Abstract
Riboswitches are cis-acting elements that regulate gene expression by affecting transcriptional termination or translational initiation in response to binding of a metabolite. A typical riboswitch is made of an upstream aptamer domain and a downstream expression platform. Both domains participate in the folding and structural rearrangement in the absence or presence of its cognate metabolite. RNA polymerase pausing is a fundamental property of transcription that can influence RNA folding. Here we show that pausing plays an important role in the folding and conformational rearrangement of the Escherichia coli btuB riboswitch during transcription by the E. coli RNA polymerase. This riboswitch consists of an approximately 200 nucleotide, coenzyme B12 binding aptamer domain and an approximately 40 nucleotide expression platform that controls the ribosome access for translational initiation. We found that transcriptional pauses at strategic locations facilitate folding and structural rearrangement of the full-length riboswitch, but have minimal effect on the folding of the isolated aptamer domain. Pausing at these regulatory sites blocks the formation of alternate structures and plays a chaperoning role that couples folding of the aptamer domain and the expression platform. Pausing at strategic locations may be a general mechanism for coordinated folding and conformational rearrangements of riboswitch structures that underlie their response to environmental cues.
Keywords: gene regulation, translational control, metabolism
RNA structures fulfill important roles in the regulation of gene expression including the control of translational initiation, transcriptional termination, and alternative splicing (1–5). In many cases, an RNA conformational change is crucial to gene regulation since one RNA sequence may adopt alternate structures. Regulation of gene expression is achieved when an input domain senses varying cellular conditions, resulting in shifting the balance between two or more structural states.
A prime example of gene regulation by RNA conformational changes in bacteria is riboswitch control. Riboswitches are cis-acting elements that regulate gene expression in response to the concentration of an intracellular metabolite. The coenzyme B12 riboswitch is located in the 5′ untranslated region of the btuB gene which encodes an outer membrane transporter for B12 (6) (Fig. 1A). The btuB riboswitch consists of two domains: an upstream coenzyme B12 binding aptamer and a downstream expression platform. When the cellular concentration of coenzyme B12 is high, this metabolite binds to the aptamer, reconfiguring the expression platform to form a structure in which the ribosome binding site (RBS) is base paired and inaccessible. When the concentration of coenzyme B12 is low, the mRNA forms an alternate structure which allows for translation (6, 7).
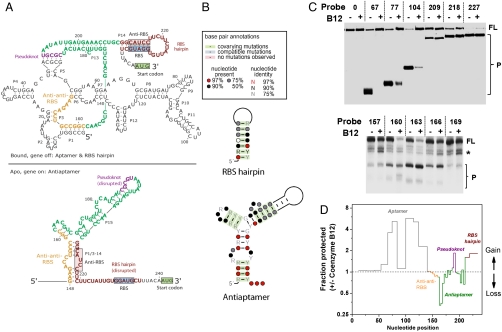
Fig. 1.
Secondary structures of the E. coli btuB riboswitch. (A) The secondary structure of the aptamer domain in the off structure encompassing residues 1–202 is the same as that derived by Breaker and coworkers (6, 7). The structurally important regions are shown in orange (anti-anti-RBS, residues 148–161), purple (pseudoknot 3′ region, residues 187–191), green (antiaptamer, residues 162–209), and brown (anti-RBS = 212–216 and RBS = 229–233, residues 210–235). In the coenzyme B12 bound off structure, the anti-anti-RBS region is part of the aptamer, allowing the pairing of the anti-RBS with the RBS regions. In the apo, or on structure, the anti-anti-RBS region pairs with the anti-RBS region to form an antiaptamer structure, leaving the RBS available for ribosome binding. (B) Phylogenetic support for the anti-RBS and RBS hairpin in the off structure, and for the anti-anti-RBS and anti-RBS interaction (the antiaptamer structure) in the on structure. (C) Structural mapping using oligonucleotide hybridization/RNase H cleavage assay in the absence and presence of 100 μM coenzyme B12. Sequences of all oligos and their complementary regions in the btuB riboswitch are listed in Fig. S2. In the first round, a total of 26 oligos were used spanning residues 12–243. Examples of the first round mapping are shown in the upper panel where the transcripts are 5′ 32P labeled. Oligo length is determined by having similar melting temperatures and varies from 6 to 26 nucleotides. In the second round, a total of 31 10-mer oligos were used spanning residues 142–242. Examples of the second round mapping are shown in the lower panel where the transcripts are internally labeled. The full-length RNA transcript also contains the first 20 codons of the btuB gene (residues 243–302); this region is not probed in our experiment. FL, full-length transcript; P, RNase H cleavage product. Star corresponds to RNAs that are either paused transcripts or randomly cleaved fragments. (D) Structural probing data are plotted as fraction protected ± coenzyme B12 versus nucleotide position. The y axis is plotted in log 2 scale. Aptamer, gray; antiaptamer, green; pseudoknot, purple; RBS hairpin, brown.
RNA folding and conformational switching occurs during transcription in vivo. Folding during transcription is particularly important for riboswitch regulation (8). Although bacterial RNA polymerases (RNAPs) synthesize RNA at tens of bases per second (9, 10), local RNA secondary structures fold at faster rates, presumably as soon as their corresponding residues are synthesized and emerge from the exit channel of the RNAP (11).
Importantly, transcription does not occur at a uniform rate. The RNAP frequently pauses at certain locations transiently during RNA synthesis (12). Previous studies demonstrated that pausing at specific sites can facilitate RNA folding during transcription in two ways. First, the 5′ to 3′ polarity of transcription implies that the 5′ region of long-range helices is transcribed prior to its 3′ partners. RNAP pauses at strategic sites located downstream of the 5′ region of all long-range helices. These pauses enable nascent RNAs to fold into beneficial nonnative structures wherein the 5′ region is sequestered and poised for subsequent folding of the native structure (13). Second, riboswitch function involves two or more competing structures; one structure directly senses cellular metabolite concentration. By pausing immediately downstream of the aptamer domain, the RNAP can block the formation of competing structures that involve residues in the expression platform (8, 14–16).
This work investigates the folding and conformational rearrangement of the Escherichia coli coenzyme B12 binding riboswitch during transcription by the cognate E. coli RNAP. This riboswitch consists of an approximately 200 nucleotide, coenzyme B12 binding aptamer domain and an approximately 40 nucleotide expression platform that controls ribosome access (Fig. 1A). We find that, when both domains are present, transcription plays an important role by enabling this riboswitch to efficiently fold into its functional structures. Several transcriptional pauses located near the end and immediately downstream of the aptamer domain are crucial for efficient folding. This stimulatory effect could be because of the paused RNAP blocking the formation of competing structures. We also find that these pauses couple folding of the tertiary structure of the aptamer domain and the downstream secondary structure in the expression platform, a result that has not been observed previously for riboswitch folding. Our results demonstrate that riboswitch folding and conformational rearrangement are strongly linked to transcription. Further, specific transcriptional pauses imposed by the RNAP enable coordinated folding between the ligand-dependent tertiary structure formation and the downstream secondary structure formation.
Results
Probing btuB Riboswitch Folding During Transcription.
The E. coli coenzyme B12 riboswitch in the 5′ UTR of the btuB gene is composed of a large aptamer domain with an intricate tertiary structure followed by a small expression platform. The consensus structure of the btuB aptamer domain has been established using phylogeny and structural mapping of the refolded RNA transcript (6). However, the complete structures in both the on and off states, including the expression platform, remain unknown. We therefore first constructed models of the secondary structure in both the apo (on) and B12 bound (off) states using phylogeny and structural probing of the nascent RNA transcript. The aptamer structure in the ligand bound, off-state encompassing residues 1–202 has already been derived by Breaker and coworkers (6, 7) and the RBS hairpin has been proposed by Nou and Kadne (17). These structures are shown in Fig. 1A, Upper. The dominant feature of our model is the antiaptamer structure in the apo (on) state (Fig. 1A). These additional elements provide a structural model of the complete btuB riboswitch, which enables testing mechanistic hypotheses on the folding and conformational switching of this large RNA.
To construct the proposed structure by covariation analysis, the standalone Basic Local Alignment Search Tool (BLAST) application was used to search for sequences homologous to the btuB region of interest in γ-proteobacterial genomes from which 71 homologous btuB genes were recovered (detailed description in SI Materials and Methods; ref. 18). The resulting hits were aligned using ClustalW2 (19) and the consensus secondary structure was predicted with RNAalifold (20, 21). This initial structure and alignment was used to create a covariance model using the Inference of RNA Alignment (INFERNAL) software package (22). This model was used to search for structural homologues in all available bacterial genomes. A total of 94 sequences were retrieved and realigned, and a new consensus structure was regenerated with RNAalifold (Fig. S1). The global statistics that support the proposed base pairings of the antiaptamer and the RBS hairpin structures are illustrated in Fig. 1B.
We used the well-established method of DNA oligonucleotide hybridization followed by RNase H digestion (23, 24) to probe the relative accessibility of the nascent btuB riboswitch transcript (Fig. 1 C and D, and Figs. S2 and S3). In the first round of structural probing, a total of 26 probes encompassing residues 12–243 were used to measure the fraction of the nascent RNA transcript protected from digestion under zero or saturating coenzyme B12 concentrations. In the presence of coenzyme B12, both the aptamer and the expression platform become more resistant to cleavage, consistent with an expected increase in structure. The anti-anti-RBS region proposed to participate in the alternate base paring responsible for switching is similarly and well protected in both the presence and absence of coenzyme B12 (Fig. 1D). This result is consistent with our model depicting this region connecting the domains being stably base paired in both the on and off states. In the second round of structural probing, a total of 31 probes encompassing residues 142–243 were used to provide higher resolution for the region spanning the anti-anti-RBS sequence to the entire expression platform. Probing at this higher resolution readily identified antiaptamer regions that show decreased protection in the presence of coenzyme B12 (Fig. 1 C and D). These results are fully consistent with our proposed structural model (Fig. 1A).
The on-off nature of the apo and the bound states is also supported by in-line probing of the RBS hairpin region in the expression platform (Fig. S4). Although single nucleotide resolution cannot be achieved in this experiment carried out in the transcription mixture, it is clear that the presence of coenzyme B12 leads to increased structure of the RBS stem and decreased structure of the RBS loop, consistent with the proposed structural model.
Interestingly, when we performed the oligonucleotide hybridization assay using gel purified then refolded btuB riboswitch transcript, a significantly different result was obtained (Fig. S5). The difference between the coenzyme B12 bound versus the apo states had become much smaller in the refolded RNA, although the pattern of protected regions remained similar between the refolded transcript and the native E. coli RNAP transcript. This result suggests that transcription is a crucial parameter in efficient folding and conformational change of the btuB riboswitch.
We used one oligo probe that shows a significant gain in protection in the presence of coenzyme B12 to determine the ligand binding affinity of the E. coli RNAP transcript (Fig. S6). We obtained an apparent binding constant of approximately 100 nM, which is comparable to the binding affinity of the btuB aptamer RNA alone (ca. 300 nM; refs. 6 and 7). This result indicates that the btuB riboswitch construct we are working with is functionally similar to those used in previous studies.
We also characterized the folding behavior of two btuB riboswitch mutants and the results are again consistent with our proposed structural model (Fig. S7). Both mutants are derived from those described previously in the studies of ligand binding to the aptamer domain (6, 7). Based on the previous results, both mutants are expected to bind coenzyme B12 at a weaker affinity by more than 15-fold. In the apo state, both mutants show little difference compared to the wild type in regard to their accessibility to the three oligo probes used. In contrast to the wild type, however, both mutants lost their ability to respond to coenzyme B12.
Transcriptional Pause Mapping.
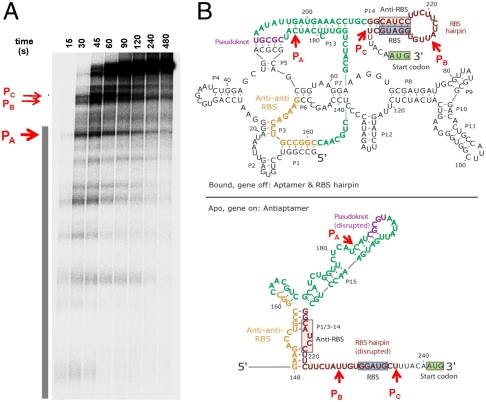
Many transcriptional pauses were observed during transcription of the btuB riboswitch by the wild-type E. coli RNAP (Fig. 2 and Fig. S8). Three of the most prominent pauses, designated as pauses A, B, and C, occur at the end and immediately downstream of the aptamer domain. Both the location and the duration of these pause sites were independent of the presence of coenzyme B12 in the transcription reaction (Fig. S8).
Fig. 2.
Pausing of btuB riboswitch during transcription. (A) A large number of pauses are present. All transcripts are 5′ 32P labeled using initiating dinucleotide [32P]pAU. The aptamer domain is indicated by a vertical bar. Most prominent pauses occur near the end (PA) and downstream (PB and PC) of the aptamer domain. Mapping of the sites A–C is shown in Fig. S8. (B) Location of the pause sites A, B, and C. Site A is between the anti-anti-RBS region and the pseudoknot region. Site B is in the RBS hairpin loop between the RBS and its pairing partners—i.e., the anti-RBS region. When RNAP pauses at sites A and B, the anti-anti-RBS region is outside, whereas the anti-RBS region is in the RNAP exit channel. In the paused complex C, the anti-RBS region is available, but the RBS region is still in the exit channel. Both the aptamer and the antiaptamer structures can form in paused complex C.
The location of the pause sites A–C suggests that pausing at these sites could result in the folding of alternate structures. When the RNAP pauses at sites A and B, the anti-anti-RBS region has already emerged from the polymerase, but the nascent RNA is long enough that only the aptamer domain can fold (Fig. 2B, Upper). For pausing at site A, the antiribosome binding site (anti-RBS) RNA is unavailable to compete with the anti-anti-RBS region for base pairing, and hence, the formation of the antiaptamer structure in the apo state is not possible (Fig. 2B, Lower). For pausing at site B, the anti-RBS region has been transcribed, but it is still buried in the paused RNAP, which covers approximately 15 nascent RNA residues (25, 26). Finally, pausing at site C enables the anti-RBS region to emerge from the RNAP, so that the antiaptamer structure in the apo state can also form. However, the anti-RBS and RBS hairpin cannot form in the paused complex C because the RBS region is still in the exit channel of the RNAP.
Correlating Pausing and Riboswitch Folding.
We next examined the correlation between pausing and riboswitch folding. We selected two oligonucleotide probes that show marked differences in protection in the absence and presence of coenzyme B12. Probes 104 and 227a are complementary to nucleotides 104–113 and 227–236, and are used as a diagnostic of conformational changes in the aptamer domain and the expression platform, respectively. The aptamer domain probe 104 showed increased protection in the presence of saturating concentration of coenzyme B12 by 5–10-fold (Fig. 1C). For the expression platform probe 227a, probing for folding was performed under milder cleavage conditions because the condition used in the structural mapping led to almost complete cleavage of the RBS region in the coenzyme B12 bound state. Under this mild condition, the increased fraction protected was approximately twofold in the presence of saturating concentration of coenzyme B12.
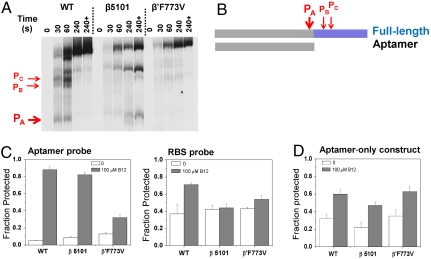
We applied two distinct strategies to demonstrate that pauses at site A and beyond make the most important contribution on btuB riboswitch folding, as compared to the pause sites in the aptamer domain upstream of site A (Fig. 3). For this purpose, one would ideally mutate the RNA transcript to eliminate individual pause sites. We tested this strategy and found it was difficult to accomplish because of the difficulty of eliminating low-strength pause sites by a mutation; further, the presence of multiple pause sites suggests possible overlapping roles, so folding results from eliminating a single pause site may be inconclusive. Rather, we used two well-characterized “fast” E. coli RNAP variants that are defective in pausing. The β5101 mutant contains two substitutions in the β-subunit (P560S, T563I) (27), whereas β′F773V contains a substitution in the catalytic bridge helix (28). Both enzymes fail to efficiently pause at strong regulatory sites (e.g., hairpin-dependent hisP and backtracked opsP sites) and bypass numerous weak pause sites more readily (29). In btuB riboswitch transcription, both mutant enzymes show significantly different pausing behavior at sites A–C as compared to the wild-type RNAP (Fig. 3A). The β5101 polymerase appears to stall at site A under the low-NTP conditions used to map the pause sites, and pauses much more weakly at sites B and C; β′F773V polymerase pauses much more weakly at all three sites.
Fig. 3.
Pausing-deficient, mutant RNAP transcription affects folding fraction of the btuB riboswitch. (A) Both mutant RNAPs significantly affect pausing at sites A–C. Lane 240+ corresponds to high [NTP] chase after 4 min. (B) Schematic representation of the two RNA constructs. The full-length construct has 302 residues and contains the entire aptamer domain, the expression platform, and the first 20 codons of the BtuB gene. The aptamer-only construct has 202 residues and contains just the aptamer domain as defined in the literature (6). The location of the pause sites A–C is also indicated. (C) Transcription by the fast RNAPs reduces the ratio of protection in the absence and presence of coenzyme B12 for both the aptamer domain and the expression platform. The cleavage reaction with the RBS probe was carried out with 25% the amount of RNase H, as compared to the cleavage reaction with the aptamer probe. (D) Transcription by fast RNAPs has little effect on differential fraction folded of the aptamer-only construct.
To assess the effect of pausing on folding of different domains, we used two different btuB riboswitch constructs (Fig. 3B). The “full-length” construct contains the aptamer domain, the entire expression platform, and the first 20 codons of the btuB gene. The “aptamer-only” construct is the same as used in previous refolding studies and ends at residue 202 (6); it does not contain any residues from the expression platform that could interfere with the folding of the anti-anti-RBS region (residues 148–161).
The extent of protection for both the full-length and the aptamer-only construct during transcription by the wild type and the two fast polymerases was examined. In the full-length construct, the extent of protection with or without coenzyme B12 was reduced by two- to fivefold when transcription was conducted using the fast enzymes (Fig. 3C). For the aptamer probe, the reduction of fraction protected was particularly severe with the faster β′F773V RNAP. For the RBS probe, the reduction was also significant for both fast polymerases. However, the aptamer-only construct displayed similar folding behavior when transcribed by all three RNAPs (Fig. 3D). The lack of effects of the fast RNAP substitutions on the aptamer-only construct indicates that pausing within the aptamer domain (at sites upstream of site A) is not required for its folding. Because the major pause sites A, B, and C are the only ones located between the anti-anti-RBS region and the btuB coding region, this result strongly suggests that pause sites A–C play a major role in btuB riboswitch folding and conformational change.
Coordinating Aptamer Domain and the Expression Platform Folding.
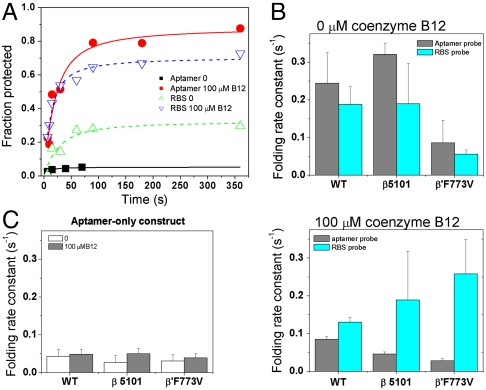
We also determined how the folding rates of the aptamer domain and the expression platform are coupled during transcription. These two domains contain folded structures that are very different from each other. The large aptamer domain folds into a complex tertiary structure capable of binding to the coenzyme B12 ligand, a metallo-organic compound of approximately 1,000 Da. In contrast, the expression platform is less than one-fifth the size of the aptamer domain and folds into simple secondary structures—a hairpin loop in the off state or part of a three-way junction followed by a single stranded region in the on state. It is the folded aptamer together with yes-or-no coenzyme B12 binding that dictates whether the expression platform folds into one or the other structure. In general, RNA tertiary structures tend to fold much slower than local secondary structures (11). Therefore, the timing of tertiary structure folding of the aptamer domain and of secondary structure folding of the expression platform may need to be coordinated to enable proper control of yes-or-no coenzyme B12 binding, which drives the structural transition of the expression platform.
Indeed, the folding rate constants of the aptamer and the expression platform were quite similar when transcription was carried out by the wild-type RNAP (Fig. 4A). In the absence of coenzyme B12, folding rates for both domains were approximately 0.2 s-1 and differed by a ratio of 1.3-fold (Fig. 4B). In the presence of saturating concentration of coenzyme B12, folding rates for both domains were approximately 0.1 s-1 and differed by a ratio of 1.5-fold. However, these rates began to diverge when transcription was carried out with the two fast RNAP mutants. This divergence is particularly striking when coenzyme B12 is present in the transcription reaction (Fig. 4B). For both fast RNAPs, folding of the aptamer domain becomes slower, whereas folding of the expression platform becomes faster, so that the ratio of the domain folding rates increases to four- to ninefold, as compared to the 1.5-fold ratio observed with the wild-type polymerase. RNAP-dependent effect on folding rates was not observed in the aptamer-only construct, however, either in the absence or in the presence of coenzyme B12 (Fig. 4C). Because only the folding of the full-length, but not of the aptamer-only construct involves conformational rearrangement upon binding coenzyme B12, these results suggest RNAP pausing near the end or immediately after the aptamer domain (i.e., at sites A–C) facilitates coordinated folding of the two domains.
Fig. 4.
Pausing-deficient RNAP affects folding kinetics. The cleavage reaction with the RBS probe was carried out with 25% the amount of RNase H, as compared to the cleavage reaction with the aptamer probe. (A) Folding kinetics of the aptamer and expression platform during transcription by the wild-type RNAP. (B) The folding rate constants of the aptamer domain and the expression platform are within 1.5-fold to each other when transcribed by the wild-type enzyme, both in the absence and presence of coenzyme B12 ligand. In contrast, transcription by the pausing-deficient, fast RNAPs leads to increased divergence of the folding rates, especially in the presence of coenzyme B12. (C) Folding rates of the aptamer-only construct do not depend on the RNAP used in the transcription.
Discussion
Folding Model of the btuB Riboswitch.
The location of the pause sites A–C in regard to the anti-anti-RBS region seems to play a crucial role in directing the btuB riboswitch folding into on or off states during transcription. The anti-anti-RBS region can be part of the aptamer domain or part of the expression platform. When RNAP pauses at site A or B, the anti-anti-RBS region has already emerged from the exit channel, and the tertiary structure of the aptamer may fold. If coenzyme B12 is present, the aptamer structure is solidified so that the anti-anti-RBS region is unavailable for interacting with downstream regions once RNA chain elongation resumes. If coenzyme B12 is absent, the aptamer structure can unfold to allow the anti-anti-RBS region to pair with the downstream anti-RBS once elongation resumes; alternatively, the aptamer structure may be sufficiently labile to allow invasion of the anti-RBS region once it emerges from the RNAP exit channel.
Pausing at sites A and B would confer two benefits for controlling btuB riboswitch folding. The first benefit is to block the formation of the antiaptamer structure. This block is achieved simply because pausing at these sites excludes downstream regions, which are not yet transcribed or bound in the RNAP exit channel, from competition with folding of the anti-anti-RBS region. For example, if RNAP lingers for some time at site A or sequesters the anti-RBS region in the exit channel at site B, the anti-RBS region cannot compete with the anti-anti-RBS region folding to form the antiaptamer structure, thereby allowing the anti-anti-RBS region to fold with other residues in the aptamer domain. The second benefit is to provide a crucial time window to enable coordinated folding of the aptamer tertiary structure and the expression platform hairpins. Tertiary folding of the large aptamer domain would be expected to be intrinsically slower than secondary structure folding of the expression platform. Because the nascent RNA in the paused complexes A and B already contains all the residues necessary for tertiary folding, pausing at these sites gives the aptamer domain extra time to fold prior to its commitment to the on or the off structure.
When polymerase pauses at site C, formation of either the aptamer or antiaptamer structure can occur. However, the anti-RBS + RBS hairpin still cannot form because the RBS region is still in the RNAP exit channel. This pause may provide additional time for rearrangement between the aptamer and antiaptamer structures before the RBS is transcribed and the final structural decision is made.
Pausing as a Common Mechanism of Blocking formation of Competing RNA Structures.
Several examples have been described in the literature on how transcriptional pauses at strategic locations control riboswitch folding (8, 14–16). The Bacillus subtilis FMN riboswitch consists of an upstream aptamer domain for flavin mononucleoside and a downstream transcriptional termination hairpin. Transcription of this riboswitch by the E. coli RNAP shows two prominent pause sites located near the end and immediately after the aptamer domain. Pausing at these sites could provide a time window for the aptamer to fold without the interference of competing structures involving downstream residues in the termination hairpin (8). The E. coli thiM riboswitch consists of an upstream aptamer domain for thiamin pyrophosphate and a downstream ribosome binding site. Transcription of this riboswitch by the E. coli RNAP shows a series of pause sites immediately after the aptamer domain. Pausing at these sites also seems to prevent the formation of a competing structure involving residues in the downstream region (15). The E. coli alx locus codes for an RNA that undergoes a pH-dependent conformational change. The neutral pH structure is the off state and contains two separate hairpins, C and D, which rearrange to form a single hairpin C/D in the alkaline pH on state structure. Transcription of this pH-dependent riboregulator by the E. coli RNAP shows two prominent pause sites that are accentuated at alkaline pH. Pausing at site C reduces the propensity of hairpin C formation, and pausing at site D completely blocks the formation of hairpin D, but still enables the formation of the C/D hairpin (14). These results, together with our current study of btuB riboswitch folding, demonstrate that pausing at strategic locations is a common mechanism to block the formation of structures that compete with folding of the structure located upstream of the pause sites.
Coordinating the Folding of the Aptamer and the Expression Platform.
Our work also provides additional insight on riboswitch folding during transcription. Our results indicate that pausing at the end of the aptamer domain coordinates folding of the aptamer tertiary structure and the secondary structure of the expression platform. For btuB riboswitch, the aptamer domain is a highly structured RNA that binds the 1-kDa coenzyme B12 ligand. In the absence of concurrent transcription, this complex tertiary structure likely folds at a much slower timescale than the structurally simple expression platform. For comparison, refolding half-time of similarly sized RNAs with comparably complex tertiary structures, e.g., the catalytic domain of the B. subtilis RNase P RNA or the Azoarcus group I ribozyme, is on the order of tenths of seconds at 37 °C (30, 31). The folding half-time of the btuB aptamer domain during transcription is 3–7 s under our condition. At an average transcription speed of 20–40 nucleotides per second under our conditions, the RNAP would have transcribed the entire expression platform in 1–2 s in the absence of pausing. Premature synthesis of the expression platform could interfere with aptamer domain folding due to the possibility of alternate structure formation involving the anti-anti-RBS region. By pausing at sites A–C, the folding half-time of the expression platform can be adjusted to become similar to that of the aptamer domain.
Coordinating the folding rates of the aptamer domain and the expression platform may not be limited to the btuB riboswitch, although the actual folding rate may depend on the identity of the aptamer domain. In the only other case known to us, the folding half-time of the E. coli thiM aptamer domain during transcription is approximately 2–3 s (15). For the structurally less complex thiM riboswitch aptamer, pausing of shorter durations may be sufficient to coordinate its folding with the expression platform.
An alternative interpretation of the observed folding rates of the aptamer domains in these riboswitches is that they reflect bimolecular binding reactions depending on their perspective ligand concentration. This interpretation is less likely for the btuB riboswitch where the folding rate was measured at coenzyme B12 concentrations that significantly exceed the cellular levels. For the thiM riboswitch, the maximum folding rate was obtained at saturating concentration of the thiamine pyrophosphate ligand.
Potential Chaperoning Effect of Pausing on Riboswitch Folding.
Unexpectedly, the folding behavior of the aptamer domain during transcription by the wild-type RNAP is different when the aptamer domain is connected to the expression platform. In the absence and presence of coenzyme B12, the ratio of fraction protected is > 10-fold in the full-length construct, but only approximately twofold in the aptamer-only construct (Fig. 3 C and D). Similarly, in the absence and presence of coenzyme B12, the folding rate of the aptamer domain in the full-length construct is two- to fourfold faster than in the aptamer-only construct (Fig. 4 B and C). When transcription is carried out by the pausing-deficient β′F773V RNAP, both the ratio of fraction protected and the folding rate of the aptamer domain in the full-length construct approach those of the aptamer-only construct. These results suggest that the kinetic differences arise from pausing downstream of the aptamer domain.
How could pausing accelerate the folding of the aptamer domain? We do not know at this time which pause site among A, B, and C is most relevant to folding; however, models can be proposed for pausing at each site. For instance, when the RNAP pauses at site B, the 5′ end of the nascent RNA outside of the RNAP exit channel approximately resides at nucleotide 211. In this paused complex, two different hairpin loops can form, one consisting of nucleotides 175–206 in the aptamer structure and the other consisting of nucleotides 169–209 in the antiaptamer structure. It is possible that the formation of either hairpin loop accelerates the folding of the region up to the anti-anti-RBS sequence. Neither hairpin loop structure can completely form in the aptamer-only construct.
Implications on B12 Riboswitch Folding in Vivo.
The insights obtained here should be relevant to btuB riboswitch folding in cells even though differences are to be expected between our study and in vivo folding. The pausing duration in cells may not be the same as in our study because pausing can be affected by several elongation factors that associate with the RNAP, e.g., NusA and NusG (12, 32), as well as NTP concentrations. The behavior of the paused complexes may also depend on the available ribosome and initiator tRNA that readily bind to the RBS when it is accessible (17). Finally, coenzyme B12 molecules in cells may be localized because this valuable metabolite could be synthesized at, or transported directly to, the intracellular site where it is incorporated into coenzyme B12 utilizing enzymes (33).
Nevertheless, the same fundamental issues of riboswitch folding have to be resolved in cells. Pausing at the downstream boundary of the aptamer domain is a simple mechanism that can both block the formation of alternate structures and coordinate the folding dynamics of the aptamer domain and the expression platform.
Materials and Methods
Detailed materials and methods information are described in SI Materials and Methods: structural probing of the btuB riboswitch transcript during E. coli RNAP transcription, cotranscriptional btuB RNA folding, DNA oligonucleotides and chemicals, preparation of btuB template DNA for in vitro transcription, structural probing of refolded btuB riboswitch RNA, in-line probing, transcriptional pause mapping, and structural bioinformatics and phylogenetic analysis.
Supplementary Material
Acknowledgments.
We thank Dr. Marc Parisien for help and advice on the phylogenetic analysis. This work was supported by National Institutes of Health (GM57880 to T.P. and T.R.S., GM67153 to I.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113086109/-/DCSupplemental.
References
- 1.Oxender DL, Zurawski G, Yanofsky C. Attenuation in the Escherichia coli tryptophan operon: Role of RNA secondary structure involving the tryptophan codon region. Proc Natl Acad Sci USA. 1979;76:5524–5528. doi: 10.1073/pnas.76.11.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.May GE, Olson S, McManus CJ, Graveley BR. Competing RNA secondary structures are required for mutually exclusive splicing of the Dscam exon 6 cluster. RNA. 2011;17:222–229. doi: 10.1261/rna.2521311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roland KL, Liu CG, Turnbough CL., Jr Role of the ribosome in suppressing transcriptional termination at the pyrBI attenuator of Escherichia coli K-12. Proc Natl Acad Sci USA. 1988;85:7149–7153. doi: 10.1073/pnas.85.19.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 5.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 6.Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nahvi A, et al. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 8.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 9.Vogel U, Jensen KF. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J Bacteriol. 1994;176:2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tolic-Norrelykke SF, Engh AM, Landick R, Gelles J. Diversity in the rates of transcript elongation by single RNA polymerase molecules. J Biol Chem. 2004;279:3292–3299. doi: 10.1074/jbc.M310290200. [DOI] [PubMed] [Google Scholar]
- 11.Bevilacqua PC, Blose JM. Structures, kinetics, thermodynamics, and biological functions of RNA hairpins. Annu Rev Phys Chem. 2008;59:79–103. doi: 10.1146/annurev.physchem.59.032607.093743. [DOI] [PubMed] [Google Scholar]
- 12.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong TN, Sosnick TR, Pan T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc Natl Acad Sci USA. 2007;104:17995–18000. doi: 10.1073/pnas.0705038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes Dev. 2009;23:2650–2662. doi: 10.1101/gad.552209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong TN, Pan T. RNA folding during transcription: Protocols and studies. Methods Enzymol. 2009;468:167–193. doi: 10.1016/S0076-6879(09)68009-5. [DOI] [PubMed] [Google Scholar]
- 16.Lemay JF, et al. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nou X, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci USA. 2000;97:7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Larkin MA, et al. Clustal W and Clustal X version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 20.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J Mol Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 21.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: Inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarrinkar PP, Williamson JR. Kinetic intermediates in RNA folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 24.Treiber DK, Williamson JR. Kinetic oligonucleotide hybridization for monitoring kinetic folding of large RNAs. Methods Enzymol. 2000;317:330–353. doi: 10.1016/s0076-6879(00)17023-5. [DOI] [PubMed] [Google Scholar]
- 25.Vassylyev DG, et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 26.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 27.Landick R, Stewart J, Lee DN. Amino acid changes in conserved regions of the beta-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- 28.Artsimovitch I, Chu C, Lynch AS, Landick R. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science. 2003;302:650–654. doi: 10.1126/science.1087526. [DOI] [PubMed] [Google Scholar]
- 29.Svetlov V, Belogurov GA, Shabrova E, Vassylyev DG, Artsimovitch I. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 2007;35:5694–5705. doi: 10.1093/nar/gkm600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang X, Pan T, Sosnick TR. Mg2+-dependent folding of a large ribozyme without kinetic traps. Nat Struct Biol. 1999;6:1091–1095. doi: 10.1038/70016. [DOI] [PubMed] [Google Scholar]
- 31.Rangan P, Masquida B, Westhof E, Woodson SA. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc Natl Acad Sci USA. 2003;100:1574–1579. doi: 10.1073/pnas.0337743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns CM, Richardson LV, Richardson JP. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J Mol Biol. 1998;278:307–316. doi: 10.1006/jmbi.1998.1691. [DOI] [PubMed] [Google Scholar]
- 33.Hufton SE, et al. Structure-function analysis of the vitamin B12 receptor of Escherichia coli by means of informational suppression. Mol Microbiol. 1995;15:381–393. doi: 10.1111/j.1365-2958.1995.tb02251.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.