Abstract
Climate change is a major environmental stress threatening biodiversity and human civilization. The best hope to secure staple food for humans and animal feed by future crop improvement depends on wild progenitors. We examined 10 wild emmer wheat (Triticum dicoccoides Koern.) populations and 10 wild barley (Hordeum spontaneum K. Koch) populations in Israel, sampling them in 1980 and again in 2008, and performed phenotypic and genotypic analyses on the collected samples. We witnessed the profound adaptive changes of these wild cereals in Israel over the last 28 y in flowering time and simple sequence repeat allelic turnover. The revealed evolutionary changes imply unrealized risks present in genetic resources for crop improvement and human food production.
Keywords: climate warming, phenotypic and genotypic diversity, plant genetic resources
In recent decades there have been increasing concerns about the future of food production influenced by climate change (1–7) to feed a fast growing world population reaching 9–10 billion by 2050 (8). Aridization has alarmingly increased in summer, resulting in dry regions of wheat growing areas, particularly in the Americas, Asia, and Africa, and in the area of origin and diversity of wild emmer wheat in the Near East (9). Cultivated wheat, the number one world food staple, is genetically impoverished by long-term selective breeding (10). The wild progenitors of cultivated cereals, especially wild emmer wheat Triticum dicoccoides Koern. (TD) (9) and wild barley Hordeum spontaneum K. Koch (HS) (11), have become eroded by urbanization and agriculture (12). Both progenitors are rich in genetic resources adapted to abiotic (e.g., solar radiation, temperature, drought, and mineral poverty) and biotic (e.g., pathogens and parasites) stresses. These progenitors are the best genetic hope for improving genetically impoverished cultivars for human food production (9–17). Thus, it is crucial to evaluate the evolutionary adaptation in natural populations of the progenitors under climate change and to understand the hidden risk in human food security. Here we examined 10 TD populations and 10 HS populations in Israel from 1980 and again in 2008 and performed phenotypic and genotypic analyses on the collected samples. We witnessed the profound adaptive changes of these wild cereals in Israel over the last 28 y in flowering time (FT) and simple sequence repeat (SSR) allelic turnover.
Results
Phenotypic Analysis.
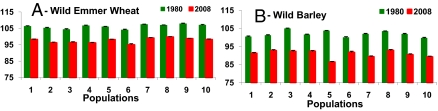
In a greenhouse experiment, we compared the time from germination until flowering of ∼800 genotypes in 20 natural populations from different habitats and climates that were subjected to 28 y of climate change. The sampled populations (16, 17) were distributed across 350 km of their Israeli range (SI Appendix, Fig. S1 and Table S1). A total of 57 independent pair-wise comparisons were made under dry (300 mm) and wet (600 mm) irrigation regimes. Dramatically, in all TD and HS populations, without exception, the 2008 populations reached FT earlier than those collected in 1980 (Sign test, P < 10−15). All populations displayed earliness (Fig. 1). Remarkably, the shortening of FT was highly significant in all 20 populations. The normal approximation for each population tested separately by a Wilcoxon rank-sum test, was −6.023 > z > −9.082 in TD and −5.939 > z > −8.855 in HS. For each population the significance was at least P < 10−8.
Fig. 1.
Differences in FT (days) of wild emmer wheat and wild barley collected in 1980 and in 2008. (A) The FT differences in 10 wild emmer wheat populations. (B) The FT differences in 10 wild barley populations. The x axis shows populations numbered from north to south. The y axis shows days from germination to flowering.
The shortening of FT was greater in HS than in TD. The average shortening for each population after the 28-y period in TD was 8.53 d (range, 7.19–10.45 d), whereas in HS it was 10.94 d (range, 8.21–17.26 d) (SI Appendix, Table S2). The difference between species was significant (Wilcoxon rank-sum test, P < 0.01). Greenhouse plants under 600 mm flowered significantly earlier than those under 300 mm. This environmental effect, about 10-times weaker than the genetic difference in FT, was caused presumably by climate change. In TD the greenhouse difference between wet and dry was −1.07 (median), ranging from −2.60 to +0.57 d (P = 0.02 by Wilcoxon matched-pairs signed-rank test, n = 19). In HS the difference was −0.775 (median), from −1.80 to +0.13 d (P < 0.01 by the same test, n = 18). The effect was weaker in HS than in TD.
We ran Spearman rank correlations of FT on environmental and demographic parameters in nature (16, 17), with the exclusion of the outlier population of Mt. Hermon because of its unique cold and rainy steppe in contrast to all other hot Israeli steppes (16). In the greenhouse, the FT was correlated in TD in both 1980 and 2008, but the interannual difference was not. In TD the FT of 1980 and 2008 was correlated with population size (rs = −0.87**, −0.87**), population marginality (−0.67**, −0.87**), soil type (−0.55, −0.82**), number of rainy days (−0.76*, −0.77*), and number of dew nights (−0.75*, −0.88**), respectively. The response to climate change during 1980–2008 showed a nonsignificant correlation with relative rainfall variation (0.55) and annual rainfall (−0.40). In HS, the FT of 2008 was correlated with rainfall (0.630@; P = 0.07) and plant formation (0.79*). The FT of 1980 and the difference were nonsignificantly correlated with mean August temperature (0.41 and 0.48, respectively). The higher environmental correlations in 2008 may suggest stronger adaptation.
SSR Analysis.
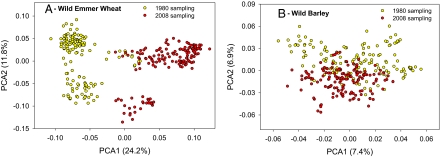
We performed a SSR marker analysis (18) of about 15 individual samples for each population of the two wild cereals in both sampling periods (SSR primers are described in SI Appendix, Table S3). Both wild cereals revealed remarkable genetic divergence, much more in TD than in HS populations (Fig. 2), in response to 28 y of climate change. Allele depletion in 2008 compared with 1980 was found for both wild cereals (SI Appendix, Table S4). In TD the total allelic count in 1980 was 318 alleles vs. 290 alleles in 2008, a highly significant reduction of 28 alleles (8.8%; P < 0.0001). Population allelic counts in 1980 were 113–173 alleles, whereas counts in 2008 were 104–157 alleles. The largest reduction was −46 alleles (Sanhedriyya). Seven populations showed reductions and three populations showed increases in allelic counts. In HS, the total allelic count in 1980 was 319 alleles and 309 in 2008, which shows a nearly significant (P = 0.082) reduction of 10 alleles and the same reduction trend as in TD. The population counts in 1980 were 94–144, whereas in 2008 it was 82–149 alleles. The largest reduction was −57 (Mt. Hermon). Six populations showed an allelic reduction and four populations showed an allelic increase. In TD, allele reduction was negatively correlated with altitude (−0.854*), humidity (−0.673*), and nearly significant with plant formation (−0.568@). In HS, without the Mt. Hermon population, the difference was positively correlated with rainfall (0.790*) but negatively correlated with evaporation (−0.692*) and plant formation (−0.867**).
Fig. 2.
Genetic associations of individual wild emmer wheat and wild barley plants, as revealed by the principle coordinates analysis of SSR markers. (A) The associations of 143 and 149 individual samples collected in 1980 and in 2008 of the 10 wild emmer wheat populations, respectively. (B) The associations of 148 and 148 individual samples collected in 1980 and in 2008 of the 10 wild barley populations, respectively.
There are sharp genetic differences in allele frequencies within and between the two wild cereals (SI Appendix, Figs. S2 and S3, and Table S5). In all TD populations there was at least one new allele that reached fixation (frequency 1) and at least one fixed allele that was lost. In contrast, in HS a new allele reached fixation in one population (Mehola) only, and a fixed allele was lost in three populations (Rosh Pinna, Eizariya, and Mehola). The average frequency of the new alleles was 0.474 (0.363–0.805) in each TD population and 0.193 (0.151–0.244) in HS. The difference between the two wild cereals was significant (P < 0.01; Wilcoxon rank-sum test). Interestingly, the lost and newly introduced alleles were widely distributed over the chromosomes, but the lost alleles became smaller in size than the new alleles for both crops (SI Appendix, Tables S6 and S7). Remarkably, the total variance present between 1980 and 2008 was 20.4% in TD against only 4.4% in HS (SI Appendix, Table S8). Overall, the SSR response of TD to climate change was much stronger than that of HS during the 28 y studied, where global warming was ongoing based on the Meteorological Service reports (6, 7).
Discussion
This comprehensive study on adaptive changes of the progenitors of cultivated wheat and barley under climate change is unique. Climate change, and its ramifications, is the only likely factor that could have caused earliness and allelic SSR turnover across the 20 populations (10 TD and 10 HS), across 350 km from north to south Israel. Notably, in Israel and other world hot and dry environments, the most important climatic factors are highly intercorrelated, with no possibility to disentangle them. The following climate scenarios were projected for Israel by the year 2100 (6, 7): (i) an increase of average temperature from 1.6 °C to 1.8 °C; specifically, 1.5 °C before 2020 and 3.5 °C in 2071–2100 were predicted compared with the years 1961–1990; (ii) a 10% reduction in precipitation by 2020 and a 20% decrease by 2050; (iii) a 10% increase in evapotranspiration; (iv) delayed winter rains; (v) increased rains intensity and shortened rainy seasons; (vi) greater seasonal temperature variability; (vii) increased frequency and severity by extreme climate events; (viii) greater spatial and temporal climatic uncertainty; and (ix) a tendency toward a more arid climate in Israel, conforming with Intergovernmental Panel on Climate Change predictions (7). These predictions were based on climatic changes during 1960–1990 (7) and on facts of increased anthropogenic CO2 emissions from 50 million tons in 1996 to 65 million tons in 2007. The constant increase in CO2 because of energy production will accelerate the sophisticated effects of all climatic factors, such as temperature, rainfall, and evaporation, as observed in Israel (6, 7). There are no specific data for climate change available on all of the studied populations, reflecting the inherit limitation of this study. However, additional assessments on the climate change trends in Israel showed a rising temperature and declining rainfall over the last 30 y (SI Appendix, Figs. S4–S6), which aligns well with the climate change predictions above. Moreover, the existing climate data in 1980 and 2008 imply that climate differences between the two years were representative of overall trends (6, 7). Greater responses to climate change in the xeric compared with mesic populations are expected and empirically confirmed here.
The assayed natural populations of wild cereals were exacerbated from 1980 to 2008, and surprisingly behaved essentially the same in phenotypic and genotypic SSR responses, presumably because of climate change in Israel. To our knowledge, no other factors, other than global warming in Israel, would induct such essentially similar behaviors for many natural populations across Israel. Without exception, the earliness across Israel, both in mesic and xeric habitats, underlines a major effect of climate change on wild cereals, paralleling earliness in other species (3) and negating neutrality. FT is selected in the major cereals. Heading date/FT is an important criterion for regional adaptations and yield in all cereals. The control of heading date is critical for reproductive success and has a major impact on grain yield in Triticeae (15). This finding also includes wild cereals that display adaptive lateness (Mount Hermon) or earliness (Samaria steppes), indicating the existence of genetic variation and selection of FT (15, 19). Total earliness in both wild cereals across Israel is a red light that may predict the future extinction of these precious genetic resources (9, 11). Climate change appears to be already affecting global wheat production, with an estimated 5.5% yield decline from warming since 1980 (4). Climate change impact on wild-crop relatives shown here necessitates the continuous efforts for in situ and ex situ conservation of these important genetic resources for future crop improvement. Total earliness in flowering indicates escape from an increasing drought (20), as is also true spatially across Israel, where xeric populations flower much earlier than mesic ones (9), thereby shortening the growth period. Eventually, grain yield will be reduced and food production decreased (4).
The SSR results are also instructive (SI Appendix, Tables S1 and S3–S8), because SSRs are substantially important in gene regulation (21, 22). The general depletion of regulatory genetic diversity (SI Appendix, Table S7) may lead to deterioration of environmental adaptation. Importantly, the SSR variance in response to climatic change was clearly higher in TD than in HS (20% vs. 4% between years and samples, respectively). Remarkably, specialist TD significantly suffered more genetic reduction than generalist wild barley, which is more adapted to climatic extremes (penetrating into the desert) and fluctuations than wild emmer (9). The former is more restricted geographically, climatically, and edaphically than the latter (16, 17). Nevertheless, some new alleles detected in both wild cereals (SI Appendix, Table S7) may be adaptive and valuable for breeding. Similarly, the genotypes/populations that displayed high yield under the stressful regime (300 mm) may prove valuable in breeding for higher drought tolerance. Although more points of comparison might be desirable, we are confident that the cereal adaptation found in Israel reveals the general pattern of wild cereals in the Near East Fertile Crescent.
The earliness of the wild cereals discovered here is not an exception for Israel, but is rather general to the growth performance of current and future cultivars around the world—as well as elsewhere—under hot and dry environmental stresses (3). Increasing risk from climate change will affect human food production (4). Thus, only the use of new adaptive evolving genetic resources may bring the best hope to secure food for humans and animals in the future. Both wild cereals in the Near East Fertile Crescent, particularly in northern Israel (9, 12, 16), are rich in adaptive genetic resources against abiotic (e.g., drought, cold, heat, salt, and mineral scarcity) and biotic (viral, bacterial, fungal, and herbicide) stresses, and with high quantity and quality storage proteins (glutenins, gliadins, and hordeins), amylases, and photosynthetic yield (9–14, 16, 17) (see also E.N.'s list of wild cereals at http://evolution.haifa.ac.il). Most of these resources, and many others, are yet untapped and are valuable for crop improvement (9–14, 16, 23). The current rich genetic map and detected quantitative trait loci of TD (24) and that of HS (25) permit the unraveling of beneficial alleles of candidate genes, but an effective marker-assisted selection could enhance the introgression of useful alleles into cultivated wheat.
The reported impact of climate change (1, 2) and the extensive and impressive fingerprint of climate change on wild animals and plants (26) alert mankind to an uncertain future. The potential risk of losing precious genetic resources should be balanced by using presumably adaptive novelties in breeding programs for earliness and drought resistance that were derived from climate change during 1980–2008 (see also ref. 20). This need for balance is particularly true in an increasing world population, where hunger is prevalent, water and fertile land resources are limited, and desertification and salinization are dramatically increasing. We can advance from the green revolution to the gene (27) and genomic revolution (28) if we appreciate both the red and green lights highlighted by the current status of our wild cereals derived from climate change.
Materials and Methods
Sampled Populations.
We compared and contrasted 10 populations of wild emmer wheat (TD) and 10 populations of wild barley (HS) spread across 350 km from Mt. Hermon in the north to Sede Boqer in the south in Israel. SI Appendix, Fig. S1 displays the geographical distribution of the populations. Seeds were collected first in 1980 and again in the same sites in 2008, after 28 y of global warming. SI Appendix, Table S1 lists the studied populations from north to south with their annual rainfall and temperature. Statistical significance is symbolized by: @, P < 0.10; *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Greenhouse Experiment.
We conducted a phenotypic assessment of 20 genotypes in each population by testing in two irrigation regimes (dry, 300 mm and wet, 600 mm) with two repeats for the dry regime and one repeat for the wet regime. All 800 genotypes were grown in a greenhouse at the Aaronshon Experimental Farm near Atlit in winter-spring 2008–2009. The genotypes of 1980 have been propagated several times to retain full germination. The greenhouse planting followed a randomized split-plot factorial (genotype × irrigation regime) block design (29). Several parameters were recorded, including flowering time from germination, biomass, and seed weight.
SSR Analysis.
About 15 individual samples for each population of emmer wheat or wild barley in either sampling period were randomly selected. Young leaf tissues were collected for DNA extraction and quantification as described previously (30). SSR analysis (18) was performed using 32 wheat and 29 barley SSR primer pairs (SI Appendix, Table S3), as described previously (30–32). These applied SSR markers should sample both transcribed and nontranscribed chromosomal regions of wild emmer wheat and wild barley genomes (SI Appendix, Table S3). Duplicated samples were also applied to each gel to minimize scoring errors, and DNA fragments were manually scored as 1 for presence or 0 for absence.
SSR Data Analysis.
SSR data of each species were analyzed essentially following those described previously (10, 33) with respect to allele polymorphism, allelic count difference, variation partition, and individual sample association. One exception is that the Shannon entropy of each marker was calculated, as previously described (34), to estimate the diversity content per locus.
Contrast and Correlation Analyses.
Phenotypic and genotypic data among populations and crops were compared and tested with respect to population, sampling period, and species, using the Wilcoxon rank-sum test (35). Correlations of phenotypic and genotypic data with environmental factors or population features were performed for each species using the Spearman rank test (35).
Supplementary Material
Acknowledgments
We thank A. Beharav for greenhouse testing designs; S. Freund, S. Raz, I. Roffman, H. Yarin, N. Daniel, O. Kossover, and A. S. Ahmad for field assistance; A. Morgenstern for administrative assistance; and G. Peterson, K. MacKay, and K. W. Richards for support in simple sequence repeat analysis and research. We are deeply grateful to Beat Keller, Robert Henry, and David Lobell for constructive comments that improved the manuscript and to Robin Permut for editing. This work was supported by Global Crop Diversity Trust Grant GS08031 and the Ancell-Teicher Research Foundation of Genetics and Molecular Evolution, which provides continuous support of all studies of E.N.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121411109/-/DCSupplemental.
References
- 1.Haughton JT, et al., editors. Climate Change 1995: The Science of Climate Change. Cambridge: Cambridge Univ. Press; 1996. [Google Scholar]
- 2.Committee to Review the IPCC . Climate Change Assessments: Review of the Processes and Procedures of the IPCC. Amsterdam: Interacademy Council; 2010. [Google Scholar]
- 3.Parmesan C. Ecological and evolutionary responses to recent climate change. Annu Rev Ecol Evol Syst. 2006;37:637–669. [Google Scholar]
- 4.Lobell DB, Schlenker W, Costa-Roberts J. Climate trends and global crop production since 1980. Science. 2011;333:616–620. doi: 10.1126/science.1204531. [DOI] [PubMed] [Google Scholar]
- 5.Chen I-C, Hill JK, Ohlemüller R, Roy DB, Thomas CD. Rapid range shifts of species associated with high levels of climate warming. Science. 2011;333:1024–1026. doi: 10.1126/science.1206432. [DOI] [PubMed] [Google Scholar]
- 6.Pe'er G, Safriel UN. Sde Boqer: Sde Boqer Campus of Ben-Gurion University of the Negev; 2000. Impact, vulnerability and adaptation to climate change in Israel. Israel's National Report on Climate Change under the United Nations Framework Convention on Climate Change. Available at http://www.bgu.ac.il/BIDR/rio/Global91-editedfinal.html. [Google Scholar]
- 7.Israel's Second National Communication on Climatic Change 2010. Submitted under the United Nations Framework Convention on Climate Change. Ministry of Environmental Protection, State of Israel, Jerusalem. Available at www.environment.gov.il Accessed February 2, 2012.
- 8.U.S. Census Bureau, International Data Base World Population: 1950-2050. June 2011 Update. 2011 Available at http://www.census.gov/population/international/data/idb/worldpopgraph.php. Accessed on February 2, 2012. [Google Scholar]
- 9.Nevo E, Korol AB, Beiles A, Fahima T. Evolution of Wild Emmer and Wheat Improvement. Population Genetics, Genetic Resources, and Genome Organization of Wheat's Progenitor, Triticum dicoccoides. Berlin: Springer; 2002. p. 364. [Google Scholar]
- 10.Fu YB, Somers DJ. Genome-wide reduction of genetic diversity in wheat breeding. Crop Sci. 2009;49:161–168. [Google Scholar]
- 11.Nevo E. Origin, evolution, population genetics and resources for breeding of wild barley, Hordeum spontaneum, in the Fertile Crescent. In: Shewry P, editor. Barley: Genetics, Molecular Biology and Biotechnology. Wallingford, UK: C.A.B. International; 1992. pp. 19–43. [Google Scholar]
- 12.Saranga Y, editor. A century of wheat research—From wild emmer discovery to genome analysis. Isr J Plant Sci. 2007;55:3–4. [Google Scholar]
- 13.Feldman M, Sears ER. The wild gene resources of wheat. Sci Am. 1981;244:102–112. [Google Scholar]
- 14.Xie W, Nevo E. Wild emmer: Genetic resources, gene mapping and potential for wheat improvement. Euphytica. 2008;164:603–614. [Google Scholar]
- 15.Peng J, Sun D, Nevo E. Wild emmer wheat, Triticum dicoccoides, occupies a pivotal position in wheat domestication process. Aust J Crop Sci. 2011;5:1127–1143. [Google Scholar]
- 16.Nevo E, Zohary D, Brown AHD, Haber M. Genetic diversity and environmental associations of wild barley, Hordeum spontaneum, in Israel. Evolution. 1979;33:815–833. doi: 10.1111/j.1558-5646.1979.tb04737.x. [DOI] [PubMed] [Google Scholar]
- 17.Nevo E, Beiles A. Genetic diversity of wild emmer wheat in Israel and Turkey: Structure, evolution and application in breeding. Theor Appl Genet. 1989;77:421–455. doi: 10.1007/BF00305839. [DOI] [PubMed] [Google Scholar]
- 18.Litt M, Luty JA. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet. 1989;44:397–401. [PMC free article] [PubMed] [Google Scholar]
- 19.Peng JH, et al. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc Natl Acad Sci USA. 2003;100:2489–2494. doi: 10.1073/pnas.252763199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams AP, et al. Forest responses to increasing aridity and warmth in the southwestern United States. Proc Natl Acad Sci USA. 2010;107:21289–21294. doi: 10.1073/pnas.0914211107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol Ecol. 2002;11:2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- 22.Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: Structure, function, and evolution. Mol Biol Evol. 2004;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- 23.Gustafson PO, Raskina O, Ma X, Nevo E. Wheat evolution, domestication and improvement. In: Carver BF, editor. Wheat: Science and Trade. Danvers, MA: Wiley Blackwell; 2009. pp. 5–30. [Google Scholar]
- 24.Nevo E. Evolution of wild wheat progenitors focusing on wild emmer. In: Kole C, editor. Wealth of Wild Species: Role in Genome Elucidation and Improvement. Berlin: Springer; 2011. [Google Scholar]
- 25.Chen G, et al. Chromosomal regions controlling seedling drought resistance in Israeli wild barley, Hordeum spontaneum C. Koch. Genet Resour Crop Evol. 2010;57:85–99. [Google Scholar]
- 26.Root TL, et al. Fingerprints of global warming on wild animals and plants. Nature. 2003;421:57–60. doi: 10.1038/nature01333. [DOI] [PubMed] [Google Scholar]
- 27.Tuberosa R, Philips RL, Gale M. 2005 In the Wake of the Double Helix from the Green Revolution to the Gene Revolution. Proceedings of the International Congress of the University of Bolognia, Italy, May 27–31, 2003. [Google Scholar]
- 28.Feuillet C, Muehlbauer GJ. Genetics and Genomics of the Triticeae. Plant Genetics and Genomics. Vol 7. Berlin: Springer; 2009. [Google Scholar]
- 29.Peleg Z, et al. Genetic diversity for drought resistance in wild emmer wheat and its ecogeographical associations. Plant Cell Environ. 2005;28:176–191. [Google Scholar]
- 30.Fu YB, et al. Allelic reduction and genetic shift in the Canadian hard red spring wheat germplasm released from 1845 to 2004. Theor Appl Genet. 2005;110:1505–1516. doi: 10.1007/s00122-005-1988-6. [DOI] [PubMed] [Google Scholar]
- 31.Fu YB, et al. Impact of plant breeding on genetic diversity of the Canadian hard red spring wheat germplasm as revealed by EST-derived SSR markers. Theor Appl Genet. 2006;112:1239–1247. doi: 10.1007/s00122-006-0225-2. [DOI] [PubMed] [Google Scholar]
- 32.Varshney RK, et al. A high density barley microsatellite consensus map with 775 SSR loci. Theor Appl Genet. 2007;114:1091–1103. doi: 10.1007/s00122-007-0503-7. [DOI] [PubMed] [Google Scholar]
- 33.Fu YB. fptest: A SAS routine for testing differences in allelic count. Mol Ecol Resour. 2010;10:389–392. doi: 10.1111/j.1755-0998.2009.02752.x. [DOI] [PubMed] [Google Scholar]
- 34.Russell JR, Hosein F, Johnson E, Waugh R, Powell W. Genetic differentiation of cocoa (Theobroma cacao L.) populations revealed by RAPD analysis. Mol Ecol. 1993;2:89–97. doi: 10.1111/j.1365-294x.1993.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 35.Zar JH. Biostatistical Analysis. 2nd Ed. Englewood Cliffs, N.J.: Prentice-Hall; 1984. p. 718. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.