Abstract
Transcription-coupled DNA repair targets DNA lesions that block progression of elongating RNA polymerases. In bacteria, the transcription-repair coupling factor (TRCF; also known as Mfd) SF2 ATPase recognizes RNA polymerase stalled at a site of DNA damage, removes the enzyme from the DNA, and recruits the Uvr(A)BC nucleotide excision repair machinery via UvrA binding. Previous studies of TRCF revealed a molecular architecture incompatible with UvrA binding, leaving its recruitment mechanism unclear. Here, we examine the UvrA recognition determinants of TRCF using X-ray crystallography of a core TRCF–UvrA complex and probe the conformational flexibility of TRCF in the absence and presence of nucleotides using small-angle X-ray scattering. We demonstrate that the C-terminal domain of TRCF is inhibitory for UvrA binding, but not RNA polymerase release, and show that nucleotide binding induces concerted multidomain motions. Our studies suggest that autoinhibition of UvrA binding in TRCF may be relieved only upon engaging the DNA damage.
Keywords: cysteine cross-linking, transcription, ATPase stimulation, UvrB
RNA polymerase (RNAP) stalled at DNA lesions on the transcribed strand elicits a preferential pathway for nucleotide excision repair (NER) called transcription-coupled repair (TCR), which is present in Bacteria and Eukarya (1). Bacterial transcription-repair coupling factor (TRCF; also known as Mfd) orchestrates this process by specific recognition of the transcription and NER assemblies, which reflects its twofold role. First, TRCF relieves transcription-dependent NER inhibition due to occlusion of the DNA lesion by RNAP (2). TRCF, an SF2 ATPase with dsDNA translocase but no helicase activity (3), approaches the stalled RNAP from behind and induces its forward translocation by stepping on dsDNA using ATP hydrolysis (4, 5). The consequent collapse of the upstream end of the transcription bubble leads to massive destabilization of the otherwise stable ternary elongation complex (TEC) and transcription termination (4–7). Rho, the only other known bacterial enzymatic terminator, induces termination by a similar forward-translocation mechanism, but translocates along the nascent RNA (8). Second, TRCF recruits the Uvr(A)BC endonuclease to the unmasked lesion by binding to UvrA (4, 9). This initiates a cascade of events resulting in lesion excision and gap filling (4, 10). TRCF also has roles beyond TCR—in the rescue of replication forks stalled by head-on collisions with RNAPs (11), in the development of antibiotic resistance (12, 13), recombination (14, 15), and transcriptional regulation (16, 17).
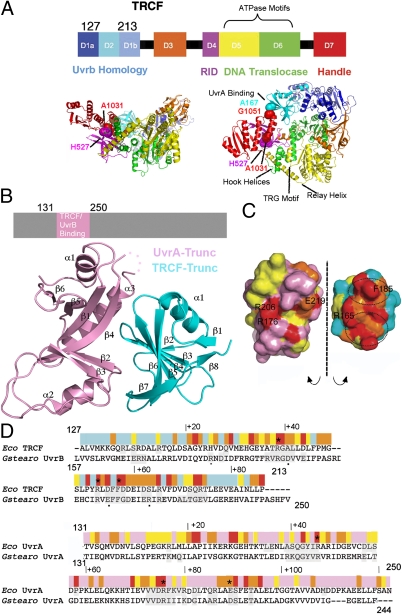
The crystal structure of apo TRCF (18) revealed a multimodular enzyme with eight domains connected by flexible linkers (Fig. 1A), an architecture that appears primed for large conformational changes, which are believed to be critical for coupling RNAP recognition to recruitment of NER enzymes. Domains D1 and D2 of TRCF are similar to the NER protein UvrB, which also binds UvrA (18, 19), suggesting that these domains serve as a platform for UvrA recruitment and possibly as a “clamp” to restrain the ATP-binding translocase domains (D5 and D6, Fig. 1A), explaining the poor ATPase function and TRCF inability to translocate on naked DNA (22).
Fig. 1.
X-ray structure of the Escherichia coli TRCF–Trunc/UvrA–Trunc complex. (A) Structure of apo E. coli TRCF (PDB ID 2EYQ). Location of engineered Cys is indicated by spheres. (B) TRCF–UvrA core complex. (C) Solvent-accessible surface of the TRCF–UvrA complex, obtained by splaying the complex open and colored by evolutionary conservation as in D. (D) E. coli TRCF-Trunc/Geobacillus stearothermophilus UvrB sequence alignment with substitutions affecting UvrA binding (20, 27) marked by an asterisk in TRCF-Trunc and by a small black dot in UvrB. Sequence conservation is indicated with a color ramp from red (invariant) to cyan (variable). Interfacing residues in the TRCF–UvrA and UvrB–UvrA core complexes (20) are shaded in gray. The UvrA sequence is annotated similarly.
Our knowledge of the mechanisms for RNAP recognition and Uvr(A)BC recruitment is rudimentary as TCR intermediates could not be detected (23). It was suggested early on that binding of TRCF/UvrB to UvrA may be competitive (4), but this hypothesis was not addressed subsequently. In addition, a structural model for ATP-bound TRCF (the state that binds DNA) is lacking, leaving details of how this “coupling” occurs unknown. However, it has been previously hypothesized, largely on the basis of in vitro studies carried out without a stalled TEC and UvrA, that the coupling occurs via a single TEC-induced conformational switch within TRCF, which synchronously enables forward translocation and UvrA recruitment (22, 24).
To better understand the mechanism of NER machinery recruitment, we combined functional studies with X-ray crystallography of a core TRCF–UvrA complex and SAXS analysis of TRCF. We show that, in apo TRCF, the UvrA-binding surface is occluded due to intramolecular contacts with domain D7 and that D7 is mobile during the catalytic cycle, but that its mobility is not important for RNAP release. Our data reveal the conformational flexibility of this macromolecular motor during the ATP hydrolysis cycle and details of TRCF mimicry of UvrB in binding to their common partner UvrA; at the same time, our data suggest that TCR relies on a fine temporal and contextual regulation of the various TRCF activities.
Results
TRCF and UvrB Share the Same Mode of UvrA Recognition.
Recent studies suggest that domain D2 of TRCF interacts with an UvrA fragment encompassing residues 131–250 (9, 18, 20, 22, 25). Furthermore, the UvrB-homology module (residues 1–349, including D2) as well as residues 131–248 of UvrA were shown to be essential for repair of the template strand in vitro and in vivo without being required for RNAP displacement (21). We have thus designed a minimal TRCF construct, TRCF-Trunc (residues 127–213, Fig. 1), which forms a core TRCF/UvrA complex in the presence of UvrA-Trunc (residues 131–250) as revealed by pull-down assays (Fig. S1A). We then determined the crystal structure of this minimal TRCF-UvrA complex, TRCF-Trunc/UvrA-Trunc (Fig. 1B and Table S1), at 2.8 Å resolution. The model was refined to R/Rfree of 23.8%/28.2%. Comparison of the two copies of the complex in the asymmetric unit did not reveal any major conformational differences (rmsdTRCF-Trunc = 0.64 Å; rmsdUvrA-Trunc = 0.53 Å).
Overall, the structure of the core UvrA–TRCF complex is similar to that of the core UvrA–UvrB complex [with 29% sequence identity between TRCF/UvrB and an rmsd of 2.5 Å over the entire backbone of Protein Data Bank (PDB) ID 3FPN] (Fig. S1B), pointing to a common mode of UvrA recognition. Interface residues are conserved between UvrB and TRCF (Fig. 1 C and D), suggesting that TRCF and UvrB binding to UvrA are mutually exclusive. In addition, severe steric clashes occur between UvrA-Trunc and D7 of TRCF (Figs. 1A and 2A). The unusual TRCF–UvrA interface is punctuated by multiple arginines (Fig. 2B) and buries a surface area of 1,796 Å2, which in the TRCF crystal structure is partially occluded due to intramolecular packing against conserved residues within D7 (Fig. 2A), specifically E1045, D1048, and R1049. Binding of UvrA to TRCF could not be detected by gel filtration (4), although an interaction could be detected by pull-down assays, which more readily capture transient interactions (9). In contrast, UvrB binding to UvrA is clearly detected by gel filtration (20). These previous results are in agreement with the small but conserved TRCF–UvrA interface that we observe. Given that TRCF participates in diverse processes beyond NER (11–14, 16, 17), its interaction with UvrA would be expected to be transient and restricted to sites of DNA damage.
Fig. 2.
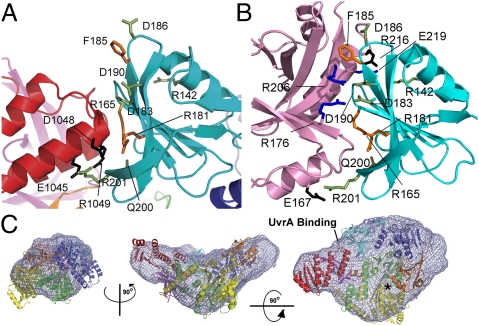
TRCF autoinhibition of UvrA binding is mediated by domain D7. (A) D2–D7 interaction seen in the crystal structure of apo TRCF (PDB ID 2EYQ). Conserved D7 residues are shown in black; interacting D2 residues are shown in green. Substitutions of orange residues are functionally important (21). (B) TRCF–UvrA interface. Residues in UvrA that bind UvrB are shown in blue (20). Other interacting residues are colored in black or as in A. (C) Crystal structure of apo E. coli TRCF (colored as in Fig. 1A) docked into the SAXS envelope. An asterisk indicates the ATP-binding site.
The TRCF–UvrA interaction is essentially bipartite, with residues in two adjacent patches contributing to binding (Fig. 1C). The patch around invariant R165 is completely occluded due to D7 interaction with the central β-sheet of TRCF-Trunc. The patch around invariant F185, which represents about half of the UvrA/TRCF–Trunc interface area, remains solvent-exposed in apo TRCF, but packs tightly against strand β4 of the central UvrA-Trunc β-sheet (Figs. 1B and 2B). A triple mutant carrying the R165A R181A F185A substitutions, shown in Fig. 2B, is defective in patch repair synthesis and does not bind UvrA in a bacterial two-hybrid system (21). Unlike wild-type TRCF, this mutant also fails to inhibit the GTPase activity of UvrA (21), consistent with an impaired interaction with UvrA. These findings strongly corroborate the physiological relevance of the observed interface. Other conserved solvent-exposed residues in TRCF-Trunc contribute to the intramolecular contacts seen in full-length TRCF. UvrA residues interacting with TRCF include conserved E219, which is important for NER (20), as well as invariant R176 and R206, which also map to the interface and are critical for UvrA–UvrB complex formation (Fig. 2B) (20). The corollary of our crystallographic study is that, for UvrA to bind, the D2–D7 contacts must be broken during the coupling process either concomitant with or subsequent to TEC binding. The downstream effect of TRCF binding to UvrA may be to promote dissociation of UvrB from UvrA to form the long-lived UvrB–DNA preincision complex that is required for the repair process (4).
TRCF Is Autoinhibited in Solution.
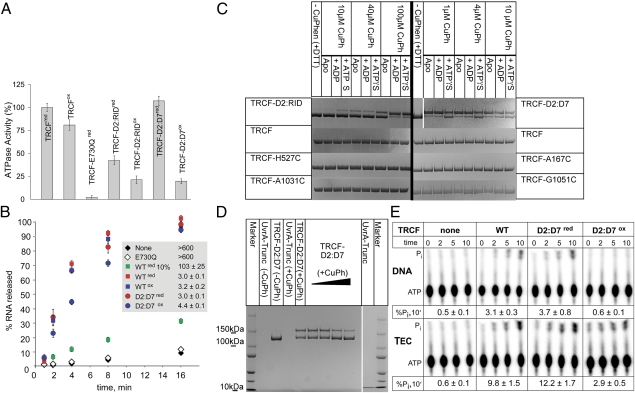
The occlusion of the UvrA-binding surface by the D7 domain in the crystal structure of apo TRCF implies that TRCF is autoinhibited for UvrA recruitment in its apo form. However, domain D7 accounts for a large number of crystal contacts (Fig. S1C) (6, 18, 26), raising the possibility that the putative inhibitory position of D7 may not be physiological and that, instead, the UvrA-binding surface is exposed as in UvrB (27). We have therefore probed TRCF using SAXS (Fig. 3). To facilitate study of the ATP-bound state, we engineered a mutant carrying the E730Q substitution in the Walker B motif. This substitution abolishes ATP hydrolysis (Fig. 4A), but not ATP, DNA, or TEC binding (Fig. S2). However, this noncatalytic TRCF is deficient in RNAP displacement (Fig. 4B), which requires ATP hydrolysis (3, 5). Limited proteolysis confirmed that the conformation of this mutant and the effects of nucleotides on its structure are similar to those of wild-type protein (Fig. S3).
Fig. 3.
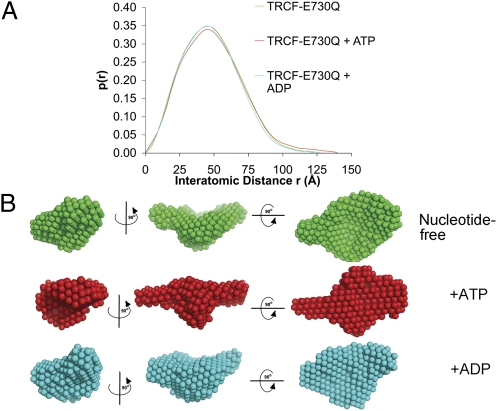
Structural flexibility in nucleotide-bound TRCF-E730Q. (A) Pair distribution functions normalized against the area under the curve. (B) Averaged filtered SAXS bead models. Views are as in Fig. 1A.
Fig. 4.
Characterization of disulfide-locked TRCF variants. (A) Steady-state ATPase activity of TRCF variants under oxidizing (“ox” superscript) and reducing (“red” superscript) conditions. Shown are averages of turnover numbers, kcat (normalized to wild type), obtained from three independent experiments ± SD. (B) Quantification of total RNA released in RNAP displacement assays. TEC half-lives were estimated from three independent experiments and are shown as the average ± SD. (C and D) SDS/PAGE of CuPh-catalyzed disulfide cross-linking with/without ADP/ATPγS (C) and UvrA-Trunc (D). (E) ATP turnover in the presence of DNA template (Upper) and TECs visualized using thin layer chromatography (Lower).
Ab initio shape reconstructions were obtained using GASBOR (28) and then aligned and filtered on the basis of occupancy. The convergence of the simulations was monitored using the normalized spatial discrepancy (NSD) criterion (29). Models displayed an excellent fit to the experimental data (Fig. S4) and low NSD values (SI Materials and Methods). As shown in Fig. 2C, there is a good fit between the TRCF crystal structure and the solution structure. This rigid-body fit was obtained using an automated exhaustive search starting from a random configuration. The central cavity and D7 “handle” are clearly reflected in the shape of the SAXS envelope. Notably, the relative position of D7 appears unchanged, confirming that TRCF is autoinhibited in solution due to the D2–D7 interaction also observed in the crystal structure. Therefore, for UvrA recruitment to occur, D7 must move to vacate the UvrA-binding site.
Nucleotide Binding and Hydrolysis Reorganize Interdomain Contacts Within TRCF.
Given the inhibitory D2–D7 interaction we observe, an important question remains unanswered: What triggers the unmasking of the binding interface and recruitment of UvrA? UvrA recruitment could be triggered by ATP binding and/or hydrolysis. To explore this question, we extended our SAXS analysis to nucleotide-bound TRCF-E730Q. Protein variants remain monomeric irrespective of nucleotide status (Fig. S5), contrasting recent reports of TRCF oligomerizing in other species (30). Comparison of the ab initio models (Fig. 3) and the SAXS-derived parameters Rg and Dmax (Table S2) revealed closely related conformations for the apo and ADP-bound states, reflecting functional similarities between apo and ADP-bound TRCF, neither of which bind DNA (9). We cannot exclude small-scale differences, especially those affecting the translocase domains that would not be discernible at the resolution of SAXS. When bound to ATP, TRCF appears to adopt a more extended conformation reflected in the longer tail of the model-independent pair distribution function (Fig. 3A). An obvious change involves D7 that appears to swing out into the solvent, thus resulting in an increase in Dmax and a modest change in Rg (Fig. 3B and Table S2). Repositioning of domains D5 and D6 observed in many other ATPases upon ATP binding and/or hydrolysis (31) is also likely to occur. Rearrangements involving the other domains also appear to take place and explain some of the apparent changes in the SAXS envelope, such as a frontal filling of the central cavity in ATP-bound TRCF. The core of the protein, however, remains as compact as in the apo state. To localize structural modules, we have also studied a TRCF variant that lacks domains D1–D3 and has been shown to display elevated ATPase and triplex displacement activity (22), which is repressed in full-length TRCF until binding RNAP (32). To prevent ATP hydrolysis, we introduced the E730Q substitution to generate TRCFΔ(D1-D3)E730Q. Comparison of the SAXS envelopes for TRCF-E730Q and TRCFΔ(D1-D3)E730Q (Fig. S6A) enabled us to assign the central protrusion (present in ATP-bound TRCF-E730Q) and the region diametrically opposed from D7 to the UvrB homology module D1–D2 and the species-specific domain of unknown function D3, respectively.
We also carried out simulations of the ATP-bound state using the coordinates of the Cα backbone of apo TRCF as input in GASBOR. These simulations converged on a solution with an NSD and χ similar to those obtained by ab initio methods. The coordinate-seeded model also features movements of D1–D3 and D7 (Fig. S6B), demonstrating the robustness and reproducibility of the simulated swinging motion of D7 and closure of the central cavity as the core domains rearrange. Placing the apo crystal structure into the SAXS envelope of ATP-bound TRCF using rigid-body fitting revealed significant discrepancies as did the comparison of the scattering profile of crystallized apo TRCF (simulated with CRYSOL) with the experimental SAXS profile of ATP-bound TRCF (Fig. S6C).
To further probe TRCF rearrangements upon ATP binding, we engineered disulfide linkages to reduce the intrinsic flexibility of the protein, lock D7, and hinder exposure of the UvrA-binding surface. We made two sets of substitutions. H527C and A1031C mutations (Fig. 1A) were introduced to create TRCF-D2:RID with a disulfide between D7 and the RNAP interaction domain RID (Fig. 1A and Fig. S1D). The other double mutant (A167C G1051C, TRCF-D2:D7) had an engineered D2–D7 disulfide across the UvrA-binding surface (Fig. 1A and Fig. S1D). Upon purification, disulfide formation became evident from the altered mobility of purified TRCF-D2:RID and TRCF-D2:D7 in SDS/PAGE analyses (Fig. S7A). Treatment with a reducing agent and subsequent alkylation caused reduction and disappearance of the slower migrating species, thus allowing the altered electrophoretic mobility to be attributed to specific Cys cross-linking. Proteins with single Cys substitutions at the aforementioned positions did not form intramolecular disulfide cross-links during their purification (Fig. S7A) or upon catalyzed oxidation (Fig. 4C), implying that the observed disulfides are due to specific linkage of C527 to C1031 and of C167 to C1051, respectively. These cross-linked mutants eluted like monomers from a size-exclusion chromatography column, with profiles comparable to that of wild-type TRCF (Fig. S5), confirming that the observed disulfides are intramolecular rather than intermolecular.
To compare disulfide-bond-formation efficiencies in the presence of ADP and ATPγS, we used a catalyst, Cu(II) (1, 10) phenanthroline (CuPh), which greatly enhances cysteine oxidation by atmospheric oxygen (33). After complete reduction and buffer exchange, we carried out oxidation at different CuPh concentrations. As shown in Fig. 4C, there was significant formation of the C527–C1013 linkage in ATPγS-bound TRCF-D2:RID and less in apo and ADP-bound TRCF-D2:RID, suggesting that the ATP-like state stabilizes D2/RID in a configuration conducive to disulfide bond formation. This trend was reversed with TRCF-D2:D7: the disulfide formed readily in the absence of nucleotide or with ADP, but not with ATPγS. These results are consistent with the TRCF crystal structure, and our SAXS data indicating nucleotide-dependent mobility of D7.
Because cysteines form a disulfide bond only if their Cβ-Cβ distance is less than about 5 Å (34), the formation of the engineered disulfides provided us with a sensitive indicator for local conformational variability. When UvrA-Trunc was titrated in the CuPh oxidation reactions without nucleotide, we consistently observed dose-dependent cross-linking in TRCF-D2:RID (Fig. S3C), suggesting that UvrA-Trunc binding promotes a relative conformation of D2 and RID that favors cross-linking, similar to ATPγS binding. This is also consistent with a modestly larger apparent Stokes radius of oxidized TRCF-D2:RID suggested by gel filtration analysis (Fig. S5). As with ATPγS, we observed the opposite trend in TRCF-D2:D7 (Fig. 4D), suggesting that, upon UvrA binding, D7 adopts a conformation incompatible with formation of the D2–D7 cross-link. When higher CuPh concentrations are used with TRCF-A167C, we also observed formation of an intramolecular cross-link of different electrophoretic mobility (Fig. S7B). This disulfide likely forms between C167 and one of the seven cysteines in wild-type TRCF. We can exclude disulfide formation between D2 and D7, as no cysteines are present in D7 (Fig. S1E). Formation of this cross-link was also nucleotide and UvrA-Trunc dependent (Fig. S7B). This suggests that the UvrB homology module may also move during the functional cycle, in agreement with our observation of the closure of the central cavity in ATP-bound TRCF (Fig. 3B).
To test if TRCF release of stalled TECs also involves a repositioning of D7 to fully activate TRCF and promote efficient binding of UvrA, we compared dissociation of TECs stalled by nucleotide deprivation by TRCF and TRCF-D2:D7. Like wild type, TRCF-D2:D7 displaced the TEC in both the reduced and the cross-linked states (Fig. 4B). In ATPase assays (Fig. 4A), we observed a reduction in the ATPase rates for both Cys mutants, but the effect was the largest for oxidized TRCF-D2:D7 even though our preparation contained a small amount (∼10%) of non-cross-linked TRCF-D2:D7 species (Fig. S7A). Thus, we can conclude that the oxidized species is greatly impaired in ATPase activity in the absence of RNAP. Fluorescence anisotropy experiments (Fig.S2 B–D) indicate that the affinity of reduced TRCF-D2:D7 for DNA is similar to wild type (Kd of 131 ± 17 nM compared with 118 ± 11 nM), whereas in oxidized TRCF-D2:D7 it is greatly reduced (Kd of 1798 ± 300 nM). This suggests that, by tethering D2 to D7, the naked DNA-binding and DNA translocation activities (in the presence of RNAP) can be uncoupled. The ability of oxidized, ATPase-deficient TRCF-D2:D7 to displace stalled TECs (Fig. 4B) implies that the ATPase function is stimulated by binding to TECs. Indeed, TECs stimulated ATP hydrolysis threefold compared with naked DNA template (Fig. 4E). Furthermore, TEC dramatically increased ATP turnover by oxidized TRCF-D2:D7, restoring it to about 30% of the total Pi released by wild type, correlating well with the more modest reduction in the RNA release by this variant. A stoichiometric titration of dsDNA in oxidation reactions revealed that naked DNA substrate does not alter the propensity to cross-link (Fig. S7C) and, consequently, the dynamic equilibrium of TRCF conformations. Thus, the robust TEC release activity of oxidized TRCF-D2:D7, which is greatly impaired in DNA binding, suggests that the initial recognition of a stalled TEC is likely provided through specific protein–protein contacts between the RID and the β-subunit of RNAP (5, 18, 35) and that protein–DNA contacts play a secondary role.
Discussion
Since the discovery of TRCF (36), the mechanistic details of UvrA recruitment, including the mode and timing of UvrA binding, have remained unknown. Here, we show how the binding of UvrA to TRCF occurs at the interface of domains D2 and D7 and closely mimics contacts between UvrA and UvrB, with residues critical for the interaction being conserved across and between the TRCF and UvrB families (Figs. 1–2). Together with functional studies of mutants in which the D2–D7 or the RID-D7 domains were locked by disulfide linkages, our study also establishes that, for UvrA to bind, the C-terminal domain D7 of TRCF has to move relative to its position in apo TRCF. SAXS analysis of the ATPase-deficient mutant TRCF-E730Q revealed that ATP binding leads to repositioning of multiple domains, including the inhibitory D7 (Fig. 3). A disulfide bond engineered across the D2–D7 interface uncouples the DNA-binding and ATPase activity of TRCF in the absence of TECs from translocation on dsDNA, RNA release, and TEC-stimulated ATP turnover (Fig. 4), suggesting that disruption of the D2–D7 interface (needed for UvrA recruitment) is not required for RNAP release. Our data allow us to take a fresh look at mechanistic models for TCR with a focus on UvrA recruitment.
TRCF Exists in a Dynamic Equilibrium of Conformations.
Our data suggest that TRCF exists in a dynamic equilibrium between two (or more) conformational states, a closed repressed conformation, and a more open state that can bind UvrA. The closed conformation predominates in solution in the absence of nucleotide and is characterized by an evolutionarily conserved D2–D7 interaction. This interaction was also observed crystallographically (18), but due to crystal contacts, its physiological relevance remained unclear. Weakening of this interaction did not abolish strand-specific repair in vitro or in vivo (21), but the effects of a tighter D2–D7 interaction on TCR were not explored.
We show that formation of the open state of TRCF is favored upon ATP binding, which results in movement of D7. UvrA binding likely also shifts the equilibrium toward the open conformation, consistent with UvrA inhibiting formation of the D2–D7 and TRCF-A167C cross-links and favoring formation of the D7-RID disulfide (Fig. 4D and Figs. S3C and S7B). However, detection of UvrA binding to full-length TRCF even in the absence of nucleotides (9) indicates that the open state is populated even in the apo protein. In addition to a breathing motion of D7, ATP also induces a repositioning of the D1–D3 module, which likely moves as a rigid body because the crystal structure of D1–D3 in isolation was shown to be identical to that in the context of the full-length protein (22). Recent work identified a protease-sensitive site in the D3-RID linker that is exacerbated in a derepressed TRCF variant, consistent with a rigid-body movement around a hinge located in this linker. The same study also revealed other protease-sensitive sites in the interdomain linkers, corroborating our conclusion that ATP (and likely also TEC) binding results in reorganization of interdomain contacts. The ATP-triggered shift toward an “open” state of TRCF could be interpreted as triggering UvrA recruitment for TCR to occur. However, with ATP, we observe on average only a partial exposure of the UvrA-binding site using SAXS, and, in pull-down assays with TRCF-E730Q (and ATP) under conditions in which we could robustly pull-down UvrA-Trunc, we could detect the TRCF–E739Q/UvrA–Trunc complex only weakly (Fig. S1A), suggesting that ATP alone is not sufficient for complete accessibility to the UvrA-binding surface. Furthermore, increased levels of UvrA are inhibitory to TCR (3), arguing that TRCF–UvrA complexes formed before TEC engagement/release may be mechanistically nonproductive.
Autoinhibition of UvrA Binding Is Likely Relieved upon Engaging the DNA Damage.
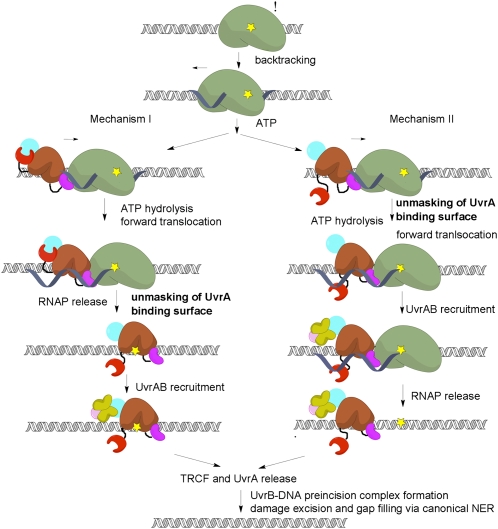
Despite previous attempts, failure to detect pathway intermediates such as TEC-TRCF-UvrAB or DNA-TRCF-UvrAB (3, 9) has left mechanistic details of the coupling largely unknown. Models for TCR differ in the timing of UvrA recruitment, occurring upon completion of RNAP release (4, 18) (mechanism I, Fig. 5) or upon engaging the RNAP (4, 18, 22) (mechanism II, Fig. 5).
Fig. 5.
TCR mechanisms. RNAP (green) stalls at DNA lesions (yellow) in the template strand and backtracks, recruiting TRCF, which promotes forward translocation of RNAP using ATP hydrolysis by the translocase module (brown) and, eventually, TEC dissociation. Next, the UvrAB complex is recruited by virtue of the unmasking of the UvrA-binding surface in D2 by motion of D7. The timing of UvrA recruitment differs in these two models, but our data argue for a sequential model (mechanism I). The pathway continues with formation of an UvrB–DNA preincision complex, subsequent DNA incisions, and gap filling (36).
In both mechanisms, the stalled TEC has a critical role. It acts as a DNA damage sensor, activates the dsDNA translocase activity of TRCF (32), and, as our data indicate (Fig. 4E), stimulates ATP turnover. Thus, TRCF appears to function akin to eukaryotic dsDNA translocases, such as chromatin-remodeling factors, which are often stimulated preferentially by nucleosome substrates over naked DNA (37).
ATPase hyperactivity can be achieved even in the absence of RNAP by substitutions weakening D2–D7 (21) or D1–D6 contacts (38, 39). Truncations lacking D7 or D1–D3, unlike full-length protein, can translocate on naked DNA (22, 32) These observations are consistent with both mechanisms in Fig. 5, in which disruption of the D2–D7 contacts enables activation in the absence of RNAP. Consequently, locking the D2–D7 interface is expected to prevent this activation, in agreement with our observation that cross-linking D2 to D7 significantly affects ATP turnover in the absence of RNAP (Fig. 4A). However, the interdomain cross-link does not abolish RNAP release (Fig. 4B), arguing that motions of D7 are not essential for TEC displacement and that the ATPase (in the absence of TEC) and the ATP-dependent translocase functions appear to be regulated differently. This agrees with recent in vivo and in vitro studies showing that triple substitutions at the D2–D7 interface result in different functional defects. D2 mutations abolish dsDNA translocation but do not compromise RNAP release, whereas substitutions in D7 affect neither dsDNA translocation nor RNAP release (21). The nature of the translocase activation in the presence of RNAP remains unknown, but our results argue against the requirement for a complete disruption of the D2–D7 contacts.
Although D7 repositioning must occur for productive UvrAB recruitment, the latter is required only at a site of damage. RNAP may stall for various reasons, and unmasking the UvrA-binding surface in the absence of RNAP-stalling lesions may have detrimental effects on cell viability. Indeed, cells expressing truncations lacking D7 are more sensitive to UV radiation, and despite efficient RNAP displacement activity in vitro (32), repair DNA less efficiently in vivo and in vitro, perhaps due to diverting rate-limiting UvrA from NER (9, 21).
What triggers productive UvrA recruitment? Among possible scenarios are the following: (i) loading/translocation on the DNA, (ii) interactions with the TEC, and (iii) binding to or near the DNA lesion coupled to/following RNAP displacement. D2–D7 interface disruption is not required for RNA release (Fig. 4) and, consequently, not required for loading/translocation on DNA. Importantly, the inhibitory effect of excess UvrA on TCR can be relieved by supplementation with TRCF (3), suggesting that UvrA-bound TRCF may be incompetent for one of the steps required for coupling, e.g., RNAP/DNA binding and/or translocation. Taken together, these observations favor the last scenario (mechanism I in Fig. 5). In this model, a fully productive complex forms only upon TRCF binding to or close to the exposed DNA lesion, which triggers complete unmasking of the UvrA interface. To gain access to the lesion, TRCF must first remove RNAP from it. This occurs through forward translocation (5, 8) during which RNAP “slides off” and releases the nascent RNA; however, it is not known whether the template DNA is released simultaneously. Following RNA release, TRCF and RNAP may maintain their interactions with each other and the DNA, allowing TRCF to slide toward the lesion. In this model, it is the recognition of the lesion (likely through local distortions in the DNA) that triggers UvrA recruitment. Once TRCF binds to UvrAB and promotes UvrB dissociation from UvrA (9), and possibly its loading onto DNA (40), TRCF likely dissociates from the DNA together with UvrA to leave behind a tight UvrB–DNA complex required for all subsequent incision events.
With TRCF engaged at the lesion, one would expect that global NER and TCR may differ in their requirements for nondamaged/damaged DNA discrimination by UvrA. The presence of TRCF does not render any of the subunits of the Uvr system redundant. Indeed, UvrA is required for UvrB loading, and UvrB residues that are important for DNA damage recognition and local strand separation are equally important for global NER and TCR (21). However, it has been shown that TCR exhibits a less stringent requirement for damage recognition by UvrA (21), likely because the lesion is recognized initially by RNAP, and perhaps later by TRCF. Together with our observation that D2–D7 cross-linking has a larger effect on UvrA binding than RNA release, these considerations also support mechanism I, which proceeds through an UvrAB-TRCF-DNA damage intermediate rather than through the TEC–TRCF–UvrAB complex of mechanism II. Further studies will be necessary to elucidate the precise sequence of events occurring during TCR, especially early on, during recruitment of NER machinery, damage detection, and preincision complex formation.
In a broader context, the D7-mediated occlusion of the D2 UvrA-binding interface that we observed may be essential for the TCR-independent functions of TRCF. In the case of head-on collisions of RNAP with replication forks (11), as well as transcriptional termination (17) and carbon catabolite repression (16), only the RNAP displacement activity of TRCF is essential, whereas the NER recruitment function is at least dispensable, if not detrimental. Our analysis thus brings insight into a general mechanism for UvrA-binding inhibition in TRCFs and suggests that TRCF function relies on a fine temporal and context-dependent tuning of its various activities.
Materials and Methods
A full description of the materials and methods used can be found in SI Materials and Methods.
X-Ray Crystallography.
Crystals of TRCF-Trunc/UvrA-Trunc were obtained via vapor diffusion and were pronouncedly anisotropic. The best diffracting crystals were obtained in 10% PEG 3350 and 4% tacsimate (pH 4.8). Cryoprotection was achieved with 14% PEG 3350, 4% tacsimate (pH 4.8), 20% ethylene glycol and by plunging the crystals in liquid nitrogen. Data were collected remotely at beamline 8.2.1 at the Advanced Light Source (ALS) at 1 Å wavelength and 100 K. Further details are included in SI Materials and Methods.
SAXS Analysis.
Data were collected at the Sibyls synchrotron beamline (ALS) at concentrations of 1–7 mg/mL protein for TRCF-E730Q and 0.3–2 mg/mL protein for TRCFΔ(D1-D3)E730Q, respectively, in triplicate or more at 10 °C. Further details are presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
A.M.D. gratefully acknowledges support from the Damon Runyon Cancer Research Foundation (DRG 1966-08). N.G. was supported by National Institutes of Health Grant P01 GM-623580 and I.A. by Grant R01 GM67153.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Structure factor amplitudes and coordinates for the Escherichia coli TRCF-Trunc/UvrA-Trunc model have been deposited in the Protein Data Bank under PDB ID 4DFC.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115105109/-/DCSupplemental.
References
- 1.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 2.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J Biol Chem. 1990;265:21330–21336. [PubMed] [Google Scholar]
- 3.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. II. Catalytic properties. J Biol Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 4.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 5.Park J-S, Marr MT, Roberts JW. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 6.Deaconescu AM, Savery N, Darst SA. The bacterial transcription repair coupling factor. Curr Opin Struct Biol. 2007;17:96–102. doi: 10.1016/j.sbi.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts J, Park JS. Mfd, the bacterial transcription repair coupling factor: Translocation, repair and termination. Curr Opin Microbiol. 2004;7:120–125. doi: 10.1016/j.mib.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J Biol Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 10.Selby CP, Sancar A. Structure and function of the (A)BC excinuclease of Escherichia coli. Mutat Res. 1990;236:203–211. doi: 10.1016/0921-8777(90)90005-p. [DOI] [PubMed] [Google Scholar]
- 11.Pomerantz RT, O'Donnell M. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science. 2010;327:590–592. doi: 10.1126/science.1179595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han J, Sahin O, Barton YW, Zhang Q. Key role of Mfd in the development of fluoroquinolone resistance in Campylobacter jejuni. PLoS Pathog. 2008;4:e1000083. doi: 10.1371/journal.ppat.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee GH, et al. The Helicobacter pylori Mfd protein is important for antibiotic resistance and DNA repair. Diagn Microbiol Infect Dis. 2009;65:454–456. doi: 10.1016/j.diagmicrobio.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Ayora S, Rojo F, Ogasawara N, Nakai S, Alonso JC. The Mfd protein of Bacillus subtilis 168 is involved in both transcription-coupled DNA repair and DNA recombination. J Mol Biol. 1996;256:301–318. doi: 10.1006/jmbi.1996.0087. [DOI] [PubMed] [Google Scholar]
- 15.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zalieckas JM, Wray LVJ, Jr, Ferson AE, Fisher SH. Transcription-repair coupling factor is involved in carbon catabolite repression of the Bacillus subtilis hut and gnt operons. Mol Microbiol. 1998;27:1031–1038. doi: 10.1046/j.1365-2958.1998.00751.x. [DOI] [PubMed] [Google Scholar]
- 17.Washburn RS, Wang Y, Gottesman ME. Role of E.coli transcription-repair coupling factor Mfd in Nun-mediated transcription termination. J Mol Biol. 2003;329:655–662. doi: 10.1016/s0022-2836(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 18.Deaconescu AM, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 19.Assenmacher N, Wenig K, Lammens A, Hopfner KP. Structural basis for transcription-coupled repair: The N terminus of Mfd resembles UvrB with degenerate ATPase motifs. J Mol Biol. 2006;355:675–683. doi: 10.1016/j.jmb.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 20.Pakotiprapha D, Liu Y, Verdine GL, Jeruzalmi D. A structural model for the damage-sensing complex in bacterial nucleotide excision repair. J Biol Chem. 2009;284:12837–12844. doi: 10.1074/jbc.M900571200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manelyte L, Kim YI, Smith AJ, Smith RM, Savery NJ. Regulation and rate enhancement during transcription-coupled DNA repair. Mol Cell. 2010;40:714–724. doi: 10.1016/j.molcel.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy MN, et al. An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd. Nucleic Acids Res. 2009;37:6042–6053. doi: 10.1093/nar/gkp680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selby CP, Sancar A. Transcription-repair coupling and mutation frequency decline. J Bacteriol. 1993;175:7509–7514. doi: 10.1128/jb.175.23.7509-7514.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava DB, Darst SA. Derepression of bacterial transcription-repair coupling factor is associated with a profound conformational change. J Mol Biol. 2011;406:275–284. doi: 10.1016/j.jmb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pakotiprapha D, et al. Crystal structure of Bacillus stearothermophilus UvrA provides insight into ATP-modulated dimerization, UvrB interaction, and DNA binding. Mol Cell. 2008;29:122–133. doi: 10.1016/j.molcel.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deaconescu AM, Darst SA. Crystallization and preliminary structure determination of Escherichia coli Mfd, the transcription-repair coupling factor. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:1062–1064. doi: 10.1107/S1744309105035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truglio JJ, et al. Interactions between UvrA and UvrB: The role of UvrB's domain 2 in nucleotide excision repair. EMBO J. 2004;23:2498–2509. doi: 10.1038/sj.emboj.7600263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volkov V, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. Biophys J. 2003;80:2946–2953. [Google Scholar]
- 30.Prabha S, Rao DN, Nagaraja V. Distinct properties of hexameric but functionally conserved Mycobacterium tuberculosis transcription-repair coupling factor. PLoS ONE. 2011;6:e19131. doi: 10.1371/journal.pone.0019131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dürr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: Differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith AJ, Szczelkun MD, Savery NJ. Controlling the motor activity of a transcription-repair coupling factor: Autoinhibition and the role of RNA polymerase. Nucleic Acids Res. 2007;35:1802–1811. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reyes N, Ginter C, Boudker O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature. 2009;462:880–885. doi: 10.1038/nature08616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Careaga CL, Falke JJ. Thermal motions of surface alpha-helices in the D-galactose chemosensory receptor. Detection by disulfide trapping. J Mol Biol. 1992;226:1219–1235. doi: 10.1016/0022-2836(92)91063-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westblade LF, et al. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 2010;38:8357–8369. doi: 10.1093/nar/gkq692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selby CP, Witkin EM, Sancar A. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: In vitro complementation with purified coupling factor. Proc Natl Acad Sci USA. 1991;88:11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hauk G, Bowman GD. Structural insights into regulation and action of SWI2/SNF2 ATPases. Curr Opin Struct Biol. 2011;21:719–727. doi: 10.1016/j.sbi.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunnewell ME. Probing the conformational changes in the repair enzyme Mfd using mutant protein constructs. 2008 MS thesis (University of Massachusetts, Amherst, MA) [Google Scholar]
- 39.Hsieh C-H. Probing the activation mechanism of transcription-coupled repair factor Mfd. 2010 MS thesis (University of Massachusetts, Amherst, MA) [Google Scholar]
- 40.Theis K, et al. The nucleotide excision repair protein UvrB, a helicase-like enzyme with a catch. Mutat Res. 2000;460:277–300. doi: 10.1016/s0921-8777(00)00032-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.