Abstract
Antigenic variation enables pathogens to avoid the host immune response by continual switching of surface proteins. The protozoan blood parasite Trypanosoma brucei causes human African trypanosomiasis (“sleeping sickness”) across sub-Saharan Africa and is a model system for antigenic variation, surviving by periodically replacing a monolayer of variant surface glycoproteins (VSG) that covers its cell surface. We compared the genome of Trypanosoma brucei with two closely related parasites Trypanosoma congolense and Trypanosoma vivax, to reveal how the variant antigen repertoire has evolved and how it might affect contemporary antigenic diversity. We reconstruct VSG diversification showing that Trypanosoma congolense uses variant antigens derived from multiple ancestral VSG lineages, whereas in Trypanosoma brucei VSG have recent origins, and ancestral gene lineages have been repeatedly co-opted to novel functions. These historical differences are reflected in fundamental differences between species in the scale and mechanism of recombination. Using phylogenetic incompatibility as a metric for genetic exchange, we show that the frequency of recombination is comparable between Trypanosoma congolense and Trypanosoma brucei but is much lower in Trypanosoma vivax. Furthermore, in showing that the C-terminal domain of Trypanosoma brucei VSG plays a crucial role in facilitating exchange, we reveal substantial species differences in the mechanism of VSG diversification. Our results demonstrate how past VSG evolution indirectly determines the ability of contemporary parasites to generate novel variant antigens through recombination and suggest that the current model for antigenic variation in Trypanosoma brucei is only one means by which these parasites maintain chronic infections.
Antigenic variation enables pathogens to evade immune responses by continual switching of surface proteins (1, 2). The African trypanosomes (Trypanosoma spp.) are vector-borne protozoan blood parasites that survive in their hosts by antigenic variation, periodically replacing a monolayer of variant surface glycoproteins (VSG) (3) that shield the cell surface from immune effectors (4, 5). Trypanosoma brucei is the cause of human African trypanosomiasis (or “sleeping sickness”), and the mechanisms for expression and dynamic replacement of VSG in this species are a model system for antigenic variation (4) as well as a classic example of adaptive evolution at the host–pathogen interface. Two related veterinary parasites, Trypanosoma congolense and Trypanosoma vivax, also use antigenic variation to cause devastating diseases in domesticated animals. Through their detrimental effects on livestock productivity, these species arguably represent greater threats to socioeconomic well-being than T. brucei does in the agrarian societies in which they are endemic. Our understanding of how antigenic diversity is organized in T. brucei was greatly improved by the T. brucei 927 reference genome sequence (6). In this paper, we present draft genome sequences for T. congolense and T. vivax; we define their global VSG repertoires in a three-way comparative analysis with T. brucei, revealing how antigenic diversity evolved in trypanosome genomes past and present.
The T. brucei genome includes many hundreds of VSG that encode a transcriptionally silent reservoir of variant antigens (6), and each cell expresses just a single gene from a specialized telomeric expression site at any time (4, 5). The parasite population collectively express multiple VSG; when the host becomes immune to the prevailing VSG, clones expressing alternative copies proliferate in a frequency-dependent manner, maintaining the infection and resulting in characteristic “waves of parasitaemia.” To survive long-term, T. brucei must generate novel VSG sequences through recombination; mechanisms may include domain shuffling (7) and gene conversion among silent, subtelomeric gene copies or possibly in situ within the expression site (8). Functional variant antigens in T. brucei consist of a- and b-type VSG (hereafter a-VSG and b-VSG), which share the cysteine-rich C-terminal domain (CTD) but are otherwise distantly related (9–11). Although VSG are known to occur in T. congolense and T. vivax (12–16), the repertoire of variant antigens in these species is uncharacterized. Consequently, the evolutionary diversification of the VSG gene family has not been examined, although it has been suggested that VSG are a source of novel genes. Two gene families, the expression site-associated genes (ESAG6/7) encoding transferrin receptor (TFR) and the VSG-related (VR) genes, are thought to have evolved from a-VSG (17, 18) and b-VSG (8, 11), respectively.
The antigenic variation phenotype is observed in all African trypanosomes, and it is assumed that this reflects a common physiological model, which has been defined in T. brucei. The aim of this study is to identify the evolutionary processes that have created contemporary VSG diversity and reveal any significant differences in how trypanosome species generate variant antigens. Despite their shared phenotype, our results show that species differ in the organization of antigenic diversity at the genome level, and they provide a basis to better understand disease progression, pathology, and host range in all African trypanosomes.
Results
VSG Gene Repertoires of T. congolense and T. vivax.
We have produced high-quality draft genome sequences for T. congolense IL3000, a sister species of T. brucei, and T. vivax Y486, a third species that branches close to the root of the African trypanosome lineage (19). These genome sequences are described in Table S1 and are accessible through GeneDB (http://www.genedb.org) or TriTrypDB (http://tritrypdb.org). Comparative analysis including the existing T. brucei 927 genome sequence shows that the principal differences in genome content relate to cell-surface architecture (Tables S2–S4). To define VSG repertoires, gene sequences with predicted cell-surface roles were extracted from all three genomes and sorted with BLASTx, resulting in 81 gene families (Materials and Methods and Table S5). Phylogenies of these families were estimated that we collectively termed the “cell-surface phylome,” (http://www.genedb.org/Page/trypanosoma_surface_phylome). The phylome contains VSG and related families already known in T. brucei but it also defines families that we believe encode the VSG repertoires of T. congolense and T. vivax.
The T. congolense VSG repertoire differs from that of T. brucei in three ways: First, there is no a-VSG subfamily of variant antigens; second, there are two b-VSG subfamilies, termed Fam13 (n = 302) and Fam16 (n = 512) by their phylome designations; and, third, unlike T. brucei VSG, which all share a relatively uniform CTD, T. congolense VSG have 15–20 different CTD types, each associated with a specific subset of Fam13 or Fam16 and none homologous to the T. brucei CTD. Hence, T. congolense b-VSG are more structurally heterogeneous than T. brucei b-VSG are. We know that both Fam13 and Fam16 contain functional variant antigens because each family encompasses both published T. congolense VSG (12–14) and VSG expressed sequence tags (15). Although there is no a-VSG variant antigen, there are homologs of the a-VSG–like TFR genes of T. brucei, i.e., ESAG6/7 (Fam15; n = 43), as well as the procyclin-associated genes (PAG) (Fam14, n = 22), which also have an a-VSG–like structure (20).
VSG structural diversity is even greater in T. vivax. We have identified four VSG subfamilies (Fam23–26) that each possess definitive patterns of conserved cysteine residues (SI Results). Fam23 (n = 540) and Fam24 (n = 279) members possess sequence motifs homologous to a-VSG and b-VSG, respectively. Fam25 (n = 227) and Fam26 (n = 87) are two subfamilies unique to T. vivax but with low (∼20%) protein sequence similarity to known VSG (Fig. S1). These specific subfamilies may have evolved in T. vivax or may represent ancestral lineages not inherited by T. brucei and T. congolense. Transcriptomic data show that multiple members of all four families are transcribed in bloodstream-stage cells (Table S6). We find no orthologs to the TFR-like genes of T. brucei and T. congolense among T. vivax VSG-like genes or indeed the numerous T. vivax-specific gene families.
Amino acid sequence homology with T. brucei VSG alone does not guarantee that putative T. vivax VSG function as variant antigens. To date, only one T. vivax VSG [ILDat 2.1 (16)] has been characterized, albeit from a dissimilar strain, and it is most closely related to Fam26. Therefore, we identified an expressed VSG in the genome strain Y486 by MS analysis of a protein specific to a relapsed infection population, peptide fragments of which are 100% identical to a predicted protein in Fam23 (TvY486_0027060; Fig. S2). Therefore, at least one a-VSG–like (i.e., Fam23) gene in T. vivax encodes a functional variant antigen.
Phylogeny of VSG Diversification.
In total, the three genome sequences yielded 1,083 a-VSG–like and 1,537 b-VSG–like full-length genes (Table S7). We estimated Bayesian and maximum likelihood phylogenies from amino acid sequence alignments (Figs. S3 and S4); however, given the large number of sequences and to enable global visualization, we also estimated a similarity network from pairwise maximum likelihood protein distances that delivered a clearer picture of relationships within the a-VSG and b-VSG lineages. The distance network includes examples of all VSG subfamilies and represents individual genes as spheres connected to others sharing identity above a threshold (Materials and Methods). The network and phylogenies are fully consistent. Fig. 1 shows the similarity network from two angles, and Movie S1 contains an animation of the 3D network; four principal features emerge.
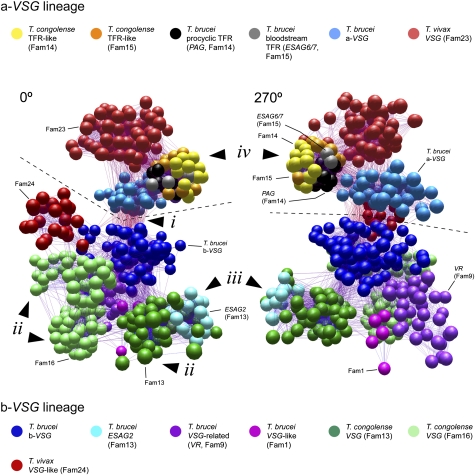
Fig. 1.
A sequence-similarity network of VSG-like sequences from African trypanosome genomes, shown from 0° and 270° angles. A 3D rendering of the network is provided as Movie S1. The network represents pairwise maximum likelihood protein distances, generated from multiple alignments of selected a-VSG–like (a-VSG, Fam23, TFR-like, and PAG-like proteins; n = 174) and b-VSG–like (b-VSG, Fam13, Fam16, Fam24, VR, ESAG2, and Fam1 proteins; n = 339) protein sequences, which are representative of global diversity. Spheres represent individual sequences shaded according to subfamily. A dashed line separates a-VSG–like subfamilies (above the line) and b-VSG subfamilies (below the line). Four significant features identified in the text are labeled: (i) sequence similarity between a-VSG and b-VSG lineages caused by the shared CTD of T. brucei VSG; (ii) diverse clusters of T. congolense b-VSG belonging to Fam13 and Fam16; (iii) the position of ESAG2 nested within Fam13; and (iv) tight cluster of TFR-like genes from both T. brucei and T. congolense.
First, the common CTD of T. brucei VSG must have evolved through horizontal transfer from one subfamily to the other. In Fig. 1, sequences cluster by lineage (a or b) rather than by species; for instance, T. vivax a-VSG (Fam23) is more similar to a-VSG–like subfamilies in T. brucei and T. congolense than to T. vivax b-VSG (Fam24). Therefore, VSG lineages are older than the genomes they occupy; indeed, they were present in the common ancestor of all African trypanosomes. The only above-threshold sequence connections occurring between a-VSG and b-VSG subfamilies (Fig. 1, i) concern T. brucei VSG and, in particular, their common CTD. This is a unique feature of T. brucei VSG and presents an interesting anomaly: despite belonging to ancient lineages separated in the ancestral trypanosome, a-VSG and b-VSG in T. brucei share a CTD that is species-specific, which can only be explained if the CTD evolved in one subfamily and was transposed to the other.
Second, b-VSG in T. brucei are derived from a single ancestral lineage, whereas T. congolense b-VSG are drawn from many lineages, suggesting that T. brucei b-VSG have passed through a “bottleneck.” In Fig. 1, all b-VSG in T. brucei (dark blue) form a cluster to the exclusion of other subfamilies. Hence, they share a recent common ancestor that evolved after the split from T. congolense. In contrast, T. congolense b-VSG comprise two lineages (Fam13 and Fam16) that originated in the T. brucei/T. congolense ancestor and form separate clusters in the network (Fig. 1, ii). We know that these lineages did not originate in T. congolense because their closest relatives are VSG-like genes in T. brucei (see below). In fact, Fam13 and Fam16 themselves split into multiple clusters in Fig. 1 (Fig. 1, ii), emphasizing the phylogenetic diversity of T. congolense VSG and relative homogeneity in T. brucei.
Third, VSG have repeatedly been a source of functional novelty on the cell surface. We know that VSG can be co-opted from variant antigen functions to novel roles, for example, the serum-resistance antigen [SRA (21)] and Trypanosoma brucei gambiense-specific glycoprotein [TgsGP (22)] proteins in Trypanosoma brucei rhodesiense and T. b. gambiense, respectively. However, SRA and tgsGP represent secondary loss of function among contemporary VSG. Fig. 1 shows that ESAG2, a gene family associated with the polycistronic VSG expression site in T. brucei, is a b-VSG–like gene, nested among T. congolense b-VSG (Fam13; Fig. 1, iii). Similarly, VR genes (purple in Fig. 1), rather than being derived from b-VSG in T. brucei, have an ancestral-type structure more akin to Fam16 in T. congolense. We have also identified another T. brucei-specific family (Fam1; pink in Fig. 1) that encodes proteins homologous to b-VSG, with a predicted GPI anchor but also a highly modified CTD. Fam1 (i.e., Tb927.6.1310) is preferentially expressed in bloodstream forms and localizes to the flagellar pocket and endosomal membranes (Fig. S5). Phylogenetic analysis clearly demonstrates that both ESAG2 and VR gene subfamilies, for which the evidence is against a variant antigen function (8, 11), are not recent derivations from T. brucei VSG (like SRA and TgsGP) but belong to ancestral VSG lineages with representatives in T. congolense that still encode functional variant antigens (Fig. S4). Hence, some of the ancestral lineages in T. congolense identified above remain in T. brucei but have been co-opted to novel roles.
Finally, the network indicates that the TFR evolved from an a-VSG gene, as suggested previously (8, 23). However, the functional transition did not occur within the T. brucei VSG expression site but instead in the T. brucei/T. congolense ancestor. A tight cluster of TFR-like genes (i.e., ESAG6/7 and PAG) from T. brucei as well as Fam14 and Fam15 sequences from T. congolense are distinct from other a-VSG subfamilies in Fig. 1 (Fig. 1, iv). The similarity network reflects their phylogeny, which shows that Fam14 and Fam15 are sister lineages to PAG and ESAG6/7, respectively, and their primary structures, which show that amino acid residues crucial for transferrin binding (18) are conserved in both species (Fig. S6). Given the absence of this entire family from T. vivax, we conclude that, rather than being loss from that species, the transferrin-receptor genes evolved before the separation of T. brucei and T. congolense but after their split from T. vivax. Our conclusion does not preclude other T. vivax-specific proteins performing a transferrin-binding function in that species.
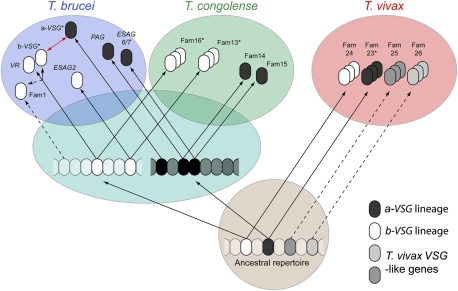
These results are summarized in a model of VSG evolution (Fig. 2). The ancestral African trypanosome possessed a-VSG and b-VSG type genes that probably formed multigene families or functioned as variant antigens. Both VSG types were inherited by T. vivax, which retains an a-VSG family that includes functional variant antigens. The T. brucei/T. congolense ancestor inherited both a-VSG and b-VSG lineages, and, at this point, one a-VSG gene was co-opted to a transferrin-binding role differentiated between insect and vertebrate life stages, founding a lineage that was inherited by both daughter species. Another a-VSG lineage retained its variant antigen function in T. brucei but was lost from T. congolense (SI Results). Of the ancestral b-VSG repertoire, two different lineages have been inherited by both species: The first has produced ESAG2 and Fam13 in T. brucei and T. congolense, respectively, whereas the second has produced b-VSG and VR in T. brucei and Fam16 in T. congolense. There is no step in this deduced scheme where a trypanosome lacks variant antigen. Clearly, these two species have adapted their common legacy differently. T. congolense VSG are drawn from multiple ancestral lineages, whereas T. brucei has relegated corresponding genes (VR, ESAG2, and perhaps Fam1) to novel roles and derives its variant antigens from single lineages derived after speciation. This difference in the phylogenetic diversity of VSG repertoires is important because it could affect the ability of the parasites to present novel antigens to their hosts and therefore to maintain infection.
Fig. 2.
A model of VSG gene family evolution in African trypanosomes. This cartoon depicts the elaboration of VSG subfamilies in contemporary and ancestral genomes. Uncertain origins are indicated by dashed lines. An asterisk indicates that a subfamily includes a proven variant antigen, although other variant antigens may occur in unmarked subfamilies. The red arrow indicates that the CTD is uniquely shared between a-VSG and b-VSG in T. brucei and has been donated from one subfamily to the other in either direction.
Tree Shape and Distribution of VSG Sequence Variation.
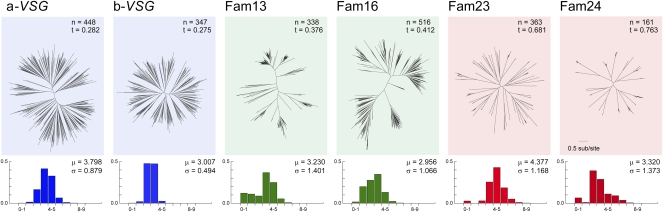
We examined the phylogenies of VSG subfamilies within species for evidence that their distinct evolutionary legacies have affected contemporary sequence evolution. Fig. 3 demonstrates how these trees have distinct topologies because of variation in the ratio of internal to terminal branches [described by “treeness” (24), T], which is low for T. brucei (T = 0.282 and 0.275), higher for T. congolense (T = 0.376 and 0.412), and highest for T. vivax (T = 0.681 and 0.763). T. congolense and T. vivax trees are more “tree-like” because they retain information about the past in basal nodes and internal branches, whereas the T. brucei tree consists mostly of long, terminal branches. Fig. 3 also compares the distribution of VSG sequence variation, showing that T. brucei distances have much narrower distributions than either T. congolense or T. vivax VSG do because both short, terminal branches and long, basal internodes are rare. Importantly, these patterns are genome-specific rather than lineage-specific effects, i.e., a-VSG and b-VSG in T. brucei display the same dynamic despite having greater identity with subfamilies in other species. They confirm that the mechanisms for antigenic variability vary between species now and likewise in the past.
Fig. 3.
Comparisons of phylogenetic tree topologies for VSG-like subfamilies. Bayesian phylogenies were estimated for six VSG subfamilies from T. brucei 927 (blue), T. congolense IL3000 (green), and T. vivax Y486 (red) with MrBayes 3.2.1 by using a WAG+Γ model. Default settings were applied, except for: Ngen = 5,000,000, Nruns = 4, samplefreq = 500, burnin = 1,000–2,500 (as required to achieve convergence). These trees contain all full-length protein sequences available (n) and include both intact genes and predicted pseudogenes. All trees are drawn to the same scale. The treeness statistic (T) describes the proportion of tree length taken up for internal branches (24) and is a measure of the phylogenetic signal-to-noise ratio. Below each tree a histogram describes the distribution of pairwise genetic distances (grouped into bins; x axis) plotted against frequency (y axis); mean average (μ) and SD (σ) are provided.
Recombination is a principal evolutionary pressure affecting T. brucei VSG (5, 8), and exchange of the unique VSG CTD is well recorded (7, 11). Recombination is also the mechanism through which VSG are transposed from subtelomeric loci into the telomeric expression site (4, 5, 8). T. brucei VSG phylogenies in Fig. 3 are consistent with frequent recombination, but the cladistic structure of T. congolense and T. vivax VSG phylogenies could only persist if recombination between clades is rare. Furthermore, the incidence of pseudogenes, which result, at least partly, from gene conversion between VSG genes (5), is much lower in T. congolense (where only 21.1% of Fam13 and 29.7% of Fam16 are predicted pseudogenes) and T. vivax (15.5% and 27.2% of Fam23 and Fam24, respectively) than in T. brucei (69.2% of a-VSG and 72.2% of b-VSG) (6). Therefore, we suspected that recombination frequency might account for species differences in sequence variation.
Contribution of Recombination to Antigenic Diversity.
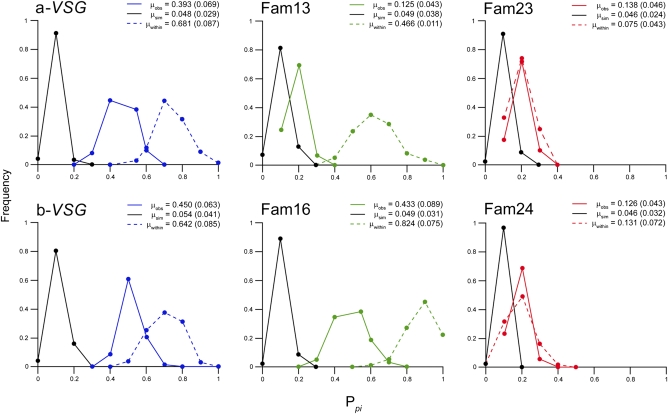
We examined VSG alignments for evidence of recombination in the form of phylogenetic incompatibility (PI) (25, 26), taking random samples of each alignment set and observing the proportion showing significant PI (Ppi; Table S8). Fig. 4 shows that Ppi (solid colored lines) was greatest for T. brucei a-VSG (0.392) and b-VSG (0.450) as well as for T. congolense Fam16 (0.433) and lower for T. congolense Fam13 (0.125) and T. vivax Fam23 (0.138) and Fam24 (0.126). In all cases, observed Ppi was significantly greater than a null distribution (black lines), confirming that PI was not solely caused by other homoplastic effects, such as rate heterogeneity (Materials and Methods). Recombination frequency is known to be proportional to sequence identity (27), and when we increased sequence identity within alignments by sampling only within crown clades, Ppi increased significantly (dashed colored lines) for T. brucei a-VSG (0.681) and b-VSG (0.642) as well as for T. congolense Fam13 (0.466) and Fam16 (0.823) but not for T. vivax. Finally, because the CTD is known to recombine in T. brucei (7, 11), we removed the CTD from T. brucei and T. congolense alignments, which resulted in a significant decrease in Ppi for T. brucei a-VSG (0.152, P < 0.0001) and b-VSG (0.234, P < 0.0001), but, in T. congolense, Ppi actually increased.
Fig. 4.
Prevalence of significant PI within VSG-like sequence alignments. PI describes conflicting phylogenetic signals caused by recombination but also rate heterogeneity or other molecular homoplasy (25). Protein sequence alignments for six VSG subfamilies were examined for PI using the pairwise homoplasy index (26). Each alignment was randomly sampled 100 times, and the proportion displaying significant PI was counted (Ppi). A distribution for Ppi was generated by creating 100 bootstrapped alignments in each case (solid colored lines). To generate a null distribution, 100 alignments were simulated from the observed Bayesian phylogeny using a maximum likelihood substitution model (WAG+Γ) that corrected for rate heterogeneity but did not consider recombination (black lines). To demonstrate the effect of genetic distance on PI, the analysis was repeated on smaller alignments of closely related sequences taken from crown clades (dashed colored lines; Materials and Methods). Mean average values, followed by SDs, are provided for observed (μobs), simulated (μsim), and within-clade (μwithin) sampling distributions.
Therefore, in T. brucei and T. congolense, the evidence for recombination is greatest among closest-related VSG but was seldom observed in T. vivax, even when sampling within clusters of highly related sequences. Although the frequency of PI is similar for T. brucei VSG and Fam16, if we compare Ppi in a global alignment of T. congolense b-VSG (0.163) with the corresponding value for T. brucei (0.450), it is clear that PI is prevalent throughout the T. brucei repertoire but only within T. congolense VSG clades. This is a sampling effect caused by their divergent evolutionary histories. Given that T. congolense VSG are phylogenetically diverse and have a wider distribution of sequence variation, they have proportionally more distant relationships and so more structural barriers to genetic exchange. In short, there are cohorts of T. congolense VSG that never recombine, as the topological differences in Fig. 3 suggest.
Discussion
The past and present evolution of VSG can now be brought together. We have shown that the composition of contemporary VSG repertoires is determined by how each species has modified the common inheritance. T. vivax has the most structurally diverse repertoire, comprising a-VSG, b-VSG, and two additional types absent elsewhere; T. congolense combines multiple ancestral b-VSG lineages, each with a distinct CTD; and T. brucei a-VSG and b-VSG are recently derived single lineages with a common CTD. It is worth remembering that sequence mosaics generated in T. brucei infections are an additional source of diversity among expressed VSG (11); it is not known whether this dynamic assortment of VSG sequences occurs in other species. Nevertheless, as a result of compositional differences, the scale of recombination varies between species, being more frequent among T. brucei and T. congolense VSG than in T. vivax, and more prevalent among T. brucei VSG than in T. congolense. Opportunities for allelic recombination among VSG may be affected by species differences in mating system; recent work indicates that sexual reproduction is frequent in T. brucei and T. congolense populations (28–29), whereas T. vivax population structure is consistent with clonal reproduction (30). However, the importance of allelic recombination to VSG diversity is debatable; because the subtelomeres of homologous chromosomes in T. brucei are frequently dissimilar, subtelomeric VSG are effectively hemizygous, and so most recombination between VSG is ectopic (11). This study shows that PI in T. brucei VSG is attributable in large part to the CTD promoting exchange throughout the repertoire, whereas the conservative CTDs of T. congolense VSG actually reduce the scale of PI and illustrate the lack of recombination between clades, emphasizing the role of gene structure in promoting ectopic recombination in mitosis rather than the frequency of allelic recombination during meiosis.
Differences in the role of the CTD indicate that, in addition to scale, the mechanism of recombination varies between species. The CTD is exchanged between T. brucei VSG but is not exposed to antibodies and therefore may not directly contribute to antigenic diversity (31). However, the CTD-encoding sequence may have a role in the duplicative transposition of VSG, which promotes the diversification of the silent archive (11) and is of paramount importance to antigenic variation because it is required to move VSG from silent, subtelomeric loci into the telomeric expression site (4, 8). It is thought that VSG transposition is facilitated by an upstream 70-bp repeat region and the conserved CTD (8), with the transposed region terminating around the 3′ end of the coding sequence (32). Hence, in 37 recent such duplicative events among silent VSG in the T. brucei 927 genome (11), 16 terminated within the CTD region. In showing that the majority of PI in T. brucei VSG alignments concerns the CTD, our data demonstrate that a major recombination breakpoint occurs between the N-terminal domains and CTDs, supporting a principal role for the CTD in promoting VSG switching. Immediately, we can see that this mechanism cannot operate in T. congolense, where the CTDs are heterogeneous and have no role in promoting exchange. Hence, we propose that the preeminence of the CTD in PI among T. brucei VSG reflects the frequent transposition of N-terminal domains, and, through its solitary CTD type, which originated uniquely through horizontal transfer between VSG lineages, T. brucei may have evolved a distinct mechanism for the VSG transpositions that are key for diversification and antigen switching.
Antigenic variation is central to the host–trypanosome relationship, intimately linked to the course and severity of disease, to parasite transmission and host range, and therefore to disease epidemiology. All African trypanosomes display antigenic variation, and, although the current T. brucei-based model might adequately describe the general phenomenon, this study shows that the genomic basis for antigenic variation has diverged among trypanosomes in a manner consistent with distinct mechanisms for generating antigenic variability. Consequently, we now have reason to expect substantial species differences beneath the general phenotype, a framework to dissect this variation, and so a basis for understanding how the enigmatic VSG connects with the wider disease.
Materials and Methods
Genome Sequencing and Annotation.
T. congolense IL3000 and T. vivax Y486 genomes were capillary-sequenced with a whole-genome shotgun strategy as described previously (6).
Annotation of VSG Genes.
T. congolense and T. vivax VSG were identified by BLASTp-based homology searches and hidden Markov models. The boundaries of all VSG ORFs were manually checked against global sequence alignments.
Comparison of Gene Content.
OrthoMCL was used to examine putative gene gains and losses. All putative losses were confirmed by examining expected genomic position and by searching unassembled sequence reads for reciprocal sequence matches by tBLASTn/BLASTx.
T. vivax Transcriptome.
T. vivax Y486 was grown from stabilate in BALB/c mice immunosuppressed with cyclophosphamide and was amplified at patent parasitaemia in three immunosuppressed mice, from which whole blood was collected. The blood was treated with the erythrocyte lysis buffer, and RNA was isolated from the pellet.
Analysis of Fam1 Gene Expression.
To determine mRNA expression levels of Fam1 family members, quantitative real-time PCR (qRT-PCR) was carried out on total RNA.
Transfection and Fam1 Protein Localization.
Ectopic expression of HA epitope-tagged Tb927.6.1310 at the N terminus (after the predicted signal peptide sequence) was carried out with constitutive and inducible expression vectors.
VSG Purification and Sequencing.
T. vivax cell extracts were run in 1D SDS/PAGE, and three bands in the estimated size range were extracted from each, trypsinized, and subjected to liquid chromatography/tandem MS analysis.
Cell-Surface Phylome.
Homologs to each T. brucei “surface” gene were identified among all T. brucei, T. congolense, T. vivax, and Trypanosoma cruzi predicted genes with wuBLAST. Gene family phylogenies were estimated with maximum likelihood and Bayesian methods.
Recombination Analysis.
PI within VSG sequence alignments was used as a measure of recombination. The pairwise homoplasy index (26) returns a single probability value for PI, which was applied to amino acid sequence alignments for seven VSG subfamilies (Table S8), subsampled 1,000 times. The proportion of subalignments with significant PI, termed Ppi, was compared between species.
Additional information on methods is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Phelix Majiwa for sample preparation and Sara Melville for advice and advocacy during project planning. Barbara Marchetti (University of Glasgow) prepared RNA for transcriptomic analysis, Richard Burchmore (University of Glasgow) performed 2D electrophoresis, and Douglas Lamont (University of Dundee) performed liquid chromatography/tandem MS analysis. Arnab Pain and Chris Newbold (Wellcome Trust Sanger Institute) provided help and advice during manuscript preparation. We thank our colleagues in the sequencing and informatics groups at the Wellcome Trust Sanger Institute. This work was funded by the Wellcome Trust Grants WT 085775/Z/08/Z, 055558/Z/98/A, and 055558/Z/98/C. The Wellcome Trust Centre for Molecular Parasitology is supported by core funding from the Wellcome Trust (Grant 085349/Z/08/Z).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The draft genome sequences reported in this paper have been deposited in the European Molecular Biology Laboratory (EMBL) Nucleotide Sequence Database [accession nos. HE575314–HE575324 and CAEQ01000352–CAEQ01002824 (Trypanosoma congolense) and HE573017–HE573027 and CAEX01000001–CAEX01008277 (Trypanosoma vivax)]. The data can be examined via GeneDB (http://www.genedb.org) and TriTrypDB (http://tritrypdb.org). T. vivax transcriptome data have been submitted to the European Bioinformatics Institute Array Express Archive (accession no. E-MTAB-475). Sequence alignments and phylogenetic trees comprising the cell-surface phylome are contained in GeneDB (http://www.genedb.org/Page/trypanosoma_surface_phylome).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117313109/-/DCSupplemental.
References
- 1.Zambrano-Villa S, Rosales-Borjas D, Carrero JC, Ortiz-Ortiz L. How protozoan parasites evade the immune response. Trends Parasitol. 2002;18(6):272–278. doi: 10.1016/s1471-4922(02)02289-4. [DOI] [PubMed] [Google Scholar]
- 2.Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borst P, Cross GA. Molecular basis for trypanosome antigenic variation. Cell. 1982;29(2):291–303. doi: 10.1016/0092-8674(82)90146-5. [DOI] [PubMed] [Google Scholar]
- 4.Pays E. Regulation of antigen gene expression in Trypanosoma brucei. Trends Parasitol. 2005;21:517–520. doi: 10.1016/j.pt.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Morrison LJ, Marcello L, McCulloch R. Antigenic variation in the African trypanosome: Molecular mechanisms and phenotypic complexity. Cell Microbiol. 2009;11:1724–1734. doi: 10.1111/j.1462-5822.2009.01383.x. [DOI] [PubMed] [Google Scholar]
- 6.Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 7.Hutchinson OC, et al. VSG structure: Similar N-terminal domains can form functional VSGs with different types of C-terminal domain. Mol Biochem Parasitol. 2003;130(2):127–131. doi: 10.1016/s0166-6851(03)00144-0. [DOI] [PubMed] [Google Scholar]
- 8.Pays E, Salmon D, Morrison LJ, Marcello L, Barry JD. Antigenic variation in Trypanosoma brucei. In: Barry JD, McCulloch R, Mottram J, Acosta-Serrano A, editors. Trypanosomes: After the Genome. Norfolk, UK: Horizon Bioscience; 2007. pp. 339–372. [Google Scholar]
- 9.Carrington M, et al. Variant specific glycoprotein of Trypanosoma brucei consists of two domains each having an independently conserved pattern of cysteine residues. J Mol Biol. 1991;221:823–835. doi: 10.1016/0022-2836(91)80178-w. [DOI] [PubMed] [Google Scholar]
- 10.Blum ML, et al. A structural motif in the variant surface glycoproteins of Trypanosoma brucei. Nature. 1993;362:603–609. doi: 10.1038/362603a0. [DOI] [PubMed] [Google Scholar]
- 11.Marcello L, Barry JD. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strickler JE, et al. Trypanosoma congolense: Structure and molecular organization of the surface glycoproteins of two early bloodstream variants. Biochemistry. 1987;26:796–805. doi: 10.1021/bi00377a021. [DOI] [PubMed] [Google Scholar]
- 13.Rausch S, Shayan P, Salnikoff J, Reinwald E. Sequence determination of three variable surface glycoproteins from Trypanosoma congolense. Conserved sequence and structural motifs. Eur J Biochem. 1994;223:813–821. doi: 10.1111/j.1432-1033.1994.tb19057.x. [DOI] [PubMed] [Google Scholar]
- 14.Eshita Y, Urakawa T, Hirumi H, Fish WR, Majiwa PA. Metacyclic form-specific variable surface glycoprotein-encoding genes of Trypanosoma (Nannomonas) congolense. Gene. 1992;113(2):139–148. doi: 10.1016/0378-1119(92)90389-7. [DOI] [PubMed] [Google Scholar]
- 15.Helm JR, et al. Analysis of expressed sequence tags from the four main developmental stages of Trypanosoma congolense. Mol Biochem Parasitol. 2009;168:34–42. doi: 10.1016/j.molbiopara.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardiner PR, et al. Characterization of a small variable surface glycoprotein from Trypanosoma vivax. Mol Biochem Parasitol. 1996;82(1):1–11. doi: 10.1016/0166-6851(96)02687-4. [DOI] [PubMed] [Google Scholar]
- 17.Schell D, et al. A transferrin-binding protein of Trypanosoma brucei is encoded by one of the genes in the variant surface glycoprotein gene expression site. EMBO J. 1991;10:1061–1066. doi: 10.1002/j.1460-2075.1991.tb08045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salmon D, et al. A novel heterodimeric transferrin receptor encoded by a pair of VSG expression site-associated genes in T. brucei. Cell. 1994;78(1):75–86. doi: 10.1016/0092-8674(94)90574-6. [DOI] [PubMed] [Google Scholar]
- 19.Adams ER, Hamilton PB, Gibson WC. African trypanosomes: Celebrating diversity. Trends Parasitol. 2010;26:324–328. doi: 10.1016/j.pt.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Koenig-Martin E, Yamage M, Roditi I. A procyclin-associated gene in Trypanosoma brucei encodes a polypeptide related to ESAG 6 and 7 proteins. Mol Biochem Parasitol. 1992;55(1-2):135–145. doi: 10.1016/0166-6851(92)90134-6. [DOI] [PubMed] [Google Scholar]
- 21.De Greef C, Hamers R. The serum resistance-associated (SRA) gene of Trypanosoma brucei rhodesiense encodes a variant surface glycoprotein-like protein. Mol Biochem Parasitol. 1994;68(2):277–284. doi: 10.1016/0166-6851(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 22.Berberof M, Pérez-Morga D, Pays EA. A receptor-like flagellar pocket glycoprotein specific to Trypanosoma brucei gambiense. Mol Biochem Parasitol. 2001;113(1):127–138. doi: 10.1016/s0166-6851(01)00208-0. [DOI] [PubMed] [Google Scholar]
- 23.Carrington M, Boothroyd JC. Implications of conserved structural motifs in disparate trypanosome surface proteins. Mol Biochem Parasitol. 1996;81(2):119–126. doi: 10.1016/0166-6851(96)02706-5. [DOI] [PubMed] [Google Scholar]
- 24.White WT, Hills SF, Gaddam R, Holland BR, Penny D. Treeness triangles: Visualizing the loss of phylogenetic signal. Mol Biol Evol. 2007;24:2029–2039. doi: 10.1093/molbev/msm139. [DOI] [PubMed] [Google Scholar]
- 25.Weiller GF. Detecting genetic recombination. Methods Mol Biol. 2008;452:471–483. doi: 10.1007/978-1-60327-159-2_22. [DOI] [PubMed] [Google Scholar]
- 26.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bell JS, McCulloch R. Mismatch repair regulates homologous recombination, but has little influence on antigenic variation, in Trypanosoma brucei. J Biol Chem. 2003;278:45182–45188. doi: 10.1074/jbc.M308123200. [DOI] [PubMed] [Google Scholar]
- 28.Morrison LJ, et al. Discovery of mating in the major African livestock pathogen Trypanosoma congolense. PLoS ONE. 2009;4:e5564. doi: 10.1371/journal.pone.0005564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peacock L, Ferris V, Bailey M, Gibson W. Intraclonal mating occurs during tsetse transmission of Trypanosoma brucei. Parasit Vectors. 2009;2:43. doi: 10.1186/1756-3305-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duffy CW, et al. Trypanosoma vivax displays a clonal population structure. Int J Parasitol. 2009;39:1475–1483. doi: 10.1016/j.ijpara.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 31.Schwede A, Jones N, Engstler M, Carrington M. The VSG C-terminal domain is inaccessible to antibodies on live trypanosomes. Mol Biochem Parasitol. 2011;175(2):201–204. doi: 10.1016/j.molbiopara.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michels PA, et al. Activation of the genes for variant surface glycoproteins 117 and 118 in Trypanosoma brucei. J Mol Biol. 1983;166:537–556. doi: 10.1016/s0022-2836(83)80283-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.