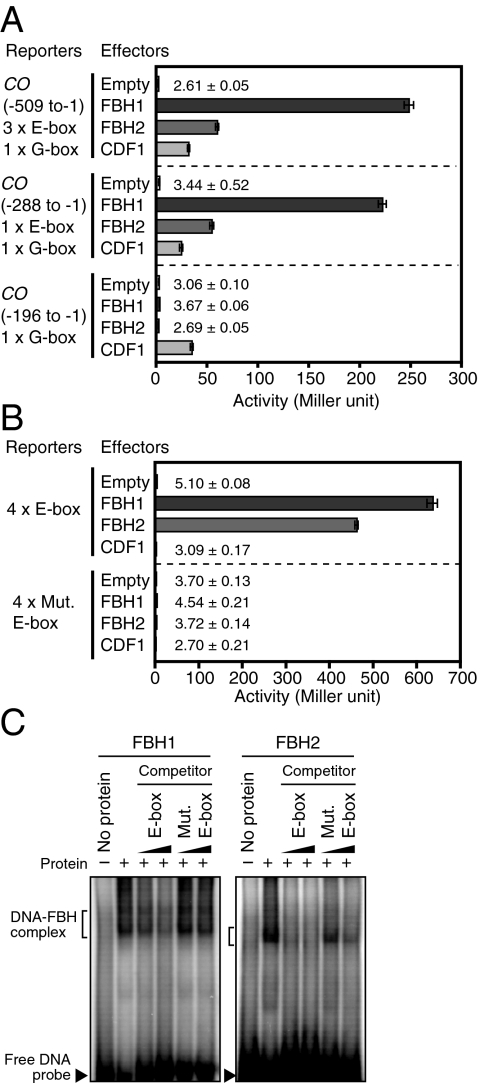
Because the expression of all known CO regulators is controlled by the circadian clock (6), we screened the clock-regulated transcription factor library using a yeast one-hybrid assay (23). Using a CO promoter fragment (500 bp), we found one transcription factor that strongly increased LacZ reporter activity (Fig. 1A). The transcription factor (At1g35460) belongs to the basic helix–loop–helix (bHLH) transcription factor family and has not been previously characterized. There is a close homolog (At4g09180) to the bHLH (74.4% identity over the entire amino acid sequences) in the Arabidopsis genome; therefore, we included the homolog in our assay. As these two genes encode bHLH proteins that affect flowering time (as shown later), we named them FLOWERING BHLH 1 (FBH1) and FBH2. Like FBH1, FBH2 increased LacZ activity, indicating that both proteins bind to the CO promoter in yeast (Fig. 1A). On the basis of the amino acid sequences of their bHLH domains, both proteins were predicted to preferentially bind to an E-box cis-element rather than a G-box (24). The CO promoter fragment that we used contains three E-box elements and one G-box element. Analysis of truncated CO promoter fragments revealed that the shorter promoter fragment (−288 to −1), which contains one E-box and one G-box element, was sufficient for the FBH-dependent induction of the LacZ reporter (Fig. 1A). However, both FBH proteins failed to induce LacZ expression when the shortest CO promoter fragment (−196 to −1) containing one G-box element and Dof-binding sites was used (Fig. 1A) (15). CDF1 could induce LacZ expression in the same yeast strain (Fig. 1A), indicating that the shortest CO promoter fragment is functional. These results suggest that FBH1 and FBH2 bind to the region that contains E-box elements. To verify that the E-box is an FBH binding site, we used a synthetic promoter that possesses four repeats of the E-box elements derived from the CO promoter (named as “4× E-box”) to control LacZ expression. Both FBH1 and FBH2 increased reporter activity (Fig. 1B). However, when the E-box elements were mutated (“4× Mut. E-box”) (24), the FBHs no longer induced reporter expression. In addition, we further confirmed the direct binding of both FBH proteins to the same E-box elements by electrophoretic mobility shift assay (EMSA) (Fig. 1C). These results suggest that FBH1 and FBH2 bind to the E-box elements in the CO promoter in vivo.
Fig. 1.
FBH1 and FBH2 bind to the CO promoter. (A) Interaction of FBH1 and FBH2 with CO promoter in yeast. Bars represent β-galactosidase enzyme activities (Miller units) controlled by CO promoter fragments. The numbers on the left denote the region of the promoter included in each reporter construct (the CO transcription start site, +1). The number of E-box and G-box elements in each fragment is indicated. CDF1 binds to the Dof-binding site (−173 to −135) on the CO promoter (15). (B) Interaction of FBH1 and FBH2 with E-box. The 20 bp of the CO promoter fragment (−239 to −219) encompassing the E-box element (with or without a mutation) was repeated four times and then fused to the minimum promoter to drive LacZ expression. All data in A and B represent means ± SEM (n = 15). (C) EMSA of FBH1 and FBH2 proteins. The four E-box-repeat fragment used in B was radioactively labeled. The same fragment and the mutated E-box-repeat fragment were used as nonlabeled competitors in 1:20 and 1:100 ratios (labeled vs. nonlabeled DNA).
FBH1 and FBH2 Are Activators in the CO/FT Photoperiodic Flowering Pathway.
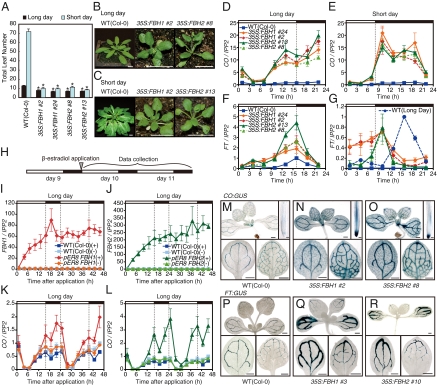
We postulated that if FBH1 and FBH2 are involved in CO transcriptional regulation in vivo, overexpression of FBHs could change CO expression levels, which consequently would alter flowering time. Therefore, we analyzed the flowering phenotype of FBH1 and FBH2 overexpressors (35S:FBH1 and 35S:FBH2, Fig. S1 A–D) under LD and short-day (SD) conditions. FBH1 and FBH2 overexpressors showed a distinct early flowering phenotype regardless of photoperiod (Fig. 2 A–C), which resembles that of the CO overexpressors (25). This result suggests that the FBH overexpressors may have increased levels of CO. As predicted, the CO expression levels were elevated in the 35S:FBH lines in LD and SD (Fig. 2 D and E and Fig. S1 O–Q), indicating that both FBH1 and FBH2 induce CO transcription. Interestingly, even though the peak CO levels in the 35S:FBH lines were almost 20 times higher than those in wild-type plants, the daily CO expression patterns in 35S:FBHs were very similar to the wild-type CO patterns in LD and SD (compare the CO patterns in Fig. 2 D and E with those in Fig. S1 E and F). Because the FBH transcripts are constitutively expressed at high levels throughout the day in 35S:FBHs, this result suggests that the transcriptional activity of FBHs may change throughout the day.
Fig. 2.
FBH1 and FBH2 control CO expression levels. (A) Flowering phenotypes of plants overexpressing FBH1 (35S:FBH1) and FBH2 (35S:FBH2) under different photoperiods. Error bars depict SEM (n = 6). Asterisks (*) denote significant difference (P < 0.001) between each overexpressor and wild-type plants. The experiment was repeated at least twice, and similar results were obtained. (B and C) Representative pictures of 35S:FBH plants in LD (B) and SD (C). The pictures were taken just after the plants bolted. (B) Wild type, 27 d old; 35S:FBH1, 18 d old; and 35S:FBH2, 18 d old in LD. (C) Wild type, 70 d old; 35S:FBH1, 28 d old; and 35S:FBH2, 28 d old in SD. (Scale bars,10 mm.) (D–G) Daily expression patterns of CO (D and E) and FT (F and G) in 35S:FBH1, 35S:FBH2, and wild-type plants in LD and SD. The FT expression pattern of wild type in LD (blue dashed line) was superimposed on SD data (G). All of the results (D–F), except G, were normalized to the highest values in the wild-type sample (the maximum value of wild type was set to 1). FT levels in SD (G) were normalized to the peak FT expression value in the wild type in LD. (H) Seedlings that possess pER8-FBH1 or pER8-FBH2 constructs were treated with β-estradiol at day 10 at the onset of light (ZT 0). The arrowhead indicates the start time point of β-estradiol application. Seedlings were harvested starting at 1 h after the onset of light (ZT 1) and then at 3-h intervals for 2 d. (I–L) FBH1, FBH2, and CO mRNA expression in wild-type plants and pER8-FBH1 and pER8-FBH2 transgenic plants after β-estradiol application. The samples treated with and without β-estradiol are indicated by (+) and (−) symbols, respectively. FBH1 and FBH2 levels were normalized to the average value in the wild-type (−) sample [the average value from all of the wild-type (−) time points was set to 1]. The CO level was normalized to the maximum value of the wild-type (−) sample [the maximum value of wild type (−) was set to 1]. Values represent means ± SEM from three biological replicates in D–G and in I–L. The bars above the graphs represent light conditions: white bars, light periods; black bars, dark periods. (M–R) Spatial expression patterns of CO and FT gene in 35S:FBH plants. Twelve-day-old wild-type (M), 35S:FBH1 (N), and 35S:FBH2 (O) plants carrying the CO:GUS reporter gene and wild-type (P), 35S:FBH1 (Q), and 35S:FBH2 (R) plants carrying the FT:GUS reporter gene were analyzed. Whole-mount staining of seedlings, cotyledons, and the first set of leaves are shown with scale bars (0.5 mm). Staining of root tips is shown with scale bars (0.1 mm).
To determine the potential contribution of other CO regulators to CO expression in the FBH overexpressors, we surveyed the daily expression patterns of known CO regulator genes, such as GI, FKF1, CDF1, and CDF2 (13, 15, 21, 22). Except for a slight reduction in the peak expression of GI, FKF1, CDF1, and CDF2 in the 35S:FBH lines, the expression patterns of these genes resembled the 35S:FBHs and wild-type plants in LD and SD (Fig. S1 G–N). Our results indicated that elevated levels of FBHs directly and specifically increased the amount of CO transcripts.
To elucidate potential causes of the early flowering phenotype of the FBH overexpressors, we investigated expression levels of the major flowering-time regulators, which function downstream of CO. The abundance of FT mRNA was also highly increased in the FBH overexpressors in LD and SD (Fig. 2 F and G and Fig. S1R). FLOWERING LOCUS C (FLC) expression was slightly reduced, and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1) expression was not altered in the 35S:FBH lines (Fig. S1 S and T) (25, 26). These results suggest that elevated FT levels may induce early flowering in the 35S:FBH lines. To genetically evaluate this possibility, we introduced the ft mutation into the 35S:FBH1 line. The 35S:FBH1 ft line showed an obvious late-flowering phenotype, which is similar to that of ft, in LD and SD (Fig. S2 A–C). This result supports the notion that the early flowering phenotype of 35S:FBH1 is mainly due to the increase in FT levels, which is likely caused by the elevated levels of CO.
We demonstrated that the elevated levels of FBH1 and FBH2 are directly associated with increased CO expression. To further analyze the FBH-dosage–dependent induction of CO, we used the estradiol-mediated FBH inducible system (pER8-FBH1 and pER8-FBH2) (27). β-Estradiol was applied to 10-d-old transgenic and wild-type seedlings, and FBH1, FBH2, and CO gene expression was analyzed for 2 d (Fig. 2H). CO expression increased only in plants in which FBH1 or FBH2 expression was induced (Fig. 2 I–L). This result further indicates that the amounts of FBH1 and FBH2 control the amplitude of daily CO oscillation.
Because CO is expressed mainly in vascular tissues (Fig. 2M) (28, 29), we analyzed whether the FBH overexpression affects the CO spatial expression pattern using the CO promoter-fused β-glucuronidase (CO:GUS) reporter (28). CO:GUS activity in the 35S:FBH seedlings was higher than that in the wild-type CO:GUS plants but was still restricted mainly to the vascular tissues (Fig. 2 M–O), even though both FBH1 and FBH2 are ubiquitously expressed (Fig. S1 A–D). In addition, ectopic GUS activity was observed in stomata in leaves and root tips (Fig. 2 M–O and Fig. S2 D and E). These results indicate that FBH1 and FBH2 activity is somehow restricted to the vascular tissue. We also analyzed the effects of FBH overexpression on the spatial pattern of FT (Fig. S2 F and G) and found that GUS activity was strongly enhanced in the 35S:FBH lines, but the tissue-specific expression pattern of FT was not altered (Fig. 2 P–R). This could be due to the increased levels of CO without a large alteration of its spatiotemporal expression pattern in these lines.
FBH1 Binds Near the Transcription Start Site of the CO Promoter in Vivo.
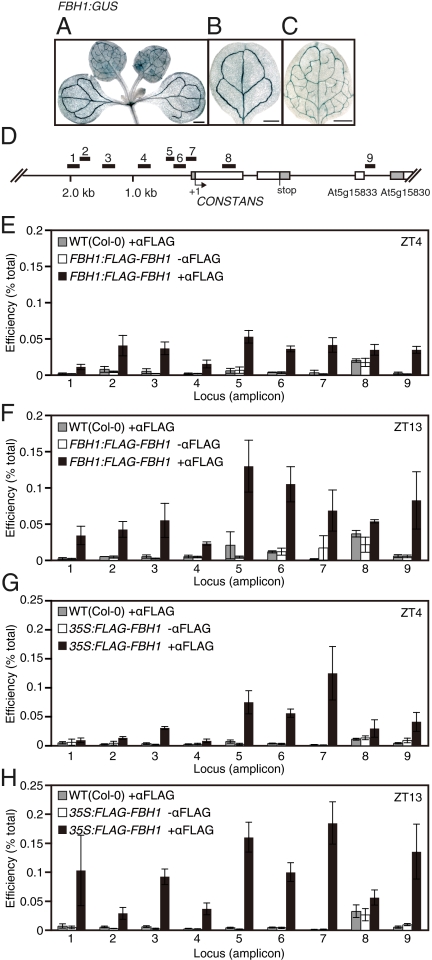
To understand the mechanism of FBH-dependent CO regulation, we examined the spatial expression pattern of FBH1 by analyzing the FBH1-promoter–controlled GUS expression pattern (FBH1:GUS). We presumed that if FBH1 is a CO regulator, its spatial expression pattern should overlap with the CO pattern. FBH1:GUS activity was predominantly detected in the vascular tissues (Fig. 3 A–C), validating our prediction.
Fig. 3.
FBH1 associates with the CO promoter. The spatial expression pattern of FBH1 was determined by histochemical staining of GUS activity in FBH1:GUS plants. Whole-mount staining of a seedling (A), a cotyledon (B), and a first leaf (C) are shown. (Scale bars, 0.5 mm.) (D) Schematic representation of the CO locus and the locations of nine amplicons for ChIP analysis. White and gray boxes represent exons and either 5′- or 3′-UTR. The At5g15833 gene encodes microRNA. (E–H) Binding of FLAG-FBH1 to the CO promoter in vivo. Two-week-old LD-grown seedlings, which possess either FBH1:FLAG-FBH1 (E and F) or 35S:FLAG-FBH1 constructs (G and H) and the wild-type plants were harvested at 4 and 13 h after the onset of light (ZT 4 and ZT 13). ChIP assays were performed using FLAG-FBH1 plants with the anti-FLAG antibody, FLAG-FBH1 plants without the antibody, and wild-type plants with the anti-FLAG antibody. The amount of immunoprecipitated DNA was quantified by qPCR using primers specific to each amplicon. Values represent the average immunoprecipitation efficiencies (%) against the total input DNA ± SEM of at least three biological replicates.
Next, we investigated whether FBH1 directly associates with the CO promoter in vivo using a chromatin immunoprecipitation (ChIP) assay. For the ChIP assay, we used transgenic plants expressing a FLAG-tagged FBH1 regulated by the FBH1 promoter (FBH1:FLAG-FBH1) and 35S:FLAG-FBH1 plants. First, we confirmed that CO levels were elevated in the FBH1:FLAG-FBH1 and 35S:FLAG-FBH1 lines in a dosage-dependent manner, indicating that the FLAG-FBH1 protein is functional (Fig. S3 A–F). To investigate FBH1 binding to the CO promoter, we harvested LD-grown plants at Zeitgeber time 4 (ZT4) when CO expression is at the trough level and at ZT 13 when daytime CO expression is at its peak. We analyzed the FLAG-FBH1–specific enrichment of DNA fragments on different CO locations (amplicons 1–9; see Table S3 for detailed information) using quantitative PCR (qPCR) (Fig. 3D). In the FBH1:FLAG-FBH1 plants, FLAG-FBH1–specific enrichment was detected from all chromatin samples harvested at ZT 4 and ZT 13 (Fig. 3 E and F) with the highest level in amplicons 5, 6, and 7, which are adjacent to the CO transcriptional start site. Amplicon 5 (position: −430 to −273) and amplicon 6 (position: −301 to −89), both of which contain one E-box, largely overlap with the region important for the FBH-dependent transcription in yeast (Fig. 1). Amplicon 7 (−89 to +66) also contains one E-box in the 5′-UTR of CO. Comparison of the results derived from both time points revealed a higher enrichment in the sample harvested at ZT 13 (Fig. 3 E and F), which coincides with up-regulation of the CO transcript (Fig. S1E). Similar trends were observed when we used the 35S:FLAG-FBH1 plants (Fig. 3 G and H). Together with our yeast one-hybrid and EMSA results, we propose that FBH1 binds to the CO chromatin to regulate CO transcription in vivo. Because FBH1 protein similarly accumulated throughout the day in LD and SD (Fig. S3 G–I), FBH1 may require some posttranslational modification or some other unknown proteins to induce CO expression.
FBH1 Homologs Have an Overlapping Function as CO Activators.
To complement our overexpression analysis, we analyzed the mutant phenotype. Because FBH1 and FBH2 have 74% amino-acid-sequence identity and the overexpressors have similar phenotypes, we aimed to obtain an fbh1 fbh2 double mutant to analyze the loss-of-function phenotype. As only the FBH2 T-DNA insertion mutant (fbh2-1) was available in public collections (Fig. S3J), we generated independent fbh1 fbh2 double-mutant lines in which FBH1 mRNA was down-regulated by two different artificial microRNA (amiRNA) constructs (amiRFBH1-1 fbh2-1 and amiRFBH1-2 fbh2-1). When we analyzed CO and FT expression in the amiRFBH1 fbh2 lines, we did not detect any differences compared with wild-type plants (Fig. S3 K–R). This result may indicate either that the 10–30% of remaining FBH1 mRNA is enough to maintain the normal mechanisms of CO regulation or that there are yet other proteins (i.e., other relatively closely related bHLH proteins) that function redundantly with FBH1 and FBH2 to compensate for the loss of both genes.
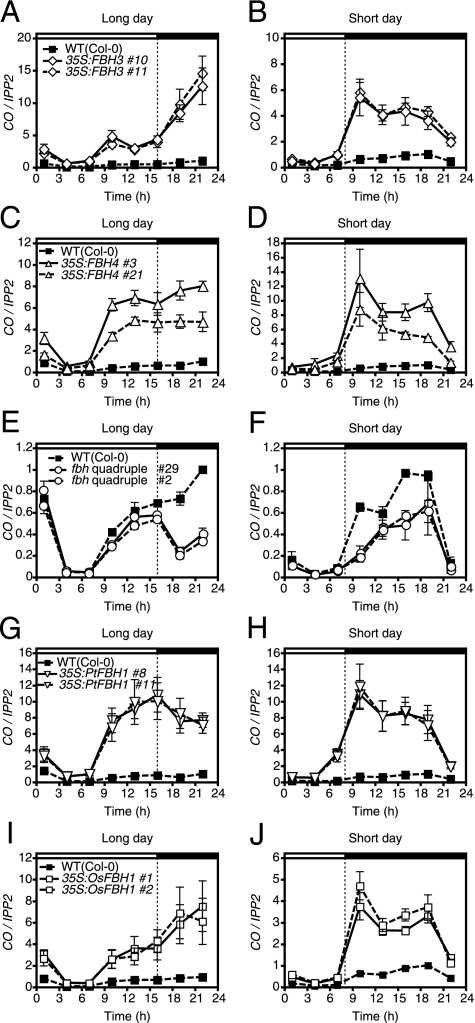
Therefore, we expanded our search for FBH1 (or FBH2) homologs. On the basis of previous phylogenetic analyses, there are four more bHLH genes in the same clade as FBH1 and FBH2 (24, 30). The deduced amino acid sequences of these four genes contain highly conserved bHLH domains; however, they have diverse sequences other than the bHLH domains. We successfully cloned three of these bHLHs (At1g51140, At2g42280, and At1g05805) and tested whether they could also induce early flowering when overexpressed. Overexpression of At1g51140 and At2g42280 (named FBH3 and FBH4) also caused an early flowering phenotype (Fig. S4 A–E). This is likely due to a high amount of FT expression (Fig. S4 F–M) caused by increased CO expression in LD and SD (Fig. 4 A–D). Similar to the FBH1 and FBH2 overexpressor phenotypes, the spatial and temporal expression patterns of CO were largely restored in the 35S:FBH3 and 35S:FBH4 lines (Fig. 4 A–D and Fig. S4 N–P). In addition, yeast one-hybrid analysis demonstrated that FBH3 and FBH4 bind to the same CO promoter regions through the E-box elements (Fig. S4 Q and R).
Fig. 4.
FBH1 homologs regulate CO transcription. (A–D) CO mRNA expression in 35S:FBH3, 35S:FBH4, and wild-type plants in LD and SD. (E and F) CO mRNA expression in two independent fbh quadruple mutants and wild-type plants in LD and SD. (G–J) CO mRNA expression in Arabidopsis plants constitutively expressing poplar FBH (35S:PtFBH1), rice FBH (35S:OsFBH1), and wild-type plants in LD and SD. All of the results were normalized to the highest value in the wild-type sample. Values represent mean ± SEM from three biological replicates for all experiments.
Temporal expression pattern analysis of all four FBH genes revealed that they are expressed throughout the day in LD and SD (Fig. S5 A–H). FBH4 (and possibly FBH1) transcription showed a diurnal oscillation pattern under constant light conditions (Fig. S5 B and H), indicating the involvement of circadian-clock regulation. Promoter:GUS analysis revealed that the FBH3 promoter is active mainly in the vascular tissues and that FBH4 is expressed in the stomata as well as in leaf vascular tissues (Fig. S5 I–K). Together with the expression pattern analyses, our results indicate that FBH3 and FBH4 have similar functions to FBH1 and FBH2 with regard to CO transcriptional regulation.
Because our results indicated that the four FBH proteins might have redundant functions, we analyzed the phenotype of fbh1 fbh2 fbh3 fbh4 quadruple mutants. To generate the fbh quadruple mutants, we used the FBH1 amiRNA construct, fbh2-1 (Fig. S3J), the FBH3 T-DNA insertion line (Fig. S6 A and B), and two FBH4 amiRNA constructs (35S:amiRFBH4-1 and 35S:amiRFBH4-3) (Fig. S6 C–F). Two independently established quadruple mutant lines [35S:amiRFBH1-2, fbh2-1, fbh3-1 and 35S:amiRFBH4-1 (#29) and 35S:amiRFBH1-2, fbh2-1, fbh3-1 and 35S:amiRFBH4-3 (#2)] were chosen for detailed analysis. CO expression analysis revealed a larger than 50% reduction of CO expression in the first 6 h of the dark periods in LD and SD in the quadruple mutants (Fig. 4 E and F), suggesting that the FBH proteins are major activators of CO especially in the beginning of the night. In LD, there is a slight reduction in afternoon CO expression (Fig. 4E). This could cause lower expression of FT and subsequently later flowering of the quadruple mutants in LD (Fig. S6 G–J). These results imply that the four FBH proteins are activators of CO transcription in Arabidopsis.
FBH Genes Are Widely Conserved Activator Genes in the CO/FT Flowering Pathway in Plants.
The CO/FT modules as well as the daily expression patterns of CO homologs are widely conserved in many plant species (11). Therefore, we hypothesized that CO transcriptional mechanisms including the FBH function might be conserved in other plants. As a primary attempt to examine this hypothesis, we analyzed the function of FBH homologs from poplar (a LD tree) and rice (a SD plant) in Arabidopsis. Two representative FBH homologs from poplar and rice (named PtFBH1 and OsFBH1, respectively) were chosen on the basis of a homology search and phylogenetic analysis (31) (see the amino acid sequence alignment of FBH1 homologs in Fig. S7 and our phylogenetic analysis in Fig. S8). Overexpression of both PtFBH1 and OsFBH1 drastically increased CO expression levels in Arabidopsis in LD and SD (Fig. 4 G–J and Fig. S9 A–F). The 35S:PtFBH1 plants showed early flowering in both LD and SD (Fig. S9G), and the 35S:OsFBH1 plants showed early flowering in LD (Fig. S9H). Because CO protein is constantly degraded in SD (32), the elevated CO levels in 35S:OsFBH1 plants may not be sufficiently high to overcome the posttranscriptional regulation of CO in SD. Nevertheless, these results imply that PtFBH1 and OsFBH1 have a similar function to Arabidopsis FBHs. In addition, there are several E-box elements in 1 kb of the promoter regions of both the poplar and rice CO ortholog genes (Fig. S9I). This evidence further indicates that PtFBH1 and OsFBH1 presumably regulate their own CO ortholog expression in poplar and rice, respectively.
Discussion
FBH Proteins Are Transcriptional Activators of CO.
It is not surprising that multiple redundant factors are involved in CO transcriptional regulation because it is the crucial mechanism in the photoperiodic flowering pathway. Interestingly, except for FKF1 and GI, all of the factors currently identified before this work are repressors of CO expression (12–21). That may indicate that CO activators are highly redundant or also involved in the processes necessary for plant survival. To overcome a potential genetic redundancy, we applied a reverse genetics approach to find additional CO regulators (23). We identified that FBH1 directly binds to the CO promoter (Figs. 1 and 3); on the basis of homology, we also identified three more bHLH proteins, FBH2, FBH3, and FBH4, which have a similar function to FBH1 (Figs. 2 and 4; and Fig. S4). Our genetic analysis revealed that all of the FBHs are transcriptional activators of CO. Ectopic overexpression of FBH drastically increased CO expression levels but did not alter the spatiotemporal expression patterns of CO (Figs. 2 and 4; Fig. S4). These results also let us infer that all of the FBHs may be posttranslationally activated at a specific time of the day mainly in the leaf vasculature and/or may work together with unidentified vascular-specific factors to regulate CO transcription.
Circadian-time–dependent activation of transcriptional activators is a conserved mechanism in mammalian, insect, and fungal clock circuits. The mammalian positive circadian regulators, CLOCK and BMAL1, and their insect counterparts, Drosophila CLOCK and CYCLE, are bHLH-domain–containing transcriptional activators that induce gene expression of negative regulators (33, 34). Their daily protein expression profiles do not show robust oscillation as negative regulators do; however, the phosphorylation states of these proteins change throughout the day and alter their binding abilities to the cis-elements (35, 36). A similar circadian change in the DNA-binding ability of the fungal clock activator WHITE COLLAR complex is also regulated by time-dependent phosphorylation (37). Therefore, one possible posttranslational mechanism that controls FBH function could be phosphorylation-dependent changes in DNA-binding abilities.
The latter possibility is also supported by our data. In the quadruple mutants in LD, two distinct peaks of CO (at around ZT 13 and at dawn) were observed (Fig. 4E). Because FBH overexpression drastically elevated CO levels from afternoon to night in LD (Fig. 2 D and E and Fig. 4 A–D), this result implies that other functionally redundant transcriptional activators contribute to the regulation of LD-specific daytime CO expression (as well as the end-of-night CO expression). Because the expression of FBH mRNAs and FBH1 protein do not show robust daily oscillation (Figs. S3 and S5), time-dependent changes in FBH activity could also be regulated by the potential spatiotemporal expression of the coactivators. Our next challenges will be to identify other coactivators of CO and also to decipher the molecular relationship between multiple CO regulators and FBH function in terms of controlling the precise timing of daily CO expression.
Our results indicate that FBH levels regulate the amplitude of daily CO oscillation. Even changing the amplitude of CO expression altered overall FT levels (Fig. 2). This implies that plants can regulate not only the timing of CO expression but also the amount of CO levels to control the overall amount of FT. Having redundant FBH proteins may enable Arabidopsis plants to accurately tune the expression level of CO as well as to increase a dynamic range of CO expression levels by regulating four different FBH expressions, so that plants can respond to various internal and external conditions more precisely and robustly for flowering.
FBH Homologs May Regulate CO Orthologs in Other Plant Species.
Our study also suggests that FBH homologs may function as transcriptional activators of CO homologs in other plants. The daily expression patterns of CO orthologs are very similar (11), indicating that transcriptional regulatory mechanisms may be also conserved. We demonstrated that PtFBH1 (poplar FBH) and OsFBH1 (rice FBH) have a similar function to FBHs in Arabidopsis (Fig. 4). Our phylogenetic analysis indicated that there is at least one (usually more) bHLH that belongs to the same clade of FBH (designated as IX, Fig. S8) in all angiosperms (Arabidopsis, poplar, rice, tomato, maize, and grape) examined. In addition, we found that the multiple E-box elements (but not G-boxes) exist on 1-kb upstream regions of the PtCO2 and Hd1 promoters (Fig. S9I). These results also indicate that E-box–binding factors (possibly bHLHs in the FBH clade) may participate in the CO transcriptional regulation. Although it is beyond the scope of this current analysis, it would be intriguing to test the function of PtFBH1 and OsFBH1 in poplar and rice, respectively.
In summary, our data indicate that, together with circadian-clock–regulated repressors, plants may possess overlapping mechanisms to regulate the expression levels of CO (and CO orthologs) by a group of related transcriptional activators to precisely regulate the timing of expression for successful reproduction.
Materials and Methods
The Colombia-0 accession was used as wild type for all experiments. The ft-101 mutant was described previously (28). Procedures for A. thaliana husbandry; yeast one-hybrid, EMSA, and ChIP assays; and the GUS-staining experiment were described previously (38–41) and were carried out with modifications detailed in the SI Materials and Methods. FBH1, FBH2, FBH3, FBH4, PtFBH1, and OsFBH1 coding regions were cloned into the pB7WG2 binary vector to generate each overexpressor line. For making the amiRNA constructs that specifically reduce the amount of FBH1 and FBH4 mRNA, specific FBH1- and FBH4-targeted amiRNA sequences were introduced into the miR319 backbone plasmid (pRS300). The resulting 35S-promoter–driven FBH1 and FBH4 amiRNA expression cassettes were cloned into pPZP221 or pH7WG2 binary vectors, respectively. FBH1 and FBH2 β-estradiol–inducible lines were generated by transformation with the pER8 plasmid containing the FBH1 and FBH2 coding regions. For expression analysis, seedlings were grown on plates containing 1× Linsmaier and Skoog media (Caisson) containing 3% sucrose under LD, SD, or 12 h light/12 h dark conditions for 10 d and harvested. The gene expression levels were measured by qPCR analyses. Detailed information is provided in SI Materials and Methods. All primer sequences used in this project are listed in Tables S1–S3.
Supplementary Material
Acknowledgments
We thank S. Kay for constant encouragement, initial support, and the transcription factor library; E. Farré and J. Pruneda-Paz for critical reading of the manuscript; J. Pruneda-Paz and S. Kay for sharing unpublished results; K. Goto for Arabidopsis lines; and N.-H. Chua and K. Torii for plasmids. S.I. was supported by a Japan Society for the Promotion of Science Postdoctoral Fellowship. Y.H.S. is partly supported by the Next Generation Biogreen 21 Program (PJ008109). This work was supported by National Institutes of Health Grant GM079712 (to T.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118876109/-/DCSupplemental.
References
- 1.Thomas B, Vince-Prue D. Photoperiodism in Plants. New York: Academic Press; 1996. [Google Scholar]
- 2.Yanovsky MJ, Kay SA. Living by the calendar: How plants know when to flower. Nat Rev Mol Cell Biol. 2003;4:265–275. doi: 10.1038/nrm1077. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi Y, Weigel D. Move on up, it's time for change—Mobile signals controlling photoperiod-dependent flowering. Genes Dev. 2007;21:2371–2384. doi: 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- 4.Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010;61:1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- 5.de Montaigu A, Tóth R, Coupland G. Plant development goes like clockwork. Trends Genet. 2010;26:296–306. doi: 10.1016/j.tig.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Imaizumi T. Arabidopsis circadian clock and photoperiodism: Time to think about location. Curr Opin Plant Biol. 2010;13:83–89. doi: 10.1016/j.pbi.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 8.Abe M, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 9.Wigge PA, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 10.Böhlenius H, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 11.Song YH, Ito S, Imaizumi T. Similarities in the circadian clock and photoperiodism in plants. Curr Opin Plant Biol. 2010;13:594–603. doi: 10.1016/j.pbi.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fowler S, et al. GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suárez-López P, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 14.Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- 15.Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- 16.Mizoguchi T, et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell. 2005;17:2255–2270. doi: 10.1105/tpc.105.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M, Ni M. RFI2, a RING-domain zinc finger protein, negatively regulates CONSTANS expression and photoperiodic flowering. Plant J. 2006;46:823–833. doi: 10.1111/j.1365-313X.2006.02740.x. [DOI] [PubMed] [Google Scholar]
- 18.Yoo SY, Kim Y, Kim SY, Lee JS, Ahn JH. Control of flowering time and cold response by a NAC-domain protein in Arabidopsis. PLoS ONE. 2007;2:e642. doi: 10.1371/journal.pone.0000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, Kim Y, Yeom M, Kim JH, Nam HG. FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell. 2008;20:307–319. doi: 10.1105/tpc.107.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu JF, Wang Y, Wu SH. Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol. 2008;148:948–959. doi: 10.1104/pp.108.124917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fornara F, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 22.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onouchi H, Igeño MI, Périlleux C, Graves K, Coupland G. Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell. 2000;12:885–900. doi: 10.1105/tpc.12.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuo J, Niu QW, Chua NH. Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
- 28.Takada S, Goto K. Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An H, et al. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development. 2004;131:3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- 30.Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol. 2010;27:862–874. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carretero-Paulet L, et al. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153:1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valverde F, et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 33.Glossop NR, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- 34.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 35.Yoshitane H, et al. Roles of CLOCK phosphorylation in suppression of E-box-dependent transcription. Mol Cell Biol. 2009;29:3675–3686. doi: 10.1128/MCB.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Q, et al. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 2006;20:2552–2565. doi: 10.1101/gad.1463506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sieburth LE, Meyerowitz EM. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deplancke B, et al. A gene-centered C. elegans protein-DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 40.Song YH, et al. Isolation of CONSTANS as a TGA4/OBF4 interacting protein. Mol Cells. 2008;25:559–565. [PubMed] [Google Scholar]
- 41.Almada R, Cabrera N, Casaretto JA, Ruiz-Lara S, González Villanueva E. VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds. Plant Cell Rep. 2009;28:1193–1203. doi: 10.1007/s00299-009-0720-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.