Abstract
Imprinted gene expression associated with Prader–Willi syndrome (PWS) and Angelman syndrome (AS) is controlled by two imprinting centers (ICs), the PWS-IC and the AS-IC. The PWS-IC operates in cis to activate transcription of genes that are expressed exclusively from the paternal allele. We have created a conditional allele of the PWS-IC to investigate its developmental activity. Deletion of the paternal PWS-IC in the embryo before implantation abolishes expression of the paternal-only genes in the neonatal brain. Surprisingly, deletion of the PWS-IC in early brain progenitors does not affect the subsequent imprinted status of PWS/AS genes in the newborn brain. These results indicate that the PWS-IC functions to protect the paternal epigenotype at the epiblast stage of development but is dispensable thereafter.
Prader–Willi syndrome (PWS) and Angelman syndrome (AS) are genetic disorders resulting from the loss of expression of a cluster of imprinted genes at chromosome 15q11-q13. Individuals with PWS lack paternal gene expression from this imprinted region whereas individuals with AS lack maternal gene expression (1). In contrast to imprinted domains controlled by a single imprinting center, the PWS/AS region is regulated by a bipartite imprinting center composed of the AS imprinting center (AS-IC) and the PWS-IC (2). Imprinting in the region results from interplay between the AS-IC and the PWS-IC. Substantial evidence suggests that the PWS-IC is a positive-acting element that stimulates the expression of genes in the 2-Mb PWS/AS region. The AS-IC is thought to act in the female germline to inactivate the PWS-IC on the future maternal allele (3). Thus, a functional PWS-IC is necessary for the expression of paternal-only genes and an intact AS-IC is necessary to silence these genes on the maternal chromosome. Using a targeted deletion of a 35-kb region spanning Snrpn exons 1 to 6, we previously reported that the location and function of the PWS-IC is conserved in mice (4). More recently, we found that a 6-kb deletion surrounding Snrpn exon 1 yields an identical phenotype to the 35-kb deletion, demonstrating that all the functional elements of the PWS-IC are within this 6-kb region (5).
The PWS-IC lies within a region of allele-specific DNA methylation. The silent maternal allele is hypermethylated whereas the paternal allele is hypomethylated. These DNA methylation imprints are erased and reestablished in the germline during gametogenesis (6–10). When the PWS-IC acts in somatic tissue to regulate allele-specific gene expression is unknown. Bielinska et al. (11) found that the PWS-IC is required postzygotically in both humans and mice. An individual with minor clinical symptoms of PWS was shown to be mosaic for a PWS-IC deletion, suggesting that the PWS-IC is necessary to maintain paternal gene expression postzygotically. Mice chimeric for a PWS-IC deletion on the paternal chromosome exhibited promoter hypermethylation and loss of expression of Ndn and Mkrn3, supporting the interpretation that the PWS-IC is required postzygotically (11).
We have created a conditional allele of the PWS-IC to further investigate the temporal and spatial functions of this element. As predicted from previous work, we find that the PWS-IC is necessary before implantation. Unexpectedly, deletion of the PWS-IC early in neurogenesis does not affect subsequent expression of imprinted genes at the locus in newborn brain. Deletion early in neurogenesis does reduce Snrpn and snoRNA expression, but this effect is likely a result of the removal of the Snrpn major promoter. These pups appear to bypass the early failure to thrive seen in other PWS models but still exhibit a reduction in postweaning weight gain. These results confirm that the PWS-IC is necessary postzygotically for paternal gene expression but also demonstrate that it is not required later in development for maintenance of the paternal epigenotype.
Results
Generation of PWS-IC Conditional Deletion Allele.
We previously demonstrated that the entire murine PWS-IC lies within a 6-kb region located between −3.7 kb and +2.3 kb with reference to Snrpn exon 1. Furthermore, we generated a conditional PWS-IC allele by flanking this region with loxP sites (5). The structure of this conditional allele is shown in Fig. S1.
Deletion of PWS-IC Before Implantation Results in Reduced Birth Weight and Neonatal Lethality.
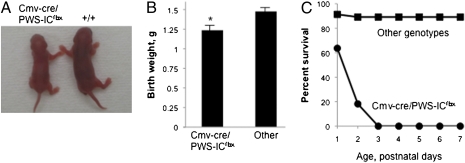
To investigate the effects of early embryonic deletion of the PWS-IC, we analyzed offspring from a mating of males heterozygous for the PWS-ICflox6kb allele with transgenic (Tg) Cmv-cre (TgCmv-cre) hemizygous females. The Cmv-cre transgene expresses Cre ubiquitously before implantation (12). Similar to paternal inheritance of a PWS-IC deletion (4, 5), PWS-IC+/flox6kb TgCmv-cre pups were visibly smaller and had significantly lower birth weights than littermates (Fig. 1 A and B). Cmv-cre–mediated deletion of the PWS-IC before implantation caused 100% postnatal lethality with no pups surviving beyond postnatal day (P) 2 (Fig. 1C). Southern blot analysis of DNA isolated from several P1 brains suggests that Cmv-cre mediated deletion is extensive (Fig. S2). As expected, mice with a somatic deletion of the PWS-IC on the maternal chromosome were normal, as were mice inheriting a maternal or paternal PWS-ICflox6kb allele in the absence of Cre expression (5).
Fig. 1.
PWS-IC+/flox6kb TgCmv-cre pups are small and have reduced survival compared with littermates of other genotypes. (A) The PWS-IC+/flox6kb TgCmv-cre neonate on the left is smaller than the WT littermate on the right (P1). (B) PWS-IC+/flox6kb TgCmv-cre mice weigh significantly less than mice of other genotypes at birth (P = 0.0007; n = 11 and n = 59, respectively). (C) PWS-IC+/flox6kb TgCmv-cre mice (circles, n = 11) exhibit postnatal lethality with no survivors beyond P2 compared with other genotypes (squares, n = 46).
Paternal Gene Expression Requires Presence of PWS-IC in Preimplantation Embryo.
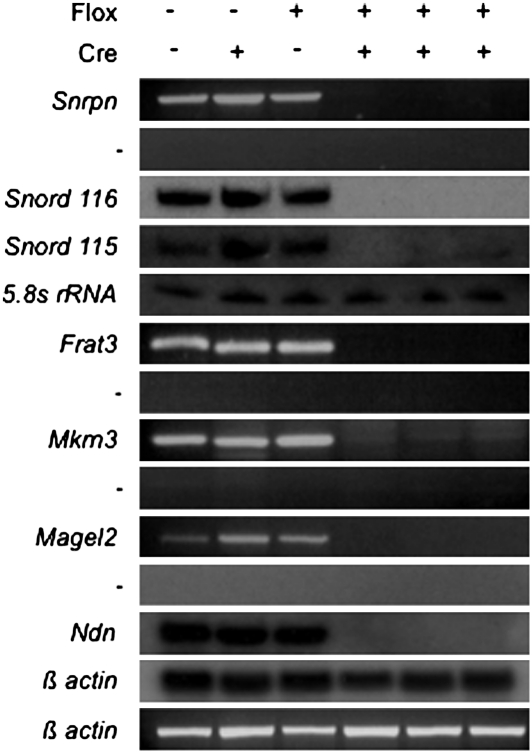
To determine the extent of the imprinting defect resulting from preimplantation deletion of the PWS-IC, we examined gene expression in RNA isolated from the brains of P1 PWS-IC+/flox6kb TgCmv-cre pups. Deletion of the PWS-IC before implantation affected the expression of all genes in the region. Snrpn, Frat3, Ndn, Magel2, Mkrn3, Snord116, and Snord115 expression was not detected in PWS-IC+/flox6kb TgCmv-cre newborn mice (Fig. 2). Gene expression patterns in PWS-IC+/flox6kb pups that did not inherit TgCmv-cre were indistinguishable from WT.
Fig. 2.
Expression of PWS genes in PWS-IC+/flox6kb TgCmv-cre P1 brains. Expression of the indicated genes was determined by either Northern blot or RT-PCR. A minus sign to the left of RT-PCR analyses indicates control samples in which reverse transcriptase was omitted during cDNA synthesis.
Deletion of PWS-IC in Neuronal Precursors Does Not Result in an Imprinting Defect.
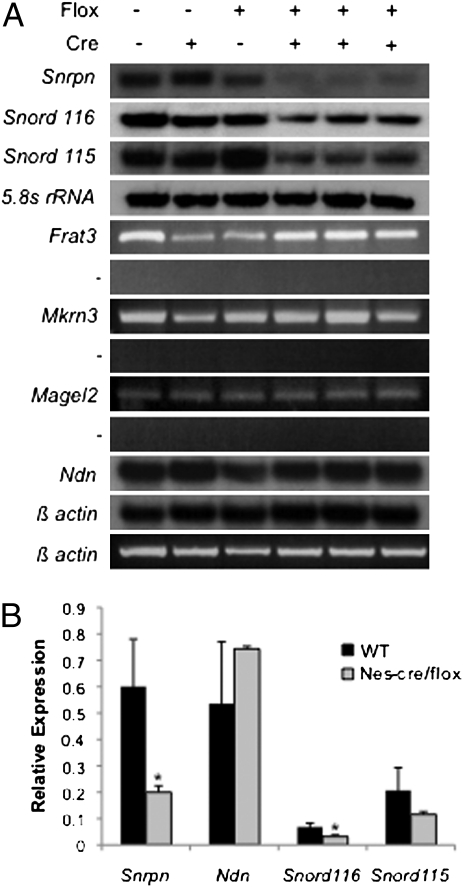
Cmv-cre–mediated deletion of the PWS-IC results in a complete imprinting defect and thereby demonstrates that the PWS-IC must be intact in the preimplantation embryo to maintain the paternal pattern of gene expression during subsequent embryonic development. We next sought to establish whether the PWS-IC is required to maintain the imprint at later times in development. PWS-ICflox6kb males were mated to transgenic Nes-cre females to mediate PWS-IC deletion in neuronal precursors and glia by embryonic day (E) 10.5 with widespread deletion by E12.5 (13, 14). The paternally expressed genes Ndn, Frat3, Magel2, and Mkrn3 were unaltered in the brains of PWS-IC+/flox6kb TgNes-cre newborns (Fig. 3). Normal expression of these genes indicates that the PWS-IC is not necessary to maintain paternal gene expression in brain precursor cells beyond E12.5.
Fig. 3.
Expression of upstream PWS genes in P1 brains is preserved whereas expression of the downstream cluster of genes is reduced in PWS-IC+/flox6kb TgNes-cre mice. (A) Gene expression assays were as indicated in Fig. 2. (B) Expression levels of Snrpn, Ndn, Snord116, and Snord115 were quantified by PhosphorImager analysis of the Northern blots. The average of the three control lanes ± SE was compared with the average of the three PWS-IC+/flox6kb TgNes-cre lanes and normalized to β-actin (Snrpn and Ndn) or 5.8S RNA (Snord115 and 116) exposures on the same blot. P values for reduction in Snrpn, Snord116, and Snord115 expression levels are 0.018, 0.017, and 0.142, respectively. Ndn level was not significantly altered (P = 0.2, Student t test).
The downstream cluster of genes, Snrpn, several snoRNAs, and Ube3a-as, are likely processed from a common 1-Mb transcript (15–17). Deletion of the PWS-IC also removes the major promoter for this transcript. Snrpn expression in the brains of P1 PWS-IC+/flox6kb TgNes-cre mice is reduced by almost 70%, whereas the snoRNAs, Snord116 and Snord115, are reduced by approximately half (Fig. 3). Nes-cre–mediated deletion of the PWS-IC in P1 brain was extensive (Fig. S2), indicating that the observed residual gene expression was not the result of incomplete PWS-IC deletion. We investigated whether deletion of the major promoter for this transcript contributed to the reduction in expression of Snrpn and the snoRNAs. Several distant upstream exons (U exons) for Snrpn have previously been found to contribute to transcription of the locus (15, 18). We tested whether the residual expression of Snrpn and the snoRNAs could originate from the Snrpn U exons in PWS-IC+/flox6kb TgNes-cre animals. Transcripts originating at Snrpn upstream exon U1 and extending to Snrpn exon 3 were detectable in the brains of PWS-IC+/flox6kb TgNes-cre mice but not in mice inheriting the same deletion via the germline, nor in PWS-IC+/flox6kb TgCmv-cre mice (Fig. 4). These results confirm and extend previous findings that the U exons contribute to the Snrpn transcription unit and also demonstrate that U exon use requires the presence of the PWS-IC before implantation (18).
Fig. 4.
Snrpn upstream exon use in the brain. Snrpn upstream exon transcripts were detected by RT-PCR using primers in upstream exon U1 and exon 3. (A) Expression from Snrpn upstream exon 1 is not detected following germline transmission of the PWS-IC deletion. (B) Snrpn upstream exon transcripts following Cmv-cre– or Nes-cre–mediated deletion.
Paternal expression of the Snrpn transcription unit is widely thought to contribute to silencing of Ube3a on the paternal allele via antisense transcription (19–21). Elimination of the major Snrpn promoter prompted us to investigate allelic expression of Ube3a. Females heterozygous for the Nes-cre transgene and heterozygous for B6.cast.c7 (congenic for Mus musculus castaneus across a portion of chromosome 7) were crossed with PWS-IC+/flox6kb males, and the progeny were analyzed at 12 wk of age. As shown for other mutations that down-regulate Snrpn–Ube3a-as expression, we also found an increase in paternal contribution of Ube3a in PWS-IC+/flox6kb TgNes-cre mice (Fig. S3).
Paternal Epigenotype at Ndn and Mkrn3 Is Stable Postzygotically.
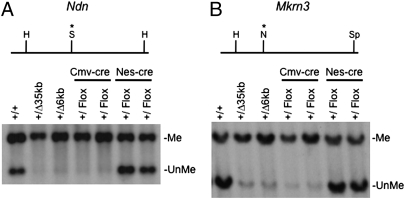
The Ndn and Mkrn3 promoters exhibit differential DNA methylation with hypermethylation of the maternal allele (22, 23). Digestion with methyl-sensitive restriction endonucleases was used to analyze the DNA methylation status of Mkrn3 and Ndn in both PWS-IC+/flox6kb TgCmv-cre and PWS-IC+/flox6kb TgNes-cre mice. Germline transmission of the 6-kb PWS-IC deletion led to the paternal allele adopting a maternal DNA methylation pattern at both Mkrn3 and Ndn (5), as did deletion with Cmv-cre. Nes-cre–mediated deletion, which does not cause an imprinting defect, resulted in a WT DNA methylation pattern at both loci (Fig. 5). Thus, the DNA methylation imprints at Ndn and Mkrn3 are stable in the brain even in the absence of the PWS-IC.
Fig. 5.
DNA methylation analysis of the Ndn and Mkrn3 differentially methylated regions. DNA from brains of WT (+/+), PWS-IC+/Δ35kb, PWS-IC+/Δ6 kb, PWS-IC+/flox6kb TgCmv-cre, and PWS-IC+/flox6kb TgNes-cre pups was digested with the indicated restriction endonucleases, Southern blotted, and probed. (A) The upper part shows a restriction map of the Ndn locus. The DNA methylation sensitive endonuclease SacII (S) is located between two HindIII (H) sites. The Ndn probe detects a 3.3-kb fragment (Me) resulting from DNA methylation at the SacII site and a 1.8-kb fragment obtained from unmethylated DNA (UnMe). (B) The upper part shows a restriction map of the Mkrn3 locus. A site recognized by the DNA methylation sensitive NotI (N) endonuclease is located between HindIII and SpeI (Sp) sites at the Mkrn3 locus. The probe detects methylated and unmethylated fragments of 6.8 and 5.0 kb, respectively.
Deletion of PWS-IC in Neuronal Precursors Results in Postweaning Growth Deficiency.
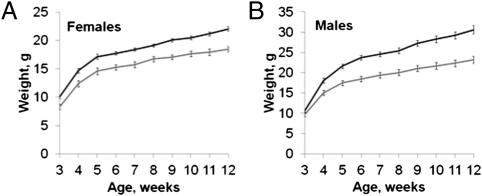
The reduction in Snrpn–Ube3a-as expression in the nervous system led us to investigate growth traits of PWS-IC+/flox6kb TgNes-cre deletion mice. Pups inheriting a PWS-ICflox6kb allele paternally and the Nes-cre transgene maternally did not exhibit a statistically significant difference in birth weight. PWS-IC+/flox6kb TgNes-cre pups regularly exhibited milk spots and lacked the pallor characteristic of PWS-IC+/flox6kb TgCmv-cre newborns or pups inheriting a germline deletion of the PWS-IC (5). PWS-IC+/flox6kb TgNes-cre pups were born with the expected Mendelian frequency (14 of 52; 26.9%) and did not show a reduction in postnatal survival compared with littermates of other genotypes. However, PWS-IC+/flox6kb TgNes-cre mice did exhibit a growth retardation that became increasingly apparent during the postweaning period (Fig. 6). By 3 wk of age, female PWS-IC+/flox6kb TgNes-cre mice displayed a significant growth deficiency (8.32 ± 0.48 vs. 10.1 ± 0.32; P = 0.022) and, although their growth curve was similar to littermates, they remained growth-restricted (Fig. 6A). Males of the same genotype also exhibited a growth deficiency that reached statistical significance by 4 wk of age (15.1 ± 0.52 vs. 18.1 ± 0.36; P = 0.012; Fig. 6B).
Fig. 6.
Postweaning growth of PWS-IC+/flox6kb TgNes-cre mice and littermates. Mice were weighed weekly after weaning. Weight curves for female (A) and male (B) PWS-IC+/flox6kb TgNes-cre mice and their littermates (n = 6 and n = 32 for females and n = 8 and n = 20 for males, PWS-IC+/flox6kb TgNes-cre and other mice, respectively). The gray line shows weight gain for PWS-IC+/flox6kb TgNes-cre mice and the black line shows the weight gain for the other genotypes. Error bars indicate SE.
Discussion
We recently demonstrated that paternal transmission of a 6-kb deletion spanning Snrpn exon 1 leads to low birth weight, neonatal lethality, and complete loss of PWS gene expression in newborn brain (5). This phenotype is indistinguishable from that of a previously described 35-kb PWS-IC deletion and indicates that the entire murine PWS-IC is included within the boundaries of the 6-kb deletion. Here, we have characterized the consequences of conditional deletion of this minimal PWS-IC.
Evidence from both a mosaic individual with some features of PWS and chimeric mice suggests that the PWS-IC is necessary postzygotically for paternal PWS gene expression and to avoid a maternal methylation imprint (11). Both the mosaic individual and chimeric mice transmitted the deletion, implying that the PWS-IC may not be necessary after separation of the germline from soma. Cmv-cre–mediated deletion of the PWS-IC before implantation results in a gene expression pattern indistinguishable from germline transmission of a PWS-IC deletion, thus indicating that the PWS-IC is necessary before or during implantation. In contrast, Nes-cre–mediated deletion of the PWS-IC in brain progenitors, where extensive deletion occurs by E12.5, does not lead to an imprinting defect. In this case, expression of Mkrn3, Magel2, Frat3, and Ndn in newborn brain is unaffected by the absence of the PWS-IC. Together, these results establish that the presence of the PWS-IC is required in the period before implantation through formation of early neural precursors. This period is characterized by dynamic epigenetic changes in somatic tissues. Fertilization is followed by DNA demethylation of the paternal and maternal genomes. The embryonic genome is then remethylated at the epiblast stage early in gastrulation (reviewed in ref. 24). This stage of genomic DNA methylation is likely a critical period for PWS-IC function. Our data are consistent with a model in which the PWS-IC prevents acquisition of a maternal imprint on the paternal allele during this developmental stage.
The finding that the paternal epigenotype can be maintained throughout much of neurogenesis in the absence of the PWS-IC was unexpected. The contribution of epigenetic modifications to neural development is poorly understood, although several reports illuminate the importance of this form of gene regulation. The human brain exhibits region specific DNA methylation patterns, suggesting that DNA methylation contributes to neural development (25). A role for epigenetic regulation in brain development is further suggested by mutations in DNA methyltransferases (reviewed in ref. 26). Our results indicate that the epigenotype of paternally expressed genes at the PWS/AS locus is dependent upon the PWS-IC during early embryonic development. However, by the time Nes-cre becomes active in neural cells at E12.5, maintenance of the paternal epigenotype no longer requires the PWS-IC.
Previous efforts to conditionally inactivate elements regulating imprinted gene expression during embryonic development have focused on the H19/Igf2 and Rasgrf1 loci. H19 is normally silenced on the paternal allele whereas Igf2 is normally silenced on the maternal chromosome. Germline transmission of a paternal IC deletion at this locus results in expression of the normally silent paternal H19 allele (27, 28). Conditional deletion of the paternal IC in zygotes also leads to biallelic H19 expression. However, H19 expression remains exclusively maternal after paternal IC deletion in terminally differentiated skeletal, cardiac, or liver cells, suggesting that the imprinted status of H19 becomes IC-independent sometime after implantation. The Rasgrf1 locus is paternally expressed. Acquisition of DNA methylation at a differentially methylated domain in the male germline is dependent upon a set of repeats located 30 kb upstream of the transcription start site. A conditional deletion strategy targeting these repeats demonstrated that, although the repeats are necessary to maintain differential methylation after fertilization, they are no longer necessary after the epiblast stage (29). Unlike these loci, the DNA methylation imprint is maternally acquired at the PWS/AS locus. The paternal PWS-IC is necessary to establish expression of paternal genes but is dispensable for paternal gene expression by the time neural precursors are formed. The Rasgrf1 study demonstrates stability of the paternal methylated state of the differentially methylated domain after implantation, whereas the H19/Igf2 studies establish that the paternal IC is dispensable for continued paternal H19 silencing.
Snrpn, Snord116, Snord115, and Ube3a-as RNAs are generated from primary transcripts with a common initiation site (15–17, 30–32). Expression of these genes is reduced following Nes-cre-mediated deletion, most likely because of the absence of the major promoter at Snrpn exon 1 rather than epigenetic modifications of the locus. As previously suggested, residual transcription of these genes is likely to initiate at several upstream promoters that splice into Snrpn exon 2 or into acceptor sites located further downstream (18).
Conditional deletion of the PWS-IC with Nes-cre results in adults that are smaller than WT littermates. The only significant alteration in PWS gene expression is a reduction in Snrpn and the snoRNAs, and an increase in paternal expression of Ube3a. In this regard, these animals resemble other models lacking expression of the Snrpn transcription unit as a result of a germline deletion of the PWS-IC or a deletion spanning from Snrpn to Ube3a (18, 33, 34). In the present PWS-ICflox model, the Snrpn U exons drive expression of all of the downstream cluster gene products indicating that the upstream exons are not exclusively dedicated to a particular processing variant. Conditional deletion of the PWS-IC with Nes-cre suggests that even modest changes in the level of the Snrpn transcription unit can significantly affect body weight. Previous studies have also shown that components of the large Snrpn to Ube3a-as transcription unit affect body weight (18, 21, 34–36). Inheritance of a Snrpn promoter that is not maternally silenced causes an increase in downstream transcription products and a significant increase in body weight (21). Unlike previous PWS-IC mutants, conditional deletion in brain progenitors of the PWS-IC by Nes-cre does not result in neonatal lethality or overt signs of neonatal failure to thrive. These results suggest that the small size of these mutants is not a result of neonatal failure to thrive but rather a reduction in the postweaning activity of the snoRNAs, as growth retardation is characteristic of other mouse models featuring reduced Snord116 expression (34–36).
Materials and Methods
Animals.
All animal procedures were previously approved by the University of Florida Institutional Animal Care and Use Committee. The PWS-ICflox6kb allele was created as previously described (5). B6.C-Tg (Cmv-cre)1Cgn/J (stock no. 006054) and B6.C-Tg(Nes-cre)1Kln/J (stock no. 006054) were obtained from the Jackson Laboratory. The Nes-cre females were used after 13 to 15 generations of backcross to C57BL/6. The B6.cast.c7 line has been previously described (20). Animal weights are expressed as averages ± SE. Statistical significance was determined by unpaired Student t test.
Gene Expression Analysis.
Northern Blot analysis and RT-PCR were performed as previously described (5). RT-PCR for the Snrpn upstream exon use was performed as described previously (18). Northern blot band intensity was digitized on the Storm 860 PhosphorImager (GE Healthcare) and analyzed with ImageQuant TL software.
DNA Methylation Analysis.
DNA was isolated from newborn brain, digested with the indicated restriction endonucleases, and analyzed by Southern blot as previously described (5). Probe details are available on request.
Supplementary Material
Acknowledgments
The authors thank Theresa Strong, Rob Nicholls, Tom Yang, and Brian Harfe for helpful comments on the manuscript; Deb Morse and Ryan Hallett for technical assistance; Ryan Fiske of the University of Florida Mouse Models Core; and Susan D'Costa, Carlos Sulsona, and Justin Bickford. This work was supported by grants from the Foundation for Prader–Willi Research and National Institute of Child Health and Human Development Grant R01 HD 037872. E.Y.S. was supported by a University of Florida Alumni Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115057109/-/DCSupplemental.
References
- 1.Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet. 2001;2:153–175. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- 2.Buiting K. Prader-Willi syndrome and Angelman syndrome. Am J Med Genet C Semin Med Genet. 2010;154C:365–376. doi: 10.1002/ajmg.c.30273. [DOI] [PubMed] [Google Scholar]
- 3.Brannan CI, Bartolomei MS. Mechanisms of genomic imprinting. Curr Opin Genet Dev. 1999;9:164–170. doi: 10.1016/S0959-437X(99)80025-2. [DOI] [PubMed] [Google Scholar]
- 4.Yang T, et al. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- 5.Dubose AJ, Smith EY, Yang TP, Johnstone KA, Resnick JL. A new deletion refines the boundaries of the murine Prader-Willi syndrome imprinting center. Hum Mol Genet. 2011;20:3461–3466. doi: 10.1093/hmg/ddr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glenn CC, Porter KA, Jong MT, Nicholls RD, Driscoll DJ. Functional imprinting and epigenetic modification of the human SNRPN gene. Hum Mol Genet. 1993;2:2001–2005. doi: 10.1093/hmg/2.12.2001. [DOI] [PubMed] [Google Scholar]
- 7.Shemer R, Birger Y, Riggs AD, Razin A. Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci USA. 1997;94:10267–10272. doi: 10.1073/pnas.94.19.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeschnigk M, et al. Imprinted segments in the human genome: Different DNA methylation patterns in the Prader-Willi/Angelman syndrome region as determined by the genomic sequencing method. Hum Mol Genet. 1997;6:387–395. doi: 10.1093/hmg/6.3.387. [DOI] [PubMed] [Google Scholar]
- 9.Hajkova P, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 10.Sutcliffe JS, et al. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- 11.Bielinska B, et al. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat Genet. 2000;25:74–78. doi: 10.1038/75629. [DOI] [PubMed] [Google Scholar]
- 12.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 14.Graus-Porta D, et al. Beta1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron. 2001;31:367–379. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 15.Landers M, et al. Regulation of the large (∼1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf. Snrpn. Nucleic Acids Res. 2004;32:3480–3492. doi: 10.1093/nar/gkh670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Meur E, et al. Dynamic developmental regulation of the large non-coding RNA associated with the mouse 7C imprinted chromosomal region. Dev Biol. 2005;286:587–600. doi: 10.1016/j.ydbio.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 17.Runte M, et al. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 18.Bressler J, et al. The SNRPN promoter is not required for genomic imprinting of the Prader-Willi/Angelman domain in mice. Nat Genet. 2001;28:232–240. doi: 10.1038/90067. [DOI] [PubMed] [Google Scholar]
- 19.Rougeulle C, Cardoso C, Fontés M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 20.Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73:316–322. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- 21.Johnstone KA, et al. A human imprinting centre demonstrates conserved acquisition but diverged maintenance of imprinting in a mouse model for Angelman syndrome imprinting defects. Hum Mol Genet. 2006;15:393–404. doi: 10.1093/hmg/ddi456. [DOI] [PubMed] [Google Scholar]
- 22.Hanel ML, Wevrick R. Establishment and maintenance of DNA methylation patterns in mouse Ndn: implications for maintenance of imprinting in target genes of the imprinting center. Mol Cell Biol. 2001;21:2384–2392. doi: 10.1128/MCB.21.7.2384-2392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jong MT, et al. A novel imprinted gene, encoding a RING zinc-finger protein, and overlapping antisense transcript in the Prader-Willi syndrome critical region. Hum Mol Genet. 1999;8:783–793. doi: 10.1093/hmg/8.5.783. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: Reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 25.Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng J, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava M, et al. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev. 2000;14:1186–1195. [PMC free article] [PubMed] [Google Scholar]
- 28.Thorvaldsen JL, Fedoriw AM, Nguyen S, Bartolomei MS. Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus. Mol Cell Biol. 2006;26:1245–1258. doi: 10.1128/MCB.26.4.1245-1258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmes R, Chang Y, Soloway PD. Timing and sequence requirements defined for embryonic maintenance of imprinted DNA methylation at Rasgrf1. Mol Cell Biol. 2006;26:9564–9570. doi: 10.1128/MCB.00058-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavaillé J, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de los Santos T, Schweizer J, Rees CA, Francke U. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which Is highly expressed in brain. Am J Hum Genet. 2000;67:1067–1082. doi: 10.1086/303106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wevrick R, Francke U. An imprinted mouse transcript homologous to the human imprinted in Prader-Willi syndrome (IPW) gene. Hum Mol Genet. 1997;6:325–332. doi: 10.1093/hmg/6.2.325. [DOI] [PubMed] [Google Scholar]
- 33.Chamberlain SJ, et al. Evidence for genetic modifiers of postnatal lethality in PWS-IC deletion mice. Hum Mol Genet. 2004;13:2971–2977. doi: 10.1093/hmg/ddh314. [DOI] [PubMed] [Google Scholar]
- 34.Tsai TF, Jiang YH, Bressler J, Armstrong D, Beaudet AL. Paternal deletion from Snrpn to Ube3a in the mouse causes hypotonia, growth retardation and partial lethality and provides evidence for a gene contributing to Prader-Willi syndrome. Hum Mol Genet. 1999;8:1357–1364. doi: 10.1093/hmg/8.8.1357. [DOI] [PubMed] [Google Scholar]
- 35.Ding F, et al. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS ONE. 2008;3:e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skryabin BV, et al. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 2007;3:e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.