Abstract
The outer membrane (OM) of Gram-negative bacteria such as Escherichia coli contains lipoproteins and integral β-barrel proteins (outer-membrane proteins, OMPs) assembled into an asymmetrical lipid bilayer. Insertion of β-barrel proteins into the OM is mediated by a protein complex that contains the OMP BamA and four associated lipoproteins (BamBCDE). The mechanism by which the Bam complex catalyzes the assembly of OMPs is not known. We report here the isolation and characterization of a temperature-sensitive lethal mutation, bamAE373K, which alters the fifth polypeptide transport-associated domain and disrupts the interaction between the BamAB and BamCDE subcomplexes. Suppressor mutations that map to codon 197 in bamD restore Bam complex function to wild-type levels. However, these suppressors do not restore the interaction between BamA and BamD; rather, they bypass the requirement for stable holocomplex formation by activating BamD. These results imply that BamA and BamD interact directly with OMP substrates.
Keywords: Omp85, membrane protein folding, conditional lethal, allele-specific
The outer membrane (OM) of Escherichia coli functions primarily as a robust permeability barrier that insulates the cell against a variety of potentially cytotoxic agents in the extracellular milieu. The cell is able to adapt to and mediate exchange with its environment through the regulated synthesis and assembly of integral OM proteins, which participate in a diverse array of cellular processes. Whereas integral proteins found in the inner membrane exclusively contain hydrophobic α-helical transmembrane domains, integral OM proteins (OMPs) instead adopt an amphipathic β-barrel conformation within the membrane (1).
Following synthesis, the aggregation-prone, unfolded precursor OMP is translocated across the inner membrane, is processed to its mature form, and is stabilized during transit through the crowded, aqueous periplasm by periplasmic chaperones, which deliver the nascent OMPs to the OM-associated β-barrel assembly machine (Bam). There, OMPs are folded and integrated into a rigid asymmetric lipid bilayer in the absence of an obvious energy source while OM integrity is maintained faithfully (1–3).
Bam is a heteropentamer (4) composed of a very highly conserved OM β-barrel (BamA) and four OM lipoproteins (BamBCDE) that dock with BamA (4–6). The hub of the physical complex is the BamA N-terminal extension, a periplasmic beaded chain comprising five structurally homologous polypeptide transport-associated (POTRA) domains that are numbered sequentially from the N terminus (7, 8). A stable association between BamA and the nonessential factor BamB requires most of the periplasmic domain of BamA. Conversely, only the POTRA 5 (P5) domain is necessary for the interaction of BamA with the essential factor BamD, which in turn mediates the apparently indirect association of BamA with BamC and BamE (4, 5, 8).
BamA and BamD are indispensable in E. coli, and depletion of either leads to rapid accumulation of unassembled OMPs in the periplasm followed by cell death (5, 6, 9, 10). Because structure–function analysis of BamA–D has the potential to yield significant insight into the general mechanism of OMP assembly [a process conserved across bacteria, chloroplasts, and mitochondria (11)], we sought to analyze the BamA–D interaction genetically and biochemically. We report here the isolation and characterization of mutations that disrupt the physical interaction between the BamAB and BamCDE subcomplexes. Suppressor analysis suggests that the P5 domain controls the activity of BamD and that the BamAB and BamCDE subcomplexes can function efficiently in the absence of a tight association provided that BamD is activated genetically.
Results
Screen for Conditional Lethal Alleles of bamA.
We sought to identify BamA residues that influence function by screening mutations in bamA that cause impaired cell growth at 37 °C. A low-copy plasmid containing bamA (pbamA+) was mutagenized (Materials and Methods) and was introduced into JCM320, a strain in which expression of only the chromosomal bamA gene can be induced or repressed by the addition of arabinose or fucose, respectively, to the growth medium (6, 8). Only the plasmid-borne, mutagenized (bamA*) allele is expressed in the JCM320/pbamA* background when arabinose is excluded.
To obtain conditional lethal alleles of bamA, pbamA* transformants of JCM320 were screened for arabinose-dependent growth at 37 °C. Uninteresting (e.g., null) mutations were avoided by simultaneously screening for arabinose-independent growth at low temperature (24 °C). We describe here one mutation identified in the screen, bamA5, which confers a temperature-sensitive (ts−) growth phenotype. Haploid bamA5 strains, such as DPR881, do not grow above 24 °C.
Mutation in the P5 Domain of BamA Is Lethal Under Conditions That Require Efficient OMP Assembly.
DNA sequence analysis reveals that the bamA5 mutation is a transition at codon 373 (GAA to AAA) causing a Glu-to-Lys substitution; accordingly, the bamA5 mutant gene henceforth is referred to as “bamAE373K,” and the mutant protein as “BamAE373K.” This residue maps to the P5 domain, the POTRA domain most proximal to the C-terminal β-barrel domain of BamA. The recently solved structure of BamA P5 shows that the Glu373 side chain is solvent-exposed (7), and phylogenetic analysis reveals a high degree of conservation at this position across Gram-negative bacteria (Fig. S1).
The conditional inducer-dependence of JCM320/pbamAE373K suggests that the BamAE373K protein is not functional at high temperature or that the functionality it retains is not sufficient to sustain rapid growth. To assess the functionality of BamAE373K in the nonpermissive condition, growth was monitored before and after depletion of inducer during incubation at 37 °C. In the presence of arabinose, the growth rate of JCM320/pbamAE373K was comparable to the growth rate of JCM320 containing pbamA+. Upon depletion of wild-type BamA, growth of the strain expressing bamAE373K slowed and stalled at a rate comparable to that observed for the empty vector control (Fig. 1A).
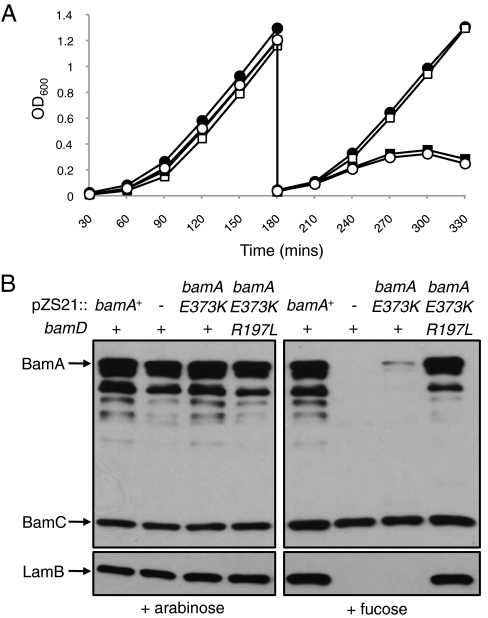
Fig. 1.
bamAE373K is a lethal mutation at 37 °C. (A) Overnight cultures of JCM320 containing pZS21 (open circles), pbamA+ (closed circles), or pbamAE373K (bamD+, closed squares; bamDR197L, open squares) were subcultured into LB medium containing d-fucose to repress expression of chromosomal bamA. Cultures were grown at 37 °C and back-diluted at 3 h. (B) Cultures of the strains shown in A were grown in the presence of arabinose (Left) or fucose (Right) to OD600 = ∼1 and subjected to SDS/PAGE followed by immunoblotting for the proteins indicated.
Depletion of bamA causes a significant decrease in the steady-state levels of all OMPs because of a combination of misassembly, misfolding, down-regulation, and degradation of Bam substrates (5, 6, 12). To determine the impact of bamAE373K on OMP assembly, the levels of several OMPs were evaluated after growth in the presence of either arabinose or fucose. Although levels of various OMPs (including LamB and BamA itself) are normal in JCM320/pbamAE373K when arabinose is present, the depletion of wild-type BamA in this background leads to a significant decrease in the steady-state levels of all OMPs tested (including BamAE373K), indicating a global defect in β-barrel assembly (Fig. 1B). This defect is comparable to that observed upon depletion of bamA+ in the presence of empty vector, suggesting that bamAE373K exhibits a null-like phenotype in the nonpermissive growth condition. Additionally, we observe a sharp reduction in OMP levels at steady state even when a haploid bamAE373K strain is grown at the permissive temperature (Fig. S2). Thus, even under permissive conditions, function of the BamAE373K mutant protein is compromised.
BamAE373K Is Properly Assembled.
A simple explanation for the observed phenotypes of bamAE373K is that perturbation of Glu373 causes BamA instability or misfolding. Because BamA is itself an OMP, it is dependent on Bam for assembly. Consequently, the significant reduction in BamAE373K levels upon depletion of bamA+ does not allow us to distinguish between a BamA instability/misfolding defect and a functional defect, because levels of folded BamAE373K would decrease in either case.
To determine whether the bamAE373K mutation affects BamA biogenesis, we sought to assess the assembly state of BamAE373K in a bamA+ background. If BamAE373K is simply folding defective or unstable, then bamA+ should complement the ts− phenotype of bamAE373K but should not cause an increase in BamAE373K levels. However, if BamAE373K is assembly competent and stable but nonfunctional, then the BamAE373K concentration should be comparable to that of BamAWT when bamA+ is supplied in trans. We find that comparable amounts of BamAWT and BamAE373K are present in bamA+ strains, suggesting that the E373K mutation does not significantly affect the biogenesis or stability of BamA. However, we consistently observe some proteolysis of BamAE373K upon purification (Fig. 2A, Upper Right, lanes 2 and 4), likely reflecting increased proteolysis of BamAE373K in cell extracts (SI Results).
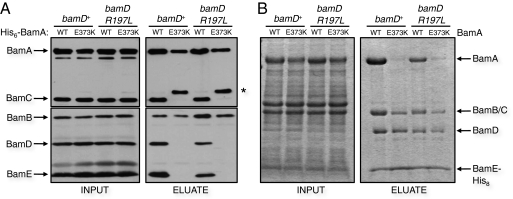
Fig. 2.
bamAE373K disrupts the BamAB–BamCDE interaction. (A) Affinity purification was performed in bamD+ and bamDR197L strains containing either pHis-BamA+ or pHis-BamAE373K. Samples were subjected to SDS/PAGE and immunoblotting for BamA–E. Protein levels are shown before (Left) and after (Right) purification. The band indicated by the asterisk corresponds to an N-terminal fragment of BamAE373K. (B) Extracts derived from solubilized E. coli membranes containing either wild-type or mutant BamAB and BamCDEHis complexes were mixed in vitro and then purified by Ni-NTA chromatography using BamE-His8. Eluates were subjected to SDS/PAGE and Coomassie blue staining.
bamAE373K Disrupts the Interaction Between BamA and BamD.
The wild-type BamABCDE complex is stable and can be purified as an intact unit (4, 6). To determine if the bamAE373K mutation affects the interactions of BamA with BamB and BamCDE, we His6-tagged and expressed the BamAWT and BamAE373K proteins. Upon purification of the tagged BamA variants, we observed no difference between the wild-type and mutant protein in the amount of coprecipitated BamB (Fig. 2A). However, we found that the amounts of coeluted BamC, BamD, and BamE are all reduced significantly by the E373K substitution (Fig. 2A). Furthermore, although purified wild-type BamAB and BamCDE subcomplexes can be combined in vitro to form five-member holocomplexes, the E373K mutation prevents holocomplex formation (Fig. 2B). These results clearly show that this P5 mutation specifically affects the physical interaction between the BamAB and BamCDE subcomplexes.
In light of the data presented in this section, we conclude that the observed destabilization of BamA–CDE in a bamAE373K background occurs because Glu373 is critically important for the physical interaction between the two Bam subcomplexes. Because the association between BamAB and BamCDE likely is stabilized by direct binding of BamA to BamD, we suggest that replacing Glu373 with Lys cripples Bam by preventing a productive association between these two essential complex components.
Suppressor Mutations in bamD Restore Bam Complex Function.
The biochemical and genetic evidence described thus far implicates Glu373 in some vital aspect of the BamA–BamD relationship. We reasoned that, if so, we potentially could isolate suppressors of the thermosensitivity of bamAE373K that map to bamD. We selected ts+ revertants of bamAE373K and mapped the affected loci. Four independent revertants were obtained. One of these four mutations was found to be a true revertant that restores Glu at position 373. The remaining three are extragenic suppressors that map to a single codon in bamD, resulting in the mutation of a highly conserved residue (Arg197) in the C-terminal domain of BamD to Leu, Ser, or His. The facts that suppressors are obtained at frequencies comparable to true reversion and that all three of them map to the same codon suggest an allele-specific relationship between the original mutation and the suppressors. The allele encoding the Leu substitution, the first to be isolated, is referred to here as “bamDR197L,” and its protein product is referred to as “BamDR197L.” Although this suppressor is discussed specifically below, it is important to note that both the Ser and His substitutions phenocopy bamDR197L by every assay we have conducted.
The bamDR197L mutation is a strong suppressor that reverses the deleterious phenotypes associated with bamAE373K: OMP levels return to normal (Fig. 1B), and the growth rate of the suppressed strain is indistinguishable from that of wild type (Fig. 1A). Additionally, the robustness of the permeability barrier in bamAE373K bamDR197L at 37 °C (see Fig. 3) is especially remarkable, given that the bamAE373K mutant does not grow at this temperature even in the absence of antibiotics. The complete suppression of bamAE373K by compensatory mutations in bamD further supports the hypothesis that bamAE373K defects are a consequence of a nonproductive interaction between BamA and BamD. This allele of bamD is a gain-of-function mutation as evidenced by the fact that it is dominant to bamD+ in a bamAE373K background; that is, it suppresses the conditional lethality of bamAE373K even in the presence of wild-type BamD.
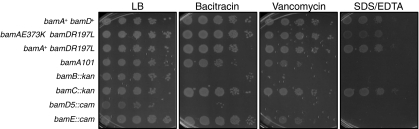
Fig. 3.
bamDR197L restores the OM permeability barrier in a bamAE373K mutant. Serial 10-fold dilutions of stationary-phase cultures of the strains indicated were prepared in 96-well plates and then were spotted onto LB medium alone or onto LB medium supplemented with 625 μg/mL bacitracin, 60 μg/mL vancomycin, or 0.5% SDS + 1.0 mM EDTA and were incubated overnight at 37 °C.
The bamD mutants described here could represent classical interactive suppressors that restore a direct physical interaction that was disrupted by the bamAE373K mutation. Additional evidence supporting such an allele-specific interaction between BamA and BamD could be obtained were it shown that the bamDR197L mutation weakens the interaction between BamD and wild-type BamA. Upon purification of His6-tagged Bam components from either whole cells or in vitro experiments using overexpressed proteins, we found that the amount of BamAWT that associates with BamDR197L compared with BamDWT is somewhat reduced (Fig. 2). Thus, the interaction between BamAWT and BamDR197L is compromised, suggesting that bamDR197L causes partial destabilization of an otherwise wild-type complex.
We suspected that bamDR197L might inhibit Bam function in a bamA+ background, perhaps causing defects similar to those observed for bamAE373K in a bamD+ context. However, when we replaced wild-type bamD of MC4100 with bamDR197L and evaluated the phenotype of the resulting bamDR197L bamA+ strain, we found this mutant to be indistinguishable from wild type with respect to growth rate, steady-state OMP levels, and OM permeability (Fig. 3 and Fig. S3). Thus, BamDR197L is functionally compatible with both BamAWT and BamAE373K. Said differently, the identity of residue 373 of BamA seems to be inconsequential in the presence of bamDR197L. In support of this conclusion, the lethality associated with a bamA mutation in which Glu373 is replaced by Arg (bamAE373R) also is suppressed completely by bamDR197L. Although bamDR197L can suppress these lethal bamA mutations, it does not bypass the requirement for bamA altogether; bamA cannot be deleted in a bamDR197L background.
bamDR197L Does Not Restore Stability of a Complex Containing BamAE373K.
If bamDR197L were indeed an allele-specific suppressor of bamAE373K, it would follow that the dissociated complex observed in bamAE373K should be restored by bamDR197L. Therefore we attempted to copurify BamCDE with BamAE373KB in either a bamD+ or bamDR197L background to determine the stability of the BamA–BamD interaction in each context. Surprisingly, the amount of BamDR197L that copurifies with BamAE373K is not appreciably different from the amount of BamDWT that copurifies with BamAE373K (Fig. 2A). Moreover, the Bam holocomplex cannot be detected after chemical crosslinking in either a bamAE373K or bamAE373K bamDR197L mutant background, suggesting that the BamA–BamD interaction is indeed dissociated and not simply weakened (SI Results and Fig. S4). These paradoxical results suggest that bamDR197L reverses all phenotypes associated with bamAE373K without stabilizing the disrupted Bam complex.
As shown in Fig. 2B, the complete Bam holocomplex can be reconstructed in vitro from overexpressed BamAB and BamCDE subcomplexes (13, 14). Consistent with the results shown in Fig. 2A, the R197L mutation in BamD does not significantly increase the amount of BamAE373KB–CDR197LE holocomplex reconstructed from mutant subcomplexes (Fig. 2B). These results show that, despite complete suppression of the bamAE373K phenotype, BamDR197L does not restore the interaction with BamAE373K. Taken together, our data suggest that restoration of Bam complex function in bamAE373K strains can be accomplished without restoring complex stability.
BamDR197L Has Increased Activity Compared with Wild-Type BamD.
We considered the formal possibility that a slight increase in complex stability, although not detectable biochemically, is sufficient to restore function to Bam. This idea is conceivable, because a ∼10-fold reduction in bamA expression causes only minor defects in OMP maturation (15), suggesting that OM biogenesis can be sustained even when the concentration of Bam holocomplex is greatly diminished. The experiments presented below argue against this possibility and suggest instead that bamDR197L is a gain-of-function mutation.
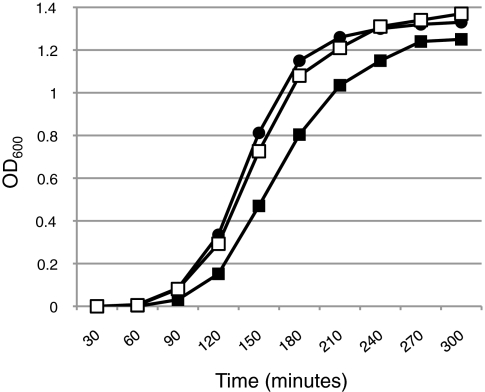
It seems likely that BamCDR197LE interacts less well with BamAWT than does BamCDE (Fig. 2). If holocomplex formation is important for function in a bamDR197L bamA+ background, then it is reasonable to expect that reducing the holocomplex levels by limiting the expression of bamA would negatively affect growth of this strain. To test the prediction that holocomplex levels are important in a bamDR197L strain, we reduced the levels of BamAWT protein by decreasing the expression level of bamA. We introduced into this strain a transposon located upstream of bamA that reduces expression of this gene by an order of magnitude; this allele is referred to as “bamA101” (15). Unexpectedly, not only is the bamDR197L bamA101 double mutant viable, the bamDR197L allele in fact suppresses the growth defect observed in a bamA101 strain (Fig. 4).
Fig. 4.
bamDR197L suppresses the growth defect caused by a reduction in bamA expression. Overnight cultures of DPR909 (circles), DPR959 (closed squares), and DPR960 (open squares) were subcultured into LB medium and grown at 37 °C. OD was measured every 30 min.
It is important to note that the OM permeability barrier is robust in both bamDR197L single and bamAE373K bamDR197L double mutants; indeed, both strains are equivalent to wild type in this respect. Although mutations known to cause modest defects in Bam function lead to increased OM permeability (5, 15, 16), the bamDR197L and bamAE373K bamDR197L mutants are as resistant to these compounds as wild type (Fig. 3). Taken together, results presented in this section argue that the mutant protein can do something the wild-type protein cannot, suggesting that suppression of bamAE373K and bamA101 by bamDR197L is the result of an increase in BamD activity.
Discussion
We describe amino acid substitutions at a single residue within the periplasmic domain of BamA that profoundly impair Bam and prevent efficient OMP assembly. This residue (Glu373) is situated on a solvent-exposed surface of P5, which is the only POTRA domain required for binding of BamA to BamD, the only essential POTRA in the BamA homolog of Neisseria meningitidis (17), and apparently the most evolutionarily ancient of the five POTRA domains (17, 18). Replacement of Glu373 with Lys (E373K) prevents growth above room temperature on rich medium and greatly destabilizes the interaction between BamA and BamD. Replacement of Glu373 with Arg (E373R) is lethal. These findings seem to indicate a critical role for Glu373 in initiating or maintaining the physical BamA–BamD association.
Compensatory mutations in BamD that suppress all defects conferred by bamAE373K in an apparently allele-specific manner do not restore the stable physical interaction between BamA and BamD. These findings suggest that the stability of BamAD is not a reliable proxy for the fidelity of the reaction catalyzed by the pair and that the defects observed in a bamAE373K background cannot be attributed solely to the physical dissociation of the two essential Bam components.
Both BamA+ and BamAE373K are fully functional in a bamDR197L background, suggesting that Glu373 is not crucial for BamA activity per se. Said differently, it appears that BamA+ and BamAE373K are equally competent to promote OMP assembly, but BamAE373K is incompatible with wild-type BamD. Because bamDR197L restores complete functionality to a machine containing BamAE373K in a manner independent of complex stability, and because the phenotype of bamDR197L is the same in the presence of either bamA+ or bamAE373K, we suggest that the bamAE373K phenotype in fact reflects the absence of BamD function rather than a lack of BamA function. In other words, the bamAE373K mutation dissociates the BamAB and BamCDE subcomplexes; this dissociation does not inactivate BamA, but it inactivates BamD. We propose that bamDR197L, a gain-of-function mutation, is a bypass suppressor that activates BamD without the need for a stable BamA–BamD interaction.
It has been established previously that the P5 domain interacts with BamCDE; however, the results presented here suggest that a stable association between the P5 domain and BamCDE is not required for OMP assembly if BamD is otherwise activated. We propose that, in the wild-type complex, BamD is “activated” via an interaction with the P5 domain of BamA. Glu373 is required for the interaction between BamD and the P5 domain and might be involved directly in BamD activation. We do not know how the BamDR197L mutation overcomes the deleterious effects of the E373K substitution, but it likely stabilizes an active form of BamD. The fact that a mutation in BamD improves OMP assembly without significantly changing how it interacts with BamAE373K implies that BamD must affect the assembly process through a direct interaction with substrates.
The fact that various unrelated substitutions at this position (R197L, R197S, R197H) suppress bamAE373K implies that the specific identity of this residue is not important for BamD activity per se. Arg197 is situated within a highly conserved patch on the surface of the BamD C-terminal domain (Fig. S5) that previously has been suggested to mediate an interaction with BamA (4, 5, 19, 20). Our findings suggest that this surface also may play a role in the activity of BamD.
Early characterization of BamD demonstrated the importance of the C-terminal domain for complex stability and Bam function. C-terminal truncations that pare the final tetratricopeptide repeat of BamD have been shown to disrupt the physical interactions between BamD and all other complex members. In addition, these truncations cause marked defects in OMP assembly at elevated temperatures, suggesting that the efficiency of β-barrel assembly is reduced in strains expressing these BamD variants (4, 5). The fact that Arg197 is found within in the C-terminal domain of BamD, which is implicated in the interaction(s) between BamA and BamD, is consistent with our suggestion that this residue is intimately involved in some aspect of the interaction and communication between these two essential factors.
In conclusion, we have identified a mutation, bamAE373K, that prevents association of the two essential Bam subcomplexes, BamAB and BamCDE, indicating that the P5 domain of BamA controls the activity of BamD in the wild-type complex. Suppressor analysis shows that the need for stable subcomplex association can be bypassed without affecting the activity of the machine. Therefore, we suggest that the functions performed by the essential members of these subcomplexes, BamA and BamD, affect the OMP assembly reaction directly. We predict that the mutant proteins described here will provide useful tools for analyzing the general β-barrel assembly mechanism.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
Strains used in this study are listed in Table S1. Media were prepared as previously described (21). XL-1 Red (Stratagene) mutagenesis and site-directed mutagenesis (QuikChange; Stratagene) were conducted according to manufacturer's specifications. Relevant oligonucleotides are listed in Table S2. In strains producing TetR, expression of bamA from pZS21 was induced with 25 μg/mL tetracycline (22).
Affinity Purification.
Bam copurification from strains DPR821, DPR822, DPR989, and DPR990 was performed essentially as described (23), except that no crosslinking agents were used, and cells were lysed in BugBuster solution (Novagen) containing lysozyme (5 μg/mL), DNase I (50 μg/mL), RNase I (50 μg/mL), and 1 mM PMSF. Western blots were prepared as previously described (23).
In Vitro Reconstruction of Mutant Bam Complexes.
Wild-type and mutant BamAB and BamCDE complexes were overexpressed as described previously using plasmids pSK38, pSK46, pCH121, pCH123, and pBamE-His (14). Cells were lysed by French press in 20 mM Tris (pH 8), 150 mM NaCl, and 1 mM PMSF. The lysate was centrifuged at 5,000 × g for 10 min at 4 °C; then the supernatant was collected and ultracentrifuged at 100,000 × g for 30 min at 4 °C. The membrane pellet was solubilized in 20 mM Tris (pH 8), 150 mM NaCl, 2% (vol/vol) TX-100, and 2 μg/mL lysozyme by incubation on a rocker at 24 °C for 1 h. The solutions then were ultracentrifuged again at 100,000 × g for 30 min at 4 °C. The clarified solutions containing mutant and wild-type BamCDE complexes were mixed with the solutions containing mutant and wild-type BamAB complexes such that the BamAB proteins were in excess. The resulting Bam complexes then were isolated by Ni-nitrilotriacetic acid (NI-NTA) affinity chromatography using the His8 tag on BamE. The Ni column was washed with 20 mM Tris (pH 8), 0.05% N-dodecyl-β-D-maltoside (DDM), and 40 mM imidazole, and the complexes were eluted in 20 mM Tris (pH 8), 0.05% DDM, and 200 mM imidazole. These eluates were run on SDS/PAGE (4–20% gradient gel) and stained with Coomassie blue.
Supplementary Material
Acknowledgments
This work was supported by National Institute of General Medical Sciences Grant GM34821 (to T.J.S.) and National Institute of Allergy and Infectious Disease Grant AI081059 (to D.K.). C.L.H. is supported by a National Science Foundation graduate research fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201362109/-/DCSupplemental.
References
- 1.Ricci DP, Silhavy TJ. The Bam machine: A molecular cooper. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.du Plessis DJF, Nouwen N, Driessen AJM. The Sec translocase. Biochim Biophys Acta. 2011;1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Sklar JG, et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malinverni JC, et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61(1):151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 6.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. Structure and flexibility of the complete periplasmic domain of BamA: The protein insertion machine of the outer membrane. Structure. 2010;18:1492–1501. doi: 10.1016/j.str.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S, et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 9.Werner J, Misra R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol Microbiol. 2005;57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- 10.Doerrler WT, Raetz CR. Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J Biol Chem. 2005;280:27679–27687. doi: 10.1074/jbc.M504796200. [DOI] [PubMed] [Google Scholar]
- 11.Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennion D, Charlson ES, Coon E, Misra R. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol Microbiol. 2010;77:1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagan CL, Kahne D. The reconstituted Escherichia coli Bam complex catalyzes multiple rounds of β-barrel assembly. Biochemistry. 2011;50:7444–7446. doi: 10.1021/bi2010784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagan CL, Kim S, Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoki SK, et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol. 2008;70:323–340. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rigel NW, Schwalm JA, Ricci DP, Silhavy TJ. BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. J Bacteriol. 2012 doi: 10.1128/JB.06426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bos MP, Robert V, Tommassen J. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 2007;8:1149–1154. doi: 10.1038/sj.embor.7401092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sánchez-Pulido L, Devos D, Genevrois S, Vicente M, Valencia A. POTRA: A conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem Sci. 2003;28:523–526. doi: 10.1016/j.tibs.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Albrecht R, Zeth K. Structural basis of outer membrane protein biogenesis in bacteria. J Biol Chem. 2011;286(31):27792–27803. doi: 10.1074/jbc.M111.238931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandoval CM, Baker SL, Jansen K, Metzner SI, Sousa MC. Crystal Structure of BamD. an essential component of the β-barrel assembly machinery of Gram negative bacteria. J Mol Biol. 2011;409(3):348–357. doi: 10.1016/j.jmb.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silhavy TJ, Berman ML, Enquist LW. Cold Spring Harbor Laboratory . Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 22.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.